Abstract
Information on the genetic control of awn development in bread wheat is currently limited to the identification of three genes that suppress awnedness, Hd, B1, and B2, and no promoters have yet been identified. Another Gramineae species, Oryza sativa, has more than ten genes involved in awn morphogenesis. This article presents results of the wheat genome sequence analysis for the search of genes orthologous to the known awn development regulators in rice, TOB1, ETT2, and DL. Using bioinformatic methods, three genes, TaTOB1, TaETT2, and TaDL, are identified in the bread wheat genome; their location is defined on the chromosomes of the second, third, and fourth homoeologous groups, respectively. The polymorphisms between homoeoalleles of the genes located on subgenomes A, B, and D are described. Identified polymorphisms include variation in the length of exons and introns in all the three genes, variation in the number of exons and introns for the TaETT2 gene homoeoalleles, inversion of TaDL-B homoeoallele relative to the TaDL-A and TaDL-D, and inversion of TaETT2-B and TaETT2-D relative to TaETT2-A. With the PCR method using primers designed for the TaTOB1 gene sequence, the homoeoalleles of this gene were identified in the genomes Au, Ab, B, G, D, SSh, M, U, and T in diploid, tetraploid, and hexaploid wheat species. The marker potential of two pairs of primers for the TaTOB1 gene for the study into the genome structure of the introgressive wheat lines in relation to this gene is shown.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
The presence of awns, needle-like extensions on the lemmas of Gramineae species, is a morphological trait that, along with several others, is used for differentiating cultivars of bread wheat (Triticum aestivum L.): among the 14 cultivars, six are awnless and eight are awned [1]. There is no natural awnedness polymorphism in durum wheat (T. durum Desf.), and all nine of its cultivars have awned ears. Existing cultivars of durum wheat without well-developed awns have bread wheat in their pedigree, from which they received genes inhibiting awn development. Genetic regulation of awnedness in wheat has been studied for a long time [2] and not least because of the apparent marker potential of this trait, especially when working with introgressive wheat genotypes [3, 4]. So far, awnedness in bread wheat is associated with the fact that its genotype lacks at least two of the three known dominant awnedness inhibitors, Hd (4AS), B1 (5AL), and B2 (6BL) [5–7]. Awns are determined structures that are considered to be modified leaves, therefore, regulators of their development, among others, should include homeotic genes. Awns are formed by the division of cells in a plane perpendicular to the direction of the apical-basal axis [8]. There is no doubt that genes involved in the regulation of this trait should include cell division promoters whose activity leads to the formation of needle-like extensions. For Oryza sativa, there are approximately ten loci of quantitative traits involved in the formation of awns [9]. Most of the identified genes encode transcription factors and, therefore, have a pleiotropic effect: the An-1 gene encodes a transcription factor from the bHLH family, the DL and TOB1 genes encode transcription factors from the YABBY family, and the OsETT2 gene encodes a transcription factor from the ARF family [9–11]. Other rice genes affecting the morphogenesis of awns, SHL2, SHO1, SHO2, and WAF1, encode enzymes in the pathway of small interfering RNA biogenesis, hence they also have a regulatory function [12–15]. It is likely that all known rice genes form a network that regulates the development of awns, but there is currently no information on molecular mechanisms and interaction of their products within this network. It has only been found that there is activation interaction between DL and OsETT2 (since dl mutants do not express OsETT2), and small RNA transcripts synthesized by the already mentioned SHL2, SHO1, SHO2, and WAF1 genes suppress the expression of OsETT2 [9, 15]. Regarding wheat, the promoter genes of awn development are not part of the current gene catalog [5]. Indeed, the Hd gene is associated with the development of awn-like elongations [6]; it has been shown that awnedness can dominate over awnlessness in diploid and tetraploid wheats [16] and the promoter of awnedness is localized on chromosome 6B in durum wheat [17] and chromosomes 6U and 6Ssh in Aegilops umbellulata L. and Ae. sharonensis Eig., respectively [3, 4].
It is known that molecular mechanisms for the regulation of ontogenesis of homologous organs, in particular leaves, are conservative in related groups of plants [18, 19]. It is logical to assume that the development of awns in wheat and rice is regulated by similar networks; that is, wheat has genes orthologous to the genes that regulate the development of awns in rice. Therefore, information on the rice genes involved in the development of awns can be used for in silico search of the orthologous sequences in the bread wheat genome. This paper presents the results of in silico exploration of the wheat genome for the TaDL, TaTOB1, and TaETT2 bread wheat genes orthologous to the respective awn development regulators in rice. The information obtained for the TaTOB1 gene was verified through a comparative study of different wheat genotypes DNA amplification spectra, which were obtained with primers designed for sequences of bread wheat.
MATERIALS AND METHODS
The bioinformatic search for orthologous sequences in the bread wheat genome was carried out using the BLAST tool in the sequences deposited in the GenBank and Plant Transcription Factor Database [20, 21]. The search was performed using the DL (AB106553), TOB1 (AK070205), and OsETT2 (AB071291) rice gene sequences. The corresponding encoding sequences for the two orthologs TaDL (AB470269) and TaETT2 (AY376129) were found in GenBank and TaTOB1 (Traes_2BL_8BEA9CE1B) in the Plant Transcription Factor Database.
To identify the chromosomal localization of genes, the coding sequences of the studied genes were subjected to the search in the sequence of the bread wheat genome at the Sequencing of the Aegilops tauschii Genome project site [22]. The exon-intron structure of the gene sequences was analyzed by comparing the nucleotide positions of the genome sequence for which the known coding sequences were aligned. The length of exons was determined by the size of the aligned cDNA fragments, and the length of introns was determined as the difference between the position of the nucleotide that is the start of the next exon and the nucleotide position that is the end of the previous exon in the genome sequence.
Based on the gene structure found in the sequence, the primers for the TaTOB1 gene were created using the Primer-BLAST [23] service so that each pair of primers was located in different exons and so that amplification products included one or more introns. The four pairs of primers created were named after the respective gene plus the sequence numbers: TaTOB1-1, TaTOB1-2, TaTOB1-3, and TaTOB1-4. Sequences of primers are provided upon request.
To identify the presence of the TaTOB1 gene in the wheat genome by PCR, the following plant material was used for DNA extraction: genotypes of bread wheat (2n = 6x = 42, AuAuBBDD) cultivars Tira, Nikoniya, Odeska 267, Yuvileyna (awned), Aurora, and Lybid (awnless); genotypes of durum wheat (2n = 4x = 28, AuAuBB) cultivars Chernomor, Leucurum (awned), Candicans, and Rubrum (awn-like elongations); Aegilops species (2n = 2x = 14): Ae. mutica (TT, awnless), Ae. sharonensis (SShSSh with awn-like elongations), Ae. comosa (MM, awned), Ae. umbellulata (UU, awned), and the hexaploid synthetic species T. migushovae (2n = 6x = 42, AbAbGGDD, awned) [24]. Genomic DNA was isolated from sprouts using the CTAB method [25]. Polymerase chain reaction was carried out by the touchdown method using primer hybridization temperatures for ten cycles starting with 60°C with decrements of 0.5°C each cycle down to 55°C [26]. The reaction mixture composition was as follows: 50 ng DNA, 0.2 mM each dNTP (Thermo Scientific, United States), 1.5 mM MgCl2, 250 nM each primer, and 1 U Taq DNA polymerase (Solis Biodyne, Estonia). The amplification products were separated in 1.5% agarose gel on SB buffer and in 6% denaturing PAGE with 6M urea [25].
RESULTS AND DISCUSSION
Bioinformatic identification of wheat genes orthologous to rice awn development regulators TOB1, DL, and OsETT2. For the three known awn development regulatory genes TOB1, DL, and OsETT2 in rice, the bread wheat genome sequence was found to have orthologs TaTOB1, TaDL, and TaETT2, for each of which homoeoalleles were localized on three subgenomes.
The TaTOB1 gene was found on the second group of chromosomes in all the three bread wheat subgenomes. Its gene structure contains six exons and five introns, some of which are polymorphic for the length among homoeoalleles (Table 1). Length polymorphisms were found for exons 1, 2, and 5 and in all introns. The difference in the length of the exons did not exceed 5 bp, while the differences were greater for the introns: the first and third introns of the TaTOB1-B gene were 20 bp longer than the two homoeologs, and the last, the fifth intron of TaTOB1-D, was shorter by 25 bp than in homoeologs.
The TaDL gene of bread wheat was localized in three subgenomes on the fourth group of chromosomes and its structure contains six exons and five introns (Table 1). The sequence of the gene located in subgenome B is inverted relative to the genes located in other subgenomes. Single-nucleotide polymorphisms of the lengths between homoeoalleles were found in exon 6 as well as in introns 1 and 4. Intron 2 in the TaDL-B gene was longer by 38 bp than in other homoeoalleles.
Differences in the number of introns and exons were revealed for homoeoalleles of the TaETT2 gene localized on the third group of chromosomes (Table 1). In particular, the TaETT2-A homoeoallele had two exons and two introns at the beginning of the sequence that were not found in the homoeoalleles of the subgenomes B and D. Accordingly, TaETT2-A consisted of 11 exons and ten introns, and two other homoeoalleles had nine exons and eight introns. Such a polymorphism may be due to the fact that only the TaETT2-A homoeoallele is functional, and the other two are pseudogenes with deletions. This assumption is supported by the inversion of the TaETT2-B and TaETT2-D sequences with respect to the TaETT2-A direction and the 3.3 kb insertion in the eighth intron of TaTT2-D (Table 1). It is known that insertions of such size are usually due to insertions of retroelements, and they often lead to altered functional activity of the gene [27].
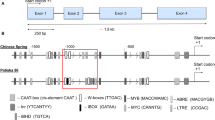
Identification of the TaTOB1 gene in the wheat genome by PCR. Four pairs of primers were designed to amplify TaTOB1 gene fragments from genomic DNA of wheat. In each of the pairs, the left and right primers were localized in the neighboring exons, thus, the entire intron and part of the encoding sequence were amplified in PCR (Fig. 1). For the pair of TaTOB1-1 primers, the left and right primers were in exons 1 and 2, with intron 1 in-between. For the second pair, the left and right primers were in exons 2 and 4 and were delimited by two introns and a third exon. For the pair of primers TaTOB1-3, the left and right primers were in exons 4 and 5, with intron 4 in-between. For the pairs of primers TaTOB1-4, the left and right primers were in exons 5 and 6, with intron 5 in-between.
Based on analysis of the bread wheat genomic sequence, the expected lengths of the amplicons were 467, 496, and 467 bp for the first pair, 1221, 1237, and 1224 bp for the second pair, 226, 234, and 247 bp for the third pair, and 369, 366, and 337 bp for the forth pair for homoeoalleles on subgenomes A (TaTOB1-A), B (TaTOB1-B), and D (TaTOB1-D), respectively. Separation of DNA amplification products of the studied genotypes in agarose gel gave us the following information. The spectra resulted from separation of amplification products obtained with each of the studied primers did not differ between T. aestivum and T. durum (Fig. 2). When separating the genomic DNA amplification products obtained with the pair TaTOB1-1, three components were observed with estimated sizes of 335, 314, and 307 bp. Electrophoretic spectra of bread and durum wheat obtained by separating the products with the second pair of primers TaTOB1-2 contained one component with estimated size of 1120 bp, and the spectra obtained using the third pair of primers TaTOB1-3 contained one component with estimated size of 375 bp. Separation of amplification products with the fourth pair of primers to the TaTOB1 gene produced two components in the wheat spectra, 337 and 251 bp. In the diploid species Ae. mutica, the spectra obtained after separating the amplification products with the first and fourth pairs of primers to the TaTOB1 gene differed from the spectra of durum and bread wheat (Fig. 2). When amplifying with the first pair of primers, instead of the three components, two components of 335 and 307 bp were obtained. In amplification with the fourth pair, the spectrum of Aegilops, in addition to the two amplicones inherent in the wheat DNA spectra, had another component, which may be the result of a nonspecific amplification. Agarose gel does not allow separating the fragments that were expected to be obtained by amplifying the homoeoalleles of the TaTOB1 gene of bread wheat due to a rather small difference in their size. Therefore, for the separation of these fragments and their subgenomic identification, the PCR products were further separated in 6% polyacrylamide gel (PAGE).
Subgenomic identification of amplification products separated by PAGE. For the purpose of subgenomic identification of the amplicons, PAGE separation was carried out for the amplification products obtained with the TaTOB1 primer and DNA of diploid wheat of the Aegilops genus, genotypes of tetraploid wheat T. durum (AuAuBB), bread hexaploid wheat cultivars (AuAuBBDD), and hexaploid synthetic species Triticum miguschovae (AbAbGGDD). The spectra of diploid, tetraploid, and hexaploid species were compared, and conclusions about the subgenomic origin of the components were made. Ae. sharonensis (Ssh) belongs to the same Sitopsis section that includes Ae. speltoides (S), the hypothetical ancestor of wheat subgenomes B and G [28]. Therefore, when DNA spectra of Ae. sharonensis, T. aestivum, T. durum, and T. miguschovae shared the amplicon, it was considered to be a product of subgenome B of the species T. aestivum and T. durum and a product of subgenome G of the species T. miguschovae. The components common to the DNA spectra of durum and bread wheat were attributed to the products of subgenome Au, and those common to the DNA of T. miguschovae and bread wheat were attributed to the products of subgenome D.
When separating the PCR products obtained with the pair of primers TaTOB1-1 in 6% PAGE, the spectra of diploid Aegilops had one component each, which is double for the DNA of Ae. sharonensis, the spectra of durum wheat cultivars had three components, the spectra of bread wheat cultivars had four components, and the spectrum of T. miguschovae had five components (Fig. 3). According to the above-described approach, the lightest double component found in the spectra of durum and bread wheat and T. miguschovae, whose mass matched with the double component of the spectrum of Ae. sharonensis, were attributed to the amplification products of homoeoalleles of subgenomes B and G. This part of the spectrum also contains amplification products of DNA from the three other Aegilops species studied. The larger component, common to bread wheat and Т. miguschovae, was attributed to the amplification product of the homoeoallele of subgenome D. The heaviest component inherent to durum and bread wheat, as well as Т. miguschovae, was attributed to amplification products of the homoeoalleles of subgenomes Au and Ab. Thus, amplification products of the TaTOB1 gene homoeoalleles are polymorphic when using primers to the 5'-part of the gene, and this pair of primers can be used in the study of the genomic structure of introgressive wheat lines with respect to the TaTOB1 gene.
Results of separating the TaTOB1-1 amplification products in PAGE. Subgenomic attribution of the amplicons: 1—Au and Ab; 2—B, G, T, U, M, and Ssh; 3—D. T. aestivum cultivars: 1—Aurora; 2—Lybid; 3—Tira; 4—Yuvileyna. T. durum genotypes: 1—Rubrum; 2—Candicans; 3—Leucurum; 4—Chernomor. Aegilops species: mut—mutica; umb—umbellulata; com—comosa; sh—sharonensis.
For the second pair of primers (Fig. 1), the size of amplification products turned out to be too large, and, therefore, it was not possible to adjust the PAGE separation conditions. When separating the PCR products with the TaTOB1-3 primers in 6% PAGE, one component was detected in each of Aegilops spectra, two in each of durum wheat cultivars, six in each bread wheat cultivars, and five in the spectrum of Т. miguschovae (Fig. 4). A component common for all spectra was identified as an amplification product of the homoeoalleles of subgenomes B and G. The lighter component found in the bread and durum wheat spectra is likely to be formed when amplifying the homoeoallele from subgenome Au common for these two species. The four heavy components in spectra of the bread wheat cultivars and the Т. miguschovae genotype are the result of amplification of homoeoalleles of subgenome D, although the components for the two wheat species differ by size. In the case of the TaTOB1-A homoeoallele for Т. miguschovae, which has subgenome Ab in contrast to subgenome Au of bread and durum wheat, it may not form an amplification product with the pair of TaTOB1-3 primers due to changes in nucleotide sequences of the fourth and/or fifth exons of the TaTOB1-Ab homoeoallele compared to the TaTOB1-B homoeoallele, based on which this pair of primers was designed. Moreover, according to the data of genomic sequencing of bread wheat for exon 5, length polymorphisms are found even between homoeoalleles of this species. However, this pair of primers can also be used in the study of the structure of introgressive wheat lines with respect to the TaTOB1 gene.
Results of separating the TaTOB1-3 amplification products in PAGE. Subgenomic attribution of the amplicons: 1—Au; 2, B, G, M, and Ssh; 3—D (for T. migushovae, Ab and D). T. aestivum cultivars: 1—Aurora; 2—Lybid; 3—Tira; 4—Yuvileyna; 5—Nikoniya. T. durum genotypes: 1—Rubrum; 2—Candicans; 3—Leucurum; 4—Chernomor. Aegilops species: com—comosa; sh—sharonensis.
The spectra obtained by separating the PCR products from DNA of the studied genotypes with the TaTOB1-4 primers revealed the following components: one in Ae. comosa, two in Ae. mutica and Ae. umbellulata, two or three in Ae. sharonensis, four in the bread and durum wheat cultivars, and four in Т. miguschovae (Fig. 5). The resulting picture is difficult to interpret. Two relatively common (in the spectrum of Ae. sharonensis amplicons, the mobility of the lightest component is higher than that in other spectra) components for all spectra were attributed to amplification products of the homoeoalleles of subgenomes B and G. Two components that lie between the TaTOB1-B and TaTOB1-G amplification products in the spectra of bread and durum wheat and Т. miguschovae were attributed to amplicons of the homoeoalleles of subgenomes Au, Ab, and D. However, this is only a preliminary assumption, which has no evidence for this material. One of the primers of the TaTOB1-4 pair is located in the fifth exon, whose variation was already suggested when considering the amplification products obtained using the pair of TaTOB1-3 primers. Therefore, the marker potential of amplification products obtained using the pair of TaTOB1-4 primers for studying the introgressive lines with respect to the TaTOB1 gene alleles is limited.
Results of separating the TaTOB1-4 amplification products in PAGE. Subgenomic attribution of the amplicons: 1—Au and Ab; 2—B, G, M, and Ssh; 3—D. T. aestivum cultivars: 1—Tira; 2—Nikoniya; 3—Odeska 267; 4—Yuvileyna; 5, 7—Lybid; 6—Aurora. T. durum genotypes: 1—Chernomor; 2—Leucurum; 3—Candicans; 4—Rubrum. Aegilops species: mut—mutica; umb—umbellulata; com—comosa; sh—sharonensis.
Comparison of the amplification spectra of DNA of awned and awnless cultivars of bread and durum wheat obtained with the use of three pairs of primers (Figs. 3–5), did not reveal any differences in the spectra of cultivars that differ for the awnedness trait. At first glance, it seems that there is no effect of the studied gene on the development of awns. Of course, one can assume that there are changes in the sequence of nucleotides that transform the functional awn development gene-promoter into a nonfunctional allele localized in that part of the TaTOB1 gene that was amplified by the pair of TaTOB1-2 primers and whose amplification products we could not analyze by separation in PAGE. However, we think that it is more correct to accept the following explanation for the absence of polymorphic products in the DNA amplification spectra of the studied genotypes polymorphic for the awnedness trait with the use of intragenic primers to the TaTOB1 gene. First, the key polymorphism could consist not in a change in the length of amplification products but in a change in the sequence, which does not lead to variation in the lengths of amplification products, such as single nucleotide substitution polymorphisms, which is why we could not identify it. Second, the polymorphism could affect the regulatory regions located upstream or downstream the coding parts of the gene, which were not investigated. Such a polymorphism may lead to a change in gene expression without altering its coding region. Therefore, as the next step, one should consider the study of this gene’s functioning in wheat through determining the level of its expression in genotypes that differ for the presence of awns.
REFERENCES
Dorofeev, V.F., Filatenko, A.A., Migushova, E.P., Udachin, R.A., and Iakubtsiner, M.M., Cultural Flora of USSR, Brezhnev, D.D., Ed., Leningrad: Kolos, 1979, pp. 239–269.
Goud, J.V. and Sadananda, A.R., Two new awn promoter genes in bread wheat, Genetics, 1978, vol. 43, pp. 12–16.
Antonyuk, M.Z., Prokopyk, D.O., Martynenko, V.S., and Ternovska, T.K., Identification of the genes promoting awnedness in the Triticum aestivum/Aegilops umbellulata inrogressive line, Cytol. Genet., 2012, vol. 46, no. 3, pp. 136–143. https://doi.org/10.3103/s009545-2712030024
Ternovska, T.K., Antonyuk, M.Z., and Martynenko, V.S., Genes promoters of awnedness in Triticinae genomes, Factory Eksper. Evol. Organ. Zb. Nauk. Prats., Kyiv: Logos, 2013, vol. 12, pp. 164–168.
McIntosh, R.A., Yamazaki, Y., Dubcovsky, J., Rogers, J., Morris, C., Appels, R., and Xia, X.C., Catalogue of Gene Symbols for Wheat, 2013.
Sourdille, P., Cadalen, T., Gay, G., Gill, B.S., and Bernard, M., Molecular and physical mapping of genes affecting awning in wheat, Plant Breed., 2002, vol. 121, pp. 320–324.
Yoshioka, M., Iehisa, J.C.M., Ohno, R., Kimura, T., Enoki, H., Nishimura, S., Nasuda, Sh., and Takumi, Sh., Three dominant awnless genes in common wheat: Fine mapping, interaction and contribution to diversity in awn shape 9and length, PLoS One, 2017, vol. 12, pp. 1–21. https://doi.org/10.1371/journal.pone.0176148
Prokopyk, D.O. and Ternovska, T.K., Homeotic genes and their role in development of morphological traits in wheat, Cytol. Genet., 2011, vol. 45, no. 1, pp. 41–54. https://doi.org/10.3103/s0095452711010099
Toriba, T. and Hirano, H.Y., The DROOPING LEAF and OsETTIN2 genes promote awn development in rice, Plant J., 2014, vol. 77, pp. 616–626. https://doi.org/10.1111/tpj.12411
Luo, J., Liu, H., Zhou, T., Gu, B., Huang, X., Shangguan, Y., Zhu, J., Li, Y., Zhao, Y., Wang, Y., Zhao, Q., Wang, A., Wang, Z., Sang, T., Wang, Z., and Han, B., An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice, Plant Cell, 2013, vol. 25, pp. 3360–3376. https://doi.org/10.1105/tpc.113.113589
Tanaka, W., Toriba, T., Ohmori, Y., Yoshida, A., Kawai, A., Mayama-Tsuchida, T., Ichikawa, H., Mitsuda, N., Ohme-Takagi, M., and Hirano, H-Y., The YABBY Gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet, Plant Cell, 2012, vol. 24, pp. 80–95. https://doi.org/10.1105/tpc.111.094797
Satoh, N., Itoh, J.I., and Nagato, Y., The SHOOTLESS2 and SHOOTLESS1 genes are involved in both initiation and maintenance of the shoot apical meristem through regulating the number of indeterminate cells, Genetics, 2003, vol. 164, no. 1, pp. 335–346. PMCID: PMC1462562.
Itoh, J.I., Kitano, H., Matsuoka, M., and Nagato, Y., Shoot organization genes regulate shoot apical meristem organization and the pattern of leaf primordium initiation in rice, Plant Cell, 2000, vol. 12, pp. 2161–2174.
Abe, M., Yoshikawa, T., Nosaka, M., Sakakibara, H., Sato, Y., Nagato, Y., and Itoh, J., WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining microRNA and transacting small interfering RNA accumulation in rice, Plant Physiol., 2010, vol. 154, pp. 1335–1346. https://doi.org/10.1104/pp.110.160234
Song, X., Wang, D., Ma, L., Chen, Z., Li, P., Cui, X., Liu, C., Cao, S., Chu, C., Tao, Y., and Cao, X., Rice RNA-dependent RNA polymerase 6 acts in small RNA biogenesis and spikelet development, Plant J., 2012, vol. 71, pp. 378–389. https://doi.org/10.1111/j.1365-313X.2012.05001.x
Goncharov, N.P., Comparative-genetic analysis—a base for wheat taxonomy revision, Czech. J. Genet. Plant Breed., 2005, vol. 41 (special issue), pp. 52–55.
Prokopyk, D.O. and Ternovska, T.K., SSR-marking of genes taking part in control of awnedness in durum wheat (Triticum durum Desf.), Visn. Ukr. Tovar. Genet. Selec., 2010, vol. 8, no. 1, pp. 31–40.
Du, F., Guan, C., and Jiao, Y., Molecular mechanisms of leaf morphogenesis, Mol. Plant, 2018, vol. 11, pp. 1117–1134.
Prunet, N. and Meyerowitz, E.M., Genetics and plant development, Comptes Rendus Biologies, 2016, vol. 339, pp. 240–246. doi.org/https://doi.org/10.1016/j.crvi.2016.05.003
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J., Basic local alignment search tool, J. Mol. Biol., 1990, vol. 215, pp. 403–410.
Jin, J., Tian, F., Yang, D.-C., Meng, Y.Q., Kong, L., Luo, J., and Gao, G., PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants, Nucleic Acids Res., 2017, vol. 45, pp. D1040–D1045. https://doi.org/10.1093/nar/gkw982
Dvorak, J., Luo, M.-C., Gu, Y.Q., et al., Sequencing the Aegilops tauschii genome. http://aegilops.wheat. ucdavis.edu.
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., and Madden, T.L., Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction, BMC Bioinform., 2012, vol. 13, no. 134. https://doi.org/10.1186/1471-2105-13-134
Zhirov, E.G., Synthesis of a new hexaploid wheat, Tr. Prikl. Bot. Genet. Sel., 1980, vol. 68, pp. 14–16.
Sambrook, J., Fritsch, E.F., and Maniatis, T., Molecular Cloning: A Laboratory Manual, New York: Cold Spring Harbor Laboratory Press, 1989.
Korbie, D.J. and Mattick, J.S., Touchdown PCR for increased specificity and sensitivity in PCR amplification, Nat. Protoc., 2008, vol. 3, pp. 1452–1456. https://doi.org/10.1038/nprot.2008.133
Kidwell, M.G. and Lisch, D., Transposable elements as sources of variation in animals and plants, Proc. Natl. Acad. Sci. U. S. A., 1997, vol. 94, pp. 7704–7711.
Petersen, G., Seberg, O., Yde, M., and Berthelsen, K., Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum), Mol. Phylogenet. Evol., 2006, vol. 39, pp. 70–82. https://doi.org/10.1016/j.ympev.2006.01.023
Funding
This study was partly supported by a grant from the International Charitable Fund of the Renaissance of the Kyiv-Mohyla Academy: Molecular Mechanisms of Development of Morphological Traits of Spike in Wheat.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by K. Lazarev
About this article
Cite this article
Navalikhina, A., Antonyuk, M., Pasichnyk, T. et al. Identification of Oryza sativa’s Awn Development Regulatory Gene Orthologs in Triticinae Accessions. Cytol. Genet. 53, 267–275 (2019). https://doi.org/10.3103/S0095452719040091
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0095452719040091