Abstract
Structural DNA nanotechnology offers the capacity to construct ultraminiaturized devices with programmed nanoscale geometry, mechanical and dynamic properties, and site-specific molecular functionalities. These features and the possibility to position and orient molecules in user-defined ways may be exploited to create custom instruments for precision measurements of molecular-scale structure, dynamics, and interactions. Such devices may help constrain molecular motion along interesting reaction coordinates and may also exert forces to probe the mechanical properties or dynamics of molecules under study. Multiple ways of reading out device states may be used, including atomic force microscopy or transmission electron microscopy imaging, single-molecule or bulk fluorescence, or ionic conductivity as in nanopore systems. Early successes with custom scientific instruments based on DNA origami underline the tremendous potential to enable new approaches to making scientific discoveries in biological and synthetic materials systems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Imaging technologies such as electron microscopy and superresolution imaging, have allowed profound insights into nanoand microscale structures and mechanisms that govern many aspects of system behavior. Advances in nano- to microscale characterization (e.g., microrheology) or single-molecule experimental methods in biophysics, have led to key insights into relating component function or local material properties to system behavior. However, the capacity to manipulate and probe biological and synthetic materials systems at the nano- and microscale remains limited due to the difficulty of engineering probes with commensurate dimensions and force scales. DNA origami provides a path to construct such probes due to the ability to design precise geometry, program mechanical and dynamic properties, and incorporate chemical functionalization in a site-specific manner in devices on the scale of ~10–1000 nm. DNA origami is uniquely suited for integration into complex materials for in situ characterization or as tools to enhance other measurement methods such as electron microscopy, atomic force microscopy (AFM), or force spectroscopy. They can also greatly enhance bulk readout methods by providing control over local parameters, such as molecular distances, within the bulk solution.
Here, we summarize current applications of DNA origami nanostructures as devices for precision measurements, and we highlight recent efforts, including implementations to enhance other imaging or measurement technologies and integration with microfabricated, synthetic, and biological materials systems.
Measurements enabled by DNA origami geometry and functionalization
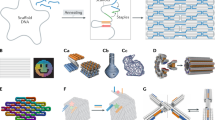
The ability to incorporate molecules or chemical functionalities in a site-specific manner into objects with complex geometry (see the article by Gothelf in this issue1) enables a unique level of control over templating molecules or materials at the nanoscale, which can be leveraged to develop measurement devices such as using DNA origami platforms to pattern motor proteins (proteins that use chemical energy to move along a substrate) to study their cooperative behavior2–4 (Figure 1a) or to pattern cellular ligands to study the effects of ligand arrangement on receptor-mediated signaling5–7 (Figure 1b–c). For example, Shaw et al. used DNA origami nanocalipers to demonstrate that signal activation and invasiveness of human breast cancer cells is directly mediated by spacing of ephrin ligands presented to Eph receptors on the cell surface by testing the response of cells to ligands positioned at a variety of separation distances.7 Templating arrangements of optically active components such as fluorophores8 (Figure 1d) or gold nanoparticles9,10 (Figure 1e) enables investigating nanoscale light transport or harvesting.11
DNA origami measurement tools that leverage geometric design. DNA origami enables templating of biomolecules or materials as a foundation for well-defined measurement systems to study (a) motor protein cooperativity using DNA origami (blue) to template defined numbers and arrangements of motors. Reprinted with permission from Reference 2. © 2012 AAAS. DNA origami has also been used to template ligands to study (b) the response of cells to ligands (red) at varying separation distances (e.g., close distance on the top schematic and transmission electron micrograph, or farther distance on the bottom schematic and transmission electron micrograph),7 or to (c) cell signaling mediated by the signaling molecule Smad 2/3 in response to clustering of receptors (green) driven by a DNA origami template (orange). Scale bar = 20 nm in (b). DNA origami templates have also been used to study (d) light transport along fluorophores and (e) along gold nanoparticles. Reprinted with permission from Reference 9. © 2016 American Chemical Society. The geometric precision of DNA origami has also enabled nanopores integrated in (f) solid-state13 or (g) synthetic membranes14 and the design of (h) nanoscale environments to study effects of confinement on the stability of G-quadruplexes. (b) Reprinted with permission from Reference 5. © 2015 Wiley. (d) Reprinted with permission from Reference 8. © 2011 American Chemical Society. (h) Reprinted with permission from Reference 17. © 2017 Macmillan Publishers Ltd. Note: dsDNA, double-stranded DNA.
The nanometer-scale resolution of DNA origami geometries can also be leveraged to create nanopores for integration with solid-state systems12,13 (Figure 1f) or lipid membranes14–16 (Figure 1g). The nanopore systems enable detection of biomolecules by measuring blockage of ion currents as molecules pass through the pore. Given the precisely controlled dimensions of just a few nanometers, DNA origami nanopores may discriminate translocation events based on size, and they can also selectively capture molecules via chemical functionalities placed directly at the pore, leading to selective detection via longer current blockages.12 In addition to biomolecule detection in nanopores, the geometrical design of DNA origami nanostructures has been leveraged to measure the effects of a confined environment on biomolecular structure and dynamics. Shrestha et al.17 studied the behavior of G-quadruplexes, a four-stranded DNA structure motif that is found in genomic DNA at the ends of chromosomes, inside square DNA origami tubes (Figure 1h). The experiments revealed that confinement stabilizes the quadruplex structure, which has implications for the function of these G-quadruplex structures in highly crowded intracellular environments.
Measurements enabled by DNA origami mechanical and dynamic properties
In addition to precise nanoscale geometry, the mechanical and dynamic properties of DNA origami devices can enable enhanced measurement capabilities such as exhibiting detectable conformational changes that are responsive to the local environment, probing interactions or conformational changes of molecular components or complexes along defined reaction coordinates, and applying forces in specific directions. For example, Liedl et al.18 and Kuzuya and co-workers19 demonstrated a single-molecule detection device consisting of two components that are pinned together (Figure 2a). Interaction with a target molecule causes a change in configuration, which can be used as an indicator of target binding. Other studies have used DNA origami nanohinges with motion constrained along a single angular degree of freedom to probe nucleosome stacking interactions20 (Figure 2b) or conformational changes21,22 (Figure 2c) where the DNA hinges function as nanoscopic calipers. Knowing the torsional stiffness of these calipers also enables quantification of mechanical response of the sample, for example, interaction forces between nucleosomes.20
DNA origami measurement tools that leverage device dynamic and mechanical properties. These measurement devices can (a) detect target biomolecules by undergoing a conformational change upon binding and (b) measure nucleosome interaction forces by analyzing the closing of a hinge with well-defined mechanical properties.20 (c) Opening or closing of a DNA origami hinge has also been used to study the conformational changes21 of nucleosomes. (a) Reprinted with permission from Reference 19. © 2011 Macmillan Publishers Ltd. Other studies have demonstrated developed dynamic devices to quantify (d) DNA stacking interactions where the stacking mediates a conformational change between closed (top) and open (bottom) states.23 (e) Similarly, depletion forces due to molecular crowding can be measured following a similar approach.24 (f) In addition, a recent study established methods to apply forces (F) to device components to study the effects of DNA tension on DNA–protein interactions. Reprinted with permission from Reference 25. © 2016 AAAS.
Kilchherr et al. determined the ultraweak forces and the lifetimes at which individual DNA base pair stacking bonds stochastically break by combining dual-beam optical tweezers with a DNA nanodevice designed to enhance the resolution of the tweezer system and to allow for controlled reversible binding and unbinding of stacking bonds23 (Figure 2d). Hudoba and co-workers developed a device that similarly converts between an expanded and compacted state24 (Figure 2e), where the distribution of states serves as a quantitative measure of depletion forces caused by molecular crowding down to the scale of 10 f N. In addition to responding to external forces, recent studies have leveraged the mechanical behavior of DNA origami devices, including the ability to apply tension forces to an internal component via the entropic elasticity of ssDNA, and the ability to design tunable component stiffness. Nickels et al. implemented entropic forces of ssDNA to test the effects of force on the interaction between DNA and a DNA-binding protein25 (Figure 2f), and Iwaki et al. developed an assay that uses a DNA origami nanospring to study the behavior of myosin under mechanical load.26
Integrating DNA origami with other measurement and material systems
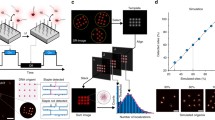
The nanoscale dimensions of DNA origami devices make them amenable to integration in a range of other experimental assays. Accordingly, a number of recent studies have leveraged the geometric and dynamic properties of DNA origami devices to enhance other existing imaging or characterization technologies, including electron microscopy, AFM, fluorescence methods, and force spectroscopy. Martin et al. used a DNA origami platform as a reference frame for structure determination in cryo-TEM (cryogenic TEM) studies, where the DNA origami structure provided a high contrast marker to identify the location and orientation of proteins incorporated in the cavity27 (Figure 3a). Other recent work has utilized a DNA origami frame to localize and aid in visualizing DNA recombination28 (Figure 3b) or formation of four-stranded G-quadruplex structures,29 and DNA origami platform structures have been widely used as measurement standards or calibration tools for super-resolution imaging30–32 (Figure 3c) or other fluorescence methods.33 The rigidity of DNA origami beams has also been leveraged to improve resolution of force spectroscopy measurements due to decreased thermal fluctuations of the handles34 (Figure 3d).
Current research in structural DNA nanotechnology has led to breakthroughs that can likely expand the scope of DNA origami measurement technologies. These include efforts to combine DNA origami devices with other materials systems such as carbon nanotubes or synthetic membranes, microfabricated systems to enable simpler control, inputs, or outputs, and integration of DNA origami devices onto biological cells. Specifically, DNA origami structures have been used to template carbon nanotubes with the ability to make electrical connections between patterned electrodes35 (Figure 3e) or to template the formation of size-controlled liposomes36 (Figure 3f). Another recent study leveraged directed assembly of DNA origami structures onto lithographically patterned sites within photonic crystal cavities37 (Figure 3g), establishing a foundation for DNA origami applications in nanophotonic devices. Lastly, Akbari et al. recently demonstrated a robust approach to attach DNA origami structures to the surfaces of live cells38 (Figure 3h), providing a foundation for applications at the cell surface. Prior implementations of smaller DNA measurement devices inside live cells39,40 and the recent development of methods to transfect DNA origami structures into cells41 also suggest potential for intracellular applications.
Conclusion
In conclusion, DNA origami offers a versatile platform to probe spatial effects on the scale from a few nm to 1000 nm. This domain is currently difficult to accurately probe with other methods. Importantly, as several of the previously mentioned studies show, DNA origami can help decouple nanoscale interactions from overall concentration dependent effects or other competing effects. Some of the examples mentioned make excellent use of the fact that origami conformational states can be coupled to individual molecular events, thus enabling single-molecule readouts by looking at the state of the individual origamis. In other previously mentioned examples, origami helps to isolate distance-dependent, small number of molecule effects while still using ensemble averaged readout methods such as fluorescence, gels, and quantitative polymerase chain reaction (in the case of biological systems).
Integrating DNA origami devices into experimental assays and with synthetic or biological materials systems. The positioning capabilities of DNA origami have been leveraged to enhance (a) cryogenic transmission electron microscope (cryo-TEM) imaging and structure determination,27 (b) atomic force microscope (AFM)-based studies of biomolecular processes such as DNA recombination mediated by the enzyme Cre recombinase, (c) calibration of super-resolution fluorescence imaging methods where fluorophores attached to DNA strands (P*) bind to specific sites on a DNA origami platform that contain the complementary strand (P), and (d) the stiffness of DNA origami nanorods, which were used as handles to enable improved resolution in force spectroscopy measurements.34 Additional work has integrated DNA origami structures with (e) carbon nanotubes to enable formation of an electrical junction shown schematically and in an AFM image (scale bar = 50 nm), (f) synthetic liposomes by using the origami structure to template the size, and (g) microfabricated optical devices where placement and orientation of the origami in a photonic crystal cavity, illustrated here by the AFM (scale bar = 250 nm) images, controls the intensity of the light output from fluorophores on the origami structure. (h) Recent work has integrated fluorescently labeled DNA origami (fluorophores in red) structures onto the surface of live cells.38 (b) Reprinted with permission from Reference 28. © 2014 American Chemical Society. (c) Reprinted with permission from Reference 30. © 2016 Macmillan Publishers Ltd. (e) Reprinted with permission from Reference 35. © 2010 Macmillan Publishers Ltd. (f) Reprinted with permission from Reference 36. © 2016 Macmillan Publishers Ltd. (g) Reprinted with permission from Reference 37. © 2016 Macmillan Publishers Ltd.
The field shows clear signs for a welcome transition from potential utility to actual utility. In the past, many studies focused on providing proof of concept of possible measurement devices, highlighting their potential capabilities and advantages, which is undoubtedly a necessary step. Now, an increasing number of studies actually use such devices to obtain new scientific insights about the molecular systems under study, which were not known or not previously accessible with other means. In addition, as suggested by recent efforts at system integration, it is an opportune time to translate DNA origami nanodevices into new material systems. Ultimately, the discoveries made with DNA origami devices, and not the devices themselves, will reward these efforts and demonstrate the transformative impact of DNA origami as instruments for precision measurement.
References
K.V. Gothelf, MRS Bull. 42 (12), 897 (2017).
N.D. Derr, B.S. Goodman, R. Jungmann, A.E. Leschziner, W.M. Shih, S.L. Reck-Peterson, Science 338 (6107), 662 (2012).
R.F. Hariadi, A.J. Appukutty, S. Sivaramakrishnan, ACS Nano 10 (9), 8281 (2016).
R.F. Hariadi, R.F. Sommese, S. Sivaramakrishnan, Elife 4, e05472 (2015).
A. Angelin, S. Weigel, R. Garrecht, R. Meyer, J. Bauer, R.K. Kumar, M. Hirtz, C.M. Niemeyer, Angew. Chem. Int. Ed. Engl. 54 (52), 15813 (2015).
R.O. Pedersen, E.G. Loboa, T.H. LaBean, Biomacromolecules 14 (12), 4157 (2013).
A. Shaw, V. Lundin, E. Petrova, F. Fordos, E. Benson, A. Al-Amin, A. Herland, A. Blokzijl, B. Högberg, A.I. Teixeira, Nat. Methods 11 (8), 841 (2014).
I.H. Stein, C. Steinhauer, P. Tinnefeld, J. Am. Chem. Soc. 133 (12), 4193 (2011).
K. Vogele, J. List, G. Pardatscher, N.B. Holland, F.C. Simmel, T. Pirzer, ACS Nano 10 (12), 11377 (2016).
G.P. Acuna, F.M. Moller, P. Holzmeister, S. Beater, B. Lalkens, P. Tinnefeld, Science 338 (6106), 506 (2012).
K. Pan, E. Boulais, L. Yang, M. Bathe, Nucleic Acids Res. 42 (4), 2159 (2014).
N.A. Bell, C.R. Engst, M. Ablay, G. Divitini, C. Ducati, T. Liedl, U.F. Keyser, Nano Lett. 12 (1), 512 (2012).
R. Wei, T.G. Martin, U. Rant, H. Dietz, Angew. Chem. Int. Ed. Engl. 51 (20), 4864 (2012).
M. Langecker, V. Arnaut, T.G. Martin, J. List, S. Renner, M. Mayer, H. Dietz, F.C. Simmel, Science 338 (6109), 932 (2012).
S. Krishnan, D. Ziegler, V. Arnaut, T.G. Martin, K. Kapsner, K. Henneberg, A.R. Bausch, H. Dietz, F.C. Simmel, Nat. Commun. 7, 12787 (2016).
K. Gopfrich, C.Y. Li, M. Ricci, S.P. Bhamidimarri, J. Yoo, B. Gyenes, A. Ohmann, M. Winterhalter, A. Aksimentiev, U.F. Keyser, ACS Nano 10 (9), 8207 (2016).
P. Shrestha, S. Jonchhe, T. Emura, K. Hidaka, M. Endo, H. Sugiyama, H. Mao, Nat. Nanotechnol. 12 (6), 582 (2017).
T. Liedl, B. Högberg, J. Tytell, D.E. Ingber, W.M. Shih, Nat. Nanotechnol. 5, 520 (2010), doi:10.1038/nnano.2010.107.
A. Kuzuya, Y. Sakai, T. Yamazaki, Y. Xu, M. Komiyama, Nat. Commun. 2, 449 (2011).
J.J. Funke, P. Ketterer, C. Lieleg, S. Schunter, P. Korber, H. Dietz, Sci. Adv. 2 (11), e1600974 (2016).
J.V. Le, Y. Luo, M.A. Darcy, C.R. Lucas, M.F. Goodwin, M.G. Poirier, C.E. Castro, ACS Nano 10 (7), 7073 (2016).
J.J. Funke, P. Ketterer, C. Lieleg, P. Korber, H. Dietz, Nano Lett. 16 (12), 7891 (2016).
F. Kilchherr, C. Wachauf, B. Pelz, M. Rief, M. Zacharias, H. Dietz, Science 353 (6304), aaf5508 (2016).
M.W. Hudoba, Y. Luo, A. Zacharias, M.G. Poirier, C.E. Castro, ACS Nano 11 (7), 6566 (2017).
P.C. Nickels, B. Wunsch, P. Holzmeister, W. Bae, L.M. Kneer, D. Grohmann, P. Tinnefeld, T. Liedl, Science 354 (6310), 305 (2016).
M. Iwaki, S.F. Wickham, K. Ikezaki, T. Yanagida, W.M. Shih, Nat. Commun. 7, 13715 (2016).
T.G. Martin, T.A. Bharat, A.C. Joerger, X.C. Bai, F. Praetorius, A.R. Fersht, H. Dietz, S.H. Scheres, Proc. Natl. Acad. Sci. U.S.A. 113 (47), E7456 (2016).
Y. Suzuki, M. Endo, Y. Katsuda, K. Ou, K. Hidaka, H. Sugiyama, J. Am. Chem. Soc. 136 (1), 211 (2014).
Y. Sannohe, M. Endo, Y. Katsuda, K. Hidaka, H. Sugiyama, J. Am. Chem. Soc. 132 (46), 16311 (2010).
R. Jungmann, M.S. Avendano, M. Dai, J.B. Woehrstein, S.S. Agasti, Z. Feiger, A. Rodal, P. Yin, Nat. Methods 13 (5), 439 (2016).
M. Dai, R. Jungmann, P. Yin, Nat. Nanotechnol. 11 (9), 798 (2016).
M. Reuss, F. Fördo˝s, H. Blom, O. Öktem, B. Högberg, H. Brismar, New J. Phys. 19, 025013 (2017).
J.J. Schmied, M. Raab, C. Forthmann, E. Pibiri, B. Wunsch, T. Dammeyer, P. Tinnefeld, Nat. Protoc. 9 (6), 1367 (2014).
E. Pfitzner, C. Wachauf, F. Kilchherr, B. Pelz, W.M. Shih, M. Rief, H. Dietz, Angew. Chem. Int. Ed. Engl. 52 (30), 7766 (2013).
H.T. Maune, S.P. Han, R.D. Barish, M. Bockrath, W.A. Goddard III, P.W. Rothemund, E. Winfree, Nat. Nanotechnol. 5 (1), 61 (2010).
Y. Yang, J. Wang, H. Shigematsu, W. Xu, W.M. Shih, J.E. Rothman, C. Lin, Nat. Chem. 8 (5), 476 (2016).
A. Gopinath, E. Miyazono, A. Faraon, P.W. Rothemund, Nature 535 (7612), 401 (2016).
E. Akbari, M.Y. Mollica, C.R. Lucas, S.M. Bushman, R.A. Patton, M. Shahhosseini, J.W. Song, C.E. Castro, Adv. Mater. 1703632 (2017), https://doi.org/10.1002/adma.201703632.
S. Modi, M.G. Swetha, D. Goswami, G.D. Gupta, S. Mayor, Y. Krishnan, Nat. Nanotechnol. 4 (5), 325 (2009).
S. Saha, V. Prakash, S. Halder, K. Chakraborty, Y. Krishnan, Nat. Nanotechnol. 10 (7), 645 (2015).
A. Chopra, S. Krishnan, F.C. Simmel, Nano Lett. 16 (10), 6683 (2016).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Castro, C.E., Dietz, H. & Högberg, B. DNA origami devices for molecular-scale precision measurements. MRS Bulletin 42, 925–929 (2017). https://doi.org/10.1557/mrs.2017.273
Published:
Issue Date:
DOI: https://doi.org/10.1557/mrs.2017.273