Abstract
The gastric cancer is one of the most common carcinomas and the second cancer-related death in the world. The risk factors for this cancer include genetic factors and environmental factors such as Helicobacter pylori infection. The protein HcpD (HP0160) of H. pylori is a member of the cysteine-rich protein family that interacts with host immune systems. One of the modern approaches to stimulate the humoral and cellular immune systems against diseases is utilization of DNA vaccines. Using qPCR method, this study aimed to evaluate the expression level of cytokines genes including IL17, IL4, and interferon gamma (IFNγ) in BALB/c mice vaccinated with pCDNA3.1(–)-hcpD DNA vaccine against H. pylori. In this study, pCDNA3.1(–)-hcpD recombinant vector was prepared and transformed into E. coli to obtain a large number of plasmids. After plasmid purification and confirmation of the transformation by digestion and PCR, the chitosan nanoparticles were synthesized using ionic gelation method. The injectable solutions containing pCDNA3.1(–)-hcpD, pCDNA3.1(–)-hcpD + nanoparticles or pCDNA3.1 (empty vector as control group) were injected into BALB/c mice, separately. Then, the blood and tissues samples from each animal were collected and the expression levels of cytokine genes were examined by a qRT-PCR method. The IL-4 expression level was significantly decreased in pcDNA3.1(–)-hcpD + nanoparticle and pcDNA3.1(–)-hcpD groups (p < 0.001). Conversely, the expression level of IFNγ gene in both groups was increased significantly (p < 0.001). The expression level of IL17 gene showed no significant difference between DNA vaccine containing nanoparticle compare with pcDNA3.1(–)-hcpD (p > 0.05). During 15, 30 and 45 post-injection days, the expression level of hcpD decreased in hip tissue of mice vaccinated with pcDNA3.1(–)-hcpD and pcDNA3.1(–)-hcpD + nanoparticle although no significant difference found between the vaccinated groups (p > 0.05). pcDNA3.1(–)-hcpD vaccine can stimulate the immune system in vaccinated mice either as the sole agent or combined with chitosan nanoparticles. Therefore this method can be an effective way for immunization against H. pylori infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Helicobacter pylori (H. pylori) is a Gram-negative, microaerophilic bacterium known as an inducer of peptic ulcers, chronic inflammation, gastric adenocarcinoma, mucosa-associated lymphoid tissue (MALT), and gastric atrophy [1, 2]. In more than 50% of the adults H. pylori infection may be acquired at an earlier age [3, 4]. Depending on the level of health, the epidemiology of H. pylori infection varies in different populations as this pathogen may infect up to 90% of the population in developing countries [3, 5]. Urease enzyme helps this dominant human pathogen to persist for a long time in the stomach and its motility help it to enter to the gastric epithelium [6].
H. pylori secrete several protein and toxins that cause tissue damage in the host. In addition to the motility features which play important roles in its pathogenicity, the virulence factors of this bacterium include the cag Pathogenicity Island (cag PAI), vacuolating cytotoxin (Vac), and urease [7, 8]. The genome of H. pylori encodes a family of eight or nine secreted proteins known as eukaryotic Sel1 regulatory proteins containing a large number of disulfide bridges [9]. For instance, Helicobacter and Campylobacter genera express Helicobacter cysteine-rich proteins (Hcp) [10, 11] from HcpD interacts with host immune systems and play a role in H. pylori pathogenicity [12, 13].
Seven days antibiotic treatment is usually suggested for patients who are positive for H. pylori infection. This treatment is typically a combination of anti-acid agents (proton-like pump inhibitors group such as omeprazole, lansoprazole, pantoprazole or H2 inhibitors including ranitidine, cimetidine, roxatidine, and famotidine) with one or two antibiotics (clarithromycin (LAC), amoxicillin or metronidazole) [14, 15].
During recent decades, vaccination is the most effective strategy for controlling the infectious diseases. In order to achieve effective vaccine against H. pylori a number of efforts have been made to use the entire cell (dead, live, and attenuated vaccines), subunit vaccines or recombinant strains (oral or injectable) as vaccines [13, 16]. Although the production of recombinant proteins are more affordable comparing to natural proteins, the production and purification of the recombinant antibodies are still costly [17].
In recent years, DNA immunization has attracted considerable attention as a promising approach for stimulation of both humoral and cellular immune responses against bacterial pathogens [18]. Typically, a DNA vaccine is a genetically engineered DNA with several advantages compared to traditional vaccines [19, 20]. Eliminating the risk of using actual infectious organisms, DNA vaccines stimulate both the antibody and cell-mediated components of the immune system. Furthermore, they are highly specific, stable at room temperature, inexpensive and easy to prepare [19, 21].
Considering the lack of appropriate and affordable therapeutic method for controlling H. pylori infections especially in developing countries and the fact that the expression of hcpD gene of H. pylori leads to stimulate the immune system of the mammalian hosts, establishing a method to produce an efficient vaccine against H. pylori seems to be necessary. The present work aimed to utilize pCDNA3.1(–)-hcpD expression construct as a DNA vaccine (either alone or in combination to chitosan nanoparticles) and evaluate the immunization efficiency against H. pylori in BALB/c mice via qPCR analyses of cytokine genes expression levels.
MATERIALS AND METHODS
Vector Preparation and Bacterial Culture
The pcDNA3.1(–)-hcpD that obtained from the previous study of Eslami and Doosti, 2017 were used as a recombinant DNA vaccine in this study [13]. Also, the pcDNA3.1(–) (Invitrogen, San Diego, CA) with 5428 bp length was used as control plasmid (without gene) for injection into BALB/c mice. The lyophilized stock of E. coli strain Top10F' was purchased from Pasteur Institute of Iran and was cultured in 5 mL of Luria–Bertani (LB) broth and then incubated for overnight at 37°C with shaking and were used for transformation and plasmid preparation.
Transformation of pcDNA3.1 (–)-hcpD Recombinant Vector
For the preparation of enough pcDNA3.1(–)-hcpD for animal injection, the recombinant vector was transformed into E. coli Top10F' using CaCl2 and heat shock (42°C for 90 s) treatment. Colony-PCR was done on each colony for identification of inserted recombinant plasmid into E. coli. After culture of the selected colony on LB broth containing 100 µg/mL of ampicillin antibiotic for overnight at 37°C, the plasmid purification was done using GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific, Freiburg, Germany) according to the recommendations of the manufacturer. The quality and quantity of the extracted plasmid were determined from the ratio of the absorbance at a wavelength of 260/280 nm using Thermo Scientific™ NanoDrop 2000 (Wilmington, DE, USA) according to the method described by Sambrook and Russell, 2001 [22]. The sequencing, PCR, and digestion with BamHI and EcoRV restriction enzymes were done on purified plasmids for confirmation of transformation. The digested products, pcDNA3.1(–)-hcpD recombinant plasmid and empty vector were electrophoresed at constant voltage (80 V) on 2% agarose gel electrophoresis and their molecular weight was approximately determined using 100 b DNA ladder. The gel was stained with 2 μg/mL of ethidium bromide and examined under UV light and a photograph was obtained under UVIdoc gel documentation systems (Uvitec, UK).
Amplification of hcpD Gene
The following specific oligonucleotide primer pairs with the accession number: FM991728 including hcpD-F: 5′-ATGATAAGAATTGGACTAAAAAG-3′ hcpD-R: 5′-TTATTGCGTATCATCTTGCGTGTC-3′ was obtained from Eslami and Doosti, 2017 study and was used for amplification of hcpD gene. The polymerase chain reaction (PCR) was performed in a final volume of 25 μL in 0.2 mL tubes containing 50 ng of purified plasmid, 1 μM of each forward and reverse primer, 2 mM MgCl2, 200 mM dNTPs, 2.5 μL of 10× reaction buffer, and 1 unit of Taq DNA polymerase (all Thermo Fisher Scientific, Freiburg, Germany). The amplifications were carried out in a Thermal Cycler (Mastercycler Gradient, Eppendrof, Germany). The temperature profiles were included an initial denaturation step at 94°C for 5 minutes, followed by 35 cycles of a denaturation step at 94°C for 1 min, a primer-annealing step at 62°C for 1 min, extension step at 72°C for 1 min, and then a final step at 72°C for 5 min. The amplified products were detected on 2% agarose gel electrophoresis as mentioned above.
Synthesis of Chitosan Nanoparticles Using Ionic Gelation Method
100 mg of chitosan powder (Sigma-Aldrich, USA) was dissolved into 50 mL of the acetic acid (2 mg/mL), and then the solution was kept at room temperature for 24 h on a magnetic stirrer at 1000 rpm. The pH of the solution was adjusted to 5.5 using 0.5 M NaOH. The entire solution was filtered through a 0.45 μm filter to filter unblemished chitosans. The sodium tri-polyphosphate (TPP) solution was prepared at a concentration of 0.7 mg/mL and passed through a 0.45 μm filter. 50 mL of acetic acid solution containing chitosan was placed on a magnetic stirrer at room temperature and 20 mL of TPP (produced in the previous step) was dropped very slowly (every 7 s, 1 drop) to the rotary solution on the stirrer.
This solution was kept on the magnetic stirrer for 1 h at 1000 rpm. The solution was centrifuged at 14 000 rpm for 15 min and the supernatant containing chitosan nanoparticles was collected. The solution was dried and powdered using freeze-dried and the physicochemical properties of chitosan nanoparticles were determined using Zeta Analyzer (Malvern Instruments, UK).
Preparation of Plasmid and Nanoparticles
Chitosan nanoparticles (1%) and plasmid solutions (2000 μg/mL in PBS) were prepared separately and then they (nanoparticle + plasmid) were mixed equally and placed at 55°C for 1 h.
Mice Grouping and Injection Method
In the present study, the ethical approval was obtained from the Ethical Committee and Research Deputy of the Islamic Azad University of Shahrekord Branch, Iran on July 22, 2016. The 6 weeks old female BALB/c mice were classified into three groups (25 mice in each group) according to different injections. These groups included injected mice by chitosan nanoparticles recombinant plasmids (DNA vaccine + nanoparticles), recombinant plasmids (DNA vaccine), and the target-free plasmids (control group). 100 μL of each solution including DNA vaccine + nanoparticles (100 μg of DNA mixed with nanoparticles), DNA vaccine (1000 μg per mL of recombinant vector in PBS), and control group (1000 μg per mL of target-free plasmids) was injected into mice’s hip muscles. The injections were carried out at intervals of 7 days during three weeks. 15, 30, and 45 days after the last injection, 7 mice from each group were killed and sampled. After anesthesia, at each sampling step, 1.5 mL of complete blood samples was obtained from the heart of the mice (the area under the jaw) and was transferred into anticoagulant (EDTA). Then, the mice’s hips were removed and the tissues at the injection site were isolated.
Quantitative Real-Time PCR (q-PCR)
For RNA isolation a total of 100 mg of each tissue or 100 μL of blood samples (buffy coat) were subjected. RNA was extracted immediately from blood and tissue samples using TRIzol reagent (Invitrogen, USA), according to manufacturer’s instructions. The quality and quantity of isolated RNA were measured by the A260/280 ratio using NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). 1 µg of each RNA sample was used for reverse transcription with a cDNA synthesis kit (Takara, Kyoto) containing oligo (dT) and specific primers according to the manufacturer’s protocol. The cDNA synthesis reaction was done at 85°C for 5 s, 42°C for 15 min, and finally the reverse transcriptase was inactivated for 5 min at 85°C.
To investigate the expression profiles of IL-17, IL-4, IFNγ cytokine genes and compared with GAPDH (as an internal control) and hcpD the specific oligonucleotide primers were designed using Gene Runner software version 3.05, Oligo Primer Analysis Software v.7 (Molecular Biology Insights Inc., Cascade, USA) and basic local alignment search tool (BLAST) of GenBank data (Table 1). After serial dilution of cDNA samples, 1 : 10 proportion was selected as appropriate. The Real-time PCR was carried out by Rotor-Gene 6000 (Corbett Research, Australia) in a final volume of 20 μL, containing 1× SYBR Green master mix (Toyobo, Japan), 1 μL of each of the primers (each 3 μM), 50 ng cDNA template, and 1 μL of cDNA sample. Amplification was performed using the following parameters: a denaturation step at 95°C for 5 min followed by 40 cycles of 15 s at 95°C (denaturation), 20 s at an annealing temperature of each primer, and 30 s at 72°C (extension). All reactions were performed at least in triplicate and the relative standard curve method was used to calculate the relative gene expression. The cycle of threshold (Ct) values was analyzed using the comparative Ct (ΔΔ Ct) method. The melting curve was generated by holding the reaction mixtures at 95°C for 60 s and then lowering the temperature to 45°C at a transition rate of 0.1°C/s and maintaining for 120 s. Then, the reaction mixture of the samples was heated slowly at a transition rate of 0.05 to 80°C with continuous collection of fluorescence at 640 nm. The melting curve and quantitative analyses were conducted in a Corbett Rotor-Gene 6000 software program (version 1.7).
Statistical Analysis
All data were collected in Statistics programs for the Social Sciences software (SPSS, Inc., Chicago, IL, USA) version 20 and variance between groups was calculated by T-test. A p-value less than 0.05 were considered as statistical significance.
RESULTS AND DISCUSSION
Confirmation of Transformation by Enzymatic Digestion
The enzymatic digestion using BamHI and EcoRV restriction enzymes was done on the pcDNA3.1(–)-hcpD recombinant expression vector (resulting in 5428 and 933 bp fragment of pcDNA3.1(–) vector and the hcpD gene respectively) and confirmed the formation of the final construct (Fig. 1).
Chitosan Nanoparticles Characteristics
The size of chitosan particles with a low molecular weight was verified by dynamic light scattering (DLS) method. The results showed that 98% of these particles had a diameter of 134.3 nm (Fig. 2).
Zeta-potential measurements were performed on chitosan nanoparticles, placed in polystyrene, using Malvern Zetasizer Nano-ZS (ZEN3600) and the dispersion intensity was measured at 25°C with an incident wavelength of 633 nm (Fig. 3).
Scanning electron microscope (SEM) (Nikon S530 Hitachi, Japan) was used to determine the appearance of chitosan nanoparticles. They exhibited a homogeneous structure with almost half-spherical shape and smooth edges (Fig. 4).
Real-Time PCR Analysis
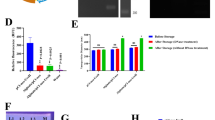
Quantitative real-time PCR analyses showed that the expression level of cytokine genes in the blood and tissue samples of vaccinated mice (the pcDNA3.1(–)-hcpD group) was changed according to details below: IFNγ gene expression was increased significantly (p-value < 0.001), IL17 gene expression was not significantly altered (p-value > 0.05), and IL4 gene expression was decreased significantly (p-value < 0.001) (Fig. 5).
In the pcDNA3.1(–)-hcpD + nanoparticle group, the expression of IFNγ and IL17 genes were increased significantly (p-value < 0.001 and p-value < 0.05, respectively). Also, the IL4 gene expression was decreased significantly (p-value < 0.001) (Fig. 6).
The comparison of the expression level of cytokine genes in the vaccinated mice by pcDNA3.1(–)-hcpD and pcDNA3.1(–)-hcpD + nanoparticle showed that IFNγ gene expression in pcDNA3.1(–)-hcpD + nanoparticle compared with pcDNA3.1(–)-hcpD group was different (statistically significant, p-value < 0.001). Moreover, the expression level of IL17 and IL4 genes in pcDNA3.1(–)-hcpD + nanoparticle group compared with pcDNA3.1(–)-hcpD were not significantly different (p-value > 0.05) (Fig. 7).
The comparison of hcpD gene expression level in tissue samples of vaccinated mice by pcDNA3.1(–)-hcpD showed significant differences in day 15 compared with day 30, day 15 compared with day 45, and day 30 compared with day 45 (p-value < 0.001) (Fig. 8). Moreover, the comparison of the target gene expression in tissue samples of vaccinated mice by pcDNA3.1(–)-hcpD + nanoparticle indicated the significant difference of hcpD gene expression level in day 15 compared with day 30, day 15 compared with day 45, and day 30 compared with day 45 (p-value < 0.001) (Fig. 8). The hcpD gene expression level in pcDNA3.1(–)-hcpD group and pcDNA3.1(–)-hcpD + nanoparticle group in each sampling time (day 15, 30, and 45 after the last injection) were not significantly different (p-value > 0.05) (Fig. 9).
H. pylori is the main cause of chronic and acute gastritis, gastric and duodenal ulcers, and prolonged inflammation of the gastric mucosa due to neutrophils, lymphocytes and plasma cells activity [23, 24]. Benefitting from the urease enzyme, this bacterium grows in the deep mucus layer of the gut epithelial cells and forms colonies. Therefore, endoscopic approaches usually fail to provide clear clinical diagnosis [25, 26]. Many studies have shown the role of this pathogen in the development of malignancy and gastric cancer; hence the World Health Organization’s (WHO) Cancer Research Agency nominated this bacterium as a carcinogen in the first class [27, 28]. Due to the antibiotic resistance and lack of a suitable vaccine against H. pylori infection, it is important to find an appropriate vaccine (especially DNA vaccine) against H. pylori for the treatment and prevention of gastrointestinal infections [28, 29]. Gene vaccines compared to the other vaccines have numerous advantages like simple production, product purity, easy storage, stimulation of both cellular and humoral immune responses, and simple maintenance at room temperature.
In the present study, after successful preparation of pcDNA3.1(–)-hcpD, this recombinant vector was used as a DNA vaccine for vaccination of BALB/c mice either alone or combined with chitosan nanoparticles and their efficiency against H. pylori was evaluated via investigation of cytokine genes expression level by q-real-time RT-PCR. The DNA vaccines including pcDNA3.1(–)-hcpD and pcDNA3.1(–)-hcpD + nanoparticle were injected into mice hip muscles. The pcDNA3.1(–) (empty vector) was used as a control group. After 15, 30, and 45 days from the last vaccination, tissues and blood samples from vaccinated mice were collected and the expression level of cytokine genes including IL-17, IL-4 and IFNγ were evaluated using real-time qPCR. The expression of the hcpD gene as a vaccine gene against H. pylori was also investigated by this method.
The results of the qPCR on the expression levels of cytokine genes showed increased levels of IFNγ in both groups (pcDNA3.1(–)-hcpD + nanoparticle and pcDNA3.1(–)-hcpD) although the expression in DNA vaccine containing nanoparticle significantly differed (p < 0.001) compared to pcDNA3.1(–)-hcpD group (without using nanoparticle). In addition, the expression level of IL17 and IL4 genes in pcDNA3.1(–)-hcpD + nanoparticle group compared to a vaccinated group by pcDNA3.1(–)-hcpD were not significantly different (p > 0.05). Moreover, the comparison of the expression levels of hcpD gene in tissue samples of vaccinated mice in both pcDNA3.1(–)-hcpD and pcDNA3.1(–)-hcpD + nanoparticle groups in each sampling day (day 15, 30, and 45 after the last injection) were significantly different (p <0.001). The comparison of the expression level of a target gene (hcpD) in tissue samples of vaccinated mice by pcDNA3.1(–)-hcpD and pcDNA3.1(–)-hcpD + nanoparticle in day 15, 30, and 45 after the last injection were not significantly different (p > 0.05).
There are many studies performed on treatment and prevention against H. pylori infection. Miyashita and colleagues in 2002 evaluated the immune response of mice after vaccination by catalase-encoding gene (pcDNA3.1-kat vaccine). They detected anti-IgG antibodies against catalase in the serum of mice which protected them against H. pylori infection and caused a severe reduction in the stomach inflammation (30). In our study, the pcDNA3.1(–)-hcpD and pcDNA3.1(–)-hcpD + nanoparticle vaccines were used and in pcDNA3.1(–)-hcpD group the IFNγ gene expression was increased and IL4 gene expression was decreased significantly. In addition, in the pcDNA3.1(–)-hcpD + nanoparticle group, the expression level of IFNγ and IL17 genes were increased and the IL4 gene expression was decreased. The findings of the present work indicated that these DNA vaccines can induce the immune response against H. pylori.
In another study, the immunization of DNA vaccine expressing the dysmotase coding gene (SOD) was evaluated by intra-muscular injection of pcDNA-SOD in BALB/c mice [31]. Their findings showed that this DNA vaccine led to the humoral and cellular immune response through stimulation of Th-1 response and IFNγ production in mice and they suggested that this vaccine can be used against Brucella abortus infection. Bai and co-workers managed to clone the BabA2 gene into pET-22b(+) expression vector and demonstrated that the recombinant BabA protein may be a potential vaccine to control and treat the H. pylori infection [32]. Another research focused on the creation of a vaccine against Brucella by targeting the extracellular protein 31 (omp31) in B. melitensis and B. ovis. Omp31 gene was cloned into a pcI expression vector and injected into BALB/c mice. The results showed that this vaccine produced cytotoxic reactions and could potentially affect the Brucella infection [33]. This research was similar to the present study, but in our study, the hcpD gene and pcDNA3.1(–) expression vector were used. Doosti and colleagues cloned Omp31 gene of Brucella melitensis into pcDNA3.1 resulting to the construction of Omp31-pcDNA3.1 vector. The immunization using this construct was evaluated in BALB/c mice and their findings showed the Th-1 cells reaction in mice [34].
In another study by Jie and co-workers in 2013, they isolated and cloned the ureI segment from the urease gene into a PCDNA3.1 expression vector to make the PCDNA3.1(+)-ureI construct. These researchers concluded that this construct was able to express the protein production of ureI and stimulate the cellular and humoral response of the immune system in the C57BL/6 mice and thus it can be used as a vaccine [35]. Their study is similar to the present study in several respects and supports our findings, although the type of the mouse and gene was different. In another research by Al-Mariri et al., 2014 the B39 and Sp41 genes from Brucella melitensis were cloned in the expression vector and used as a DNA vaccine concluding that this vaccine could be a successful method for the immunization of BALB/c mice against Brucella infection [36]. Mahmoudi Vashian and Doosti, 2016 created an ureG-pcI-neo recombinant vector and transformed it into the CHO cells by electroporation and analyzed the expression of the ureG gene on SDS-PAGE gel. Results showed that the ureG cloned gene was able to express and produce a specific protein in CHO animal cells. They showed in the animal model that this genetic construct can be considered as a suitable candidate for vaccination against H. pylori [37].
In a study by Safarpour and colleagues, a tagD gene from H. pylori was cloned into a PFLAG-CMV-3 eukaryotic expression vector. They transferred PFLAG-CMV-3-tagD into the CHO cells by electroporation method and evaluated its expression level in this cell line. They concluded that this construct gene is useful to evaluate the immunogenicity as a DNA vaccine against H. pylori infection in animal models [28]. Their study in terms of work processes was similar to the present work.
According to the findings of the present study, the expression of IFNγ gene in pcDNA3.1(–)-hcpD + nanoparticle and pcDNA3.1(–)-hcpD groups was increased, while IL-4 expression was reduced in both groups. In addition, the expression level of IL17 gene in pcDNA3.1(–)-hcpD + nanoparticle group were increased but in pcDNA3.1(–)-hcpD group was not changed significantly. Generally, these effects indicated the stimulation of the mice immune system by both vaccines with or without chitosan nanoparticles at a significant level. The comparison of the expression level of hcpD gene in the hip tissue of vaccinated mice in both pcDNA3.1(–)-hcpD and pcDNA3.1(–)-hcpD + nanoparticle groups during 15, 30 and 45 days after injection showed that the expression was decreased with the passage of time and no significant difference was observed in both vaccinated mice.
CONCLUSIONS
The pcDNA3.1(–)-hcpD along with nanoparticles produced in this study is useful for stimulation of the immune system and it can be used as a DNA vaccine against H. pylori infection in human studies in the future.
REFERENCES
Taddesse, G., Habteselassie, A., Desta, K., Esayas, S, and Bane, A., Association of dyspepsia symptoms and Helicobacter pylori infections in private higher clinic, Addis Ababa, Ethiopia, Ethiop. Med. J., 2011, vol. 49, no. 2, pp.109–116.
Mahachai, V., Vilaichone, R.K., Pittayanon, R., Roj-borwonwitaya, J., Leelakusolvong, S., Kositchaiwat, C., et al., Thailand consensus on Helicobacter pylori treatment 2015, Asian Pac. J. Cancer Prev., 2016, vol. 17, no. 5, pp. 2351–2360.
Sardarian, H., Fakheri, H., Hosseini, V., Taghvaei, T., Maleki, I., and Mokhtare, M., Comparison of hybrid and sequential therapies for Helicobacter pylori eradication in Iran: A prospective randomized trial, Helicobacter, 2013, vol. 18, pp. 129–134.
Michel, A., Pawlita, M., Boeing, H., Gissmann, L., and Waterboer, T., Helicobacter pylori antibody patterns in Germany: A cross-sectional population study, Gut Pathog., 2014, vol. 6, p. 10.
Marandi, F., Hatam, A., and Saybani, M.R., Diagnosing gastric cancer and ulcer using color domain and their classifications, J. Appl. Environ. Biol. Sci., 2015, vol. 5, no. 4, pp. 138–145.
Talebi Bezmin Abadi, A., Vaccine against Helicobacter pylori: Inevitable approach, World J. Gastroenterol., 2016, vol. 22, no. 11, pp. 3150–3157.
Salimzadeh, L., Bagheri, N., Zamanzad, B., Azadegan Dehkordi, F., Rahimian, G., Hashemzadeh-Chaleshtori, M., et al., Frequency of virulence factors in Helicobacter pylori-infected patients with gastritis, Microb. Pathog., 2015, vol. 80, pp. 67–72.
Bagheri, N., Azadegan-Dehkordi, F., Rafieian-Kopaei, M., Rahimian, G., Asadi-Samani, M., and Shirzad, H., Clinical relevance of Helicobacter pylori virulence factors in Iranian patients with gastrointestinal diseases, Microb. Pathog., 2016, vol. 100, pp. 154–162.
Ogura, M., Perez, J.C., Mittl, P.R.E., Lee, H.-K., Dailide, G., Tan, S., et al., Helicobacter pylori evolution: lineage- specific adaptations in homologs of eukaryotic sel1-like genes, PLoS Comput. Biol., 2007, vol. 3, no. 8, p. e151.
Deml, L., Aigner, M., Decker, J., Eckhardt, A., Schütz, C., Mittl, P.R.E., et al., Characterization of the Helicobacter pylori cysteine-rich protein A as a T-helper cell type 1 polarizing agent, Infect. Immun., 2005, vol. 73, no. 8, pp. 4732–4742.
Kawai, M., Furuta, Y., Yahara, K., Tsuru, T., Oshima, K., Handa, N., et al., Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes, BMC Microbiol., 2011, vol. 11, p. 104.
Mittl, P.R.E., Lüthy, L., Reinhardt, C., Joller, H. Detection of high titers of antibody against Helicobacter cysteine-rich proteins A, B, C, and E in Helicobacter pylori-infected individuals, Clin. Diagn. Lab. Immunol., 2003, vol. 10, no. 4, pp. 542–545.
Eslami, E., Doosti, A., Cloning and expression study of the hcpD gene of Helicobacter pylori, J. Ardabil Univ. Med. Sci., 2017, vol. 17, no. 1, pp. 46–57.
Li, W.Q., Ma, J.L., Zhang, L., Brown, L.M., Li, J.Y., Shen, L., et al., Effects of Helicobacter pylori treatment on gastric cancer incidence and mortality in subgroups, J. Natl. Cancer Inst., 2014, vol. 106, no. 7. https://doi.org/10.1093/jnci/dju116
O’Morain, C. and Smith, S., Helicobacter pylori treatment failure: The rationale for alternative antibiotics, Digestion, 2016, vol. 93, no. 4, pp. 309–310.
Zeng, M., Mao, X.H., Li, J.X., Tong, W.D., Wang, B., Zhang, Y.J., et al., Efficacy, safety, and immunogenicity of an oral recombinant Helicobacter pylori vaccine in children in China: A randomized, double-blind, placebo-controlled, phase 3 trial, Lancet, 2015, vol. 386, no. 10002, pp. 1457–1464.
Wang, B., Pan, X., Wang, H., Zhou, Y., Zhu, J., Yang, J., et al., Immunological response of recombinant H. pylori multi-epitope vaccine with different vaccination strategies, Int. J. Clin. Exp. Pathol., 2014, vol. 7, no. 10, pp. 6559–6566.
Flingai, S., Czerwonko, M., Goodman, J., Kudchodkar, S.B., Muthumani, K., and Weiner, D.B., Synthetic DNA vaccines: Improved vaccine potency by electroporation and co-delivered genetic adjuvants, Front. Immunol., 2013, vol. 4, p. 354.
Doosti, A., Ghasemi-Dehkordi, P., Kargar, M., and Sharifi, A., Generation of divalent DNA vaccine based on p39 and shiga-like toxin 2 (stx2) genes, Genetika, 2015, vol. 47, no. 2, pp. 499–507.
Guo, L., Yang, H., Tang, F., Yin, R., Liu, H., Xiaojuan, G., et al., Oral immunization with a multivalent epitope-based vaccine, based on NAP, urease, HSP60, and HpaA, providestherapeutic effect on H. pylori infection in Mongolian gerbils, Front. Cell. Infect. Microbiol., 2017, vol. 7, p. 349.
Chen, J., Li, N., and She, F. Helicobacter pylori outer inflammatory protein DNA vaccineloaded bacterial ghost enhances immune protective efficacy in C57BL/6 mice, Vaccine, 2014, vol. 32, no. 46, pp. 6054–6060.
Sambrook, J. and Russell, D.W., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2001, pp. 148–190.
Salama, N.R., Hartung, M.L., and Müller, A., Life in the human stomach: Persistence strategies of the bacterial pathogen Helicobacter pylori, Nat. Rev. Microbiol., 2013, vol. 11, no. 6, pp. 385–399.
Emara, M.H., Elhawari, S.A., Yousef, S., Radwan, M.I., and Abdel-Aziz, H.R., Emerging role of probiotics in the management of Helicobacter pylori infection: Histopathologic perspectives, Helicobacter, 2016, vol. 21, no. 1, pp. 3–10.
Coelho, L.G. and Coelho, M., Clinical management of Helicobacter pylori: the Latin American perspective, Dig. Dis., 2014, vol. 32, no. 3, pp. 302–309.
Patel, S.K., Pratap, C.B., Jain, A.K., Gulati, A.K., and Nath, G., Diagnosis of Helicobacter pylori: What should be the gold standard?, World J. Gastroenterol., 2014, vol. 20, no. 36, pp. 12 847–12 859.
Ferro, A., Peleteiro, B., Malvezzi, M., Bosetti, C., Bertuccio, P., Levi, F., et al., Worldwide trends in gastric cancer mortality (1980–2011), with predictions to 2015, and incidence by subtype, Eur. J. Cancer, 2014, vol. 50, no. 7, pp. 1330–1344.
Safarpour, M., Kazemi, Z., Doosti, E., and Doosti, A., Cloning tagD gene from Helicobacter pylori in PFLAG-CMV-3 eukaryotic vector to generate a DNA vaccine, Pars J. Med. Sci., 2016, vol. 14, no. 4, pp. 43–50.
Chionh, Y.T., Arulmuruganar, A., Venditti, E., Ng, G.Z., Han, J.X., Entwisle, C., et al., Heat shock protein complex vaccination induces protection against Helicobacter pylori without exogenous adjuvant, Vaccine, 2014, vol. 32, no. 20, pp. 2350–2358.
Miyashita, M., Joh, T., Watanabe, K., Todoroki, I., Seno, K., Ohara, H., et al., Immune responses in mice to intranasal and intracutaneous administration of a DNA vaccine encoding Helicobacter pylori-catalase, Vaccine, 2002, vol. 20, no. 17, pp. 2336–2342.
Onate, A.A., Céspedes, S., Cabrera, A., Rivers, R., González, A., Muñoz, C., et al., A DNA vaccine encoding Cu, Zn superoxide dismutase of Brucella abortus induces protective immunity in BALB/c mice, Infect. Immun., 2003, vol. 71, no. 9, pp. 4857–4861.
Bai, Y., Zhang, Y.L., Chen, Y., Jin, J.F., Zhang, Z.S., and Zhou, D.Y., Cloning and expression and immunogenicity of Helicobacter pylori BabA2 gene, World J. Gastroenterol., 2004, vol. 10, no. 17, p. 2560.
Cassataro, J., Velikovsky, C.A., de la Barrera, S., Estein, S.M., Bruno, L., and Bowden, R., A DNA vaccine coding for the Brucella outer membrane protein 31 confers protection against B. melitensis and B. ovis infection by eliciting a specific cytotoxic response, Infect. Immun., 2005, vol. 73, no. 10, pp. 6537–6546.
Doosti, A., Ghasemi-Dehkordi, P., Javadi, G.R., Sardari, S., and Shokrgozar, M.A., DNA vaccine encoding the Omp31 gene of Brucella melitensis induces protective immunity in BALB/c mice, Res. J. Biol. Sci., 2009, vol. 4, no. 1, pp. 126–131.
Jie, L., Wen, Y., Sheng, L., Yan, Z., Cui Ming, Z., and Yi-mou, W.U., Immunocompetence of Helicobacter pylori ureI DNA vaccine in mice, Chin. J. Zoonoses, 2013, vol. 29, no. 9, pp. 895–898.
Al-Mariri, A., Akel, R., and Abbady, A., A DNA vaccine encoding p39 and sp41 of Brucella melitensis induces protective immunity in BALB/c mice, Arch. Med. Vet., 2014, vol. 46, no. 1, pp. 53–62.
Mahmoudi Vashian, Z. and Doosti, A., Cloning and gene expression of ureG gene as a DNA vaccine candidate against Helicobacter pylori, J. Guilan Univ. Med. Sci., 2017, vol. 26, no. 102, pp. 20–29.
ACKNOWLEDGMENTS
This article was obtained from the MSc thesis. We would also like to sincere thanks from Research Deputy of Islamic Azad University of Shahrekord Branch and Biotechnology Research Center for their sincere cooperation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
About this article
Cite this article
Nasr-Esfahani, M., Doosti, A. & Jami, M.S. Chitosan Nanoparticles-Mediated pCDNA3.1(–)-hcpD DNA Vaccine against Helicobacter pylori in BALB/c Mice. Mol. Genet. Microbiol. Virol. 34, 131–139 (2019). https://doi.org/10.3103/S0891416819020083
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0891416819020083