Abstract
Protein-based materials are a powerful instrument for a new generation of biological materials, with many chemical and mechanical capabilities. Through the manipulation of DNA, researchers can design proteins at the molecular level, engineering a vast array of structural building blocks. However, our capability to rationally design and predict the properties of such materials is limited by the vastness of possible sequence space. Directed evolution has emerged as a powerful tool to improve biological systems through mutation and selection, presenting another avenue to produce novel protein materials. In this prospective review, we discuss the application of directed evolution for protein materials, reviewing current examples and developments that could facilitate the evolution of protein for material applications.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
A cornerstone of human technology has been harnessing and repurposing the materials available in our environments. Many such materials, such as wood, silk, or bone, are harvested from living organisms. These materials are exquisitely ordered structures, often with impressive properties, and are robustly grown by living systems from relatively simple feedstocks under mild conditions. For most of human history, we have grown and harvested these natural materials, however in recent decades this relationship has started to change. Increasingly, we have taken greater control over biological systems at a molecular level, programing biological materials through DNA manipulation and manufacturing using biological platforms.
Recent developments in biotechnology have facilitated huge advances in biological engineering of living systems. Breakthroughs in DNA synthesis and sequencing technology have facilitated the study and reliable construction of DNA encoding for increasingly more complex systems. The discipline of synthetic biology has emerged alongside these technologies, seeking to apply engineering principles to biology[1] and develop tools for the manipulation of these systems. Using an engineering approach, researchers have begun to characterize and develop biological systems, from genetic components,[2] to genetic circuits,[3] and towards whole organisms[4] or even microbial ecosystems.[5] The tools developed within the field of synthetic biology have been applied in many ways, from helping to understand fundamental principles of biology,[6] to the biosynthesis of high-value chemicals.[7] As the discipline grows and develops, it continues to find new and innovative applications.
The developments of synthetic biology have also become attractive to the material sciences.[8–10] Proteins in particular have become a powerful tool for materials science, capable of myriad mechanical and chemical properties due to rich complexity emerging from their sequences. The potential applications of customizable engineered and biosynthesized protein materials span many disciplines and industries, and so far implementation has been primarily medical. Engineered protein materials can be used in drug delivery,[11,12] or as tissue culture scaffolds.[13–15] In the future, materials made by engineered living systems could have sophisticated functionality, with complex spatial organization through synthetic patterning processes.[16] Furthermore, engineered living materials can have significant advantages over traditional materials, being able to grow, self-heal, and respond to their environments.[17]
Our understanding of natural materials and structural biology has advanced significantly in recent years, for example in our understanding of common structural motifs.[18] However, especially among proteins, many challenges remain in mapping amino acid sequence to structure and consequently function. While the folding and three-dimensional structure of proteins is a well-known unsolved problem,[19] the macromolecular assembly of proteins into oligomers presents an even more complex challenge. Given that protein materials require macromolecular assembly, predicting their large-scale structures and the emergent material properties reveals significant lack in knowledge.
However, our limited understanding and ability to rationally design proteins does not have to be a handicap for producing novel protein materials. If we look at nature, the rich complexity of living systems has evolved over time without requiring rational design or guidance. Evolution occurs agnostically to the system in question, with function emerging and improving through random mutation and selection.
Directed evolution has been a staple of biotechnology throughout history, primarily practiced by farmers breeding the crops and farm animals found today. However, in recent decades, this process has been harnessed in the laboratory. With the advancement of molecular biology, researchers have been able to harness evolution in a targeted manner, applying mutations to specific proteins and selecting variants with the desired molecular properties. Directed evolution has emerged as a leading technology for the creation of new and improved biological systems, and indeed such efforts have been recognized by the 2018 Nobel Prize for Chemistry. The efficacy of such methods has led to widespread use of evolutionary techniques within synthetic biology,[20] facilitating not only the improvement of synthetic biological systems, but also furthering our functional understanding through the identification of responsible mutations.
Given the vast untapped potential of protein materials, new tools are required to speed up their development. This prospective review focuses on the application of directed evolution to protein materials. We will first provide a brief overview of artificial protein materials, highlighting several classes of material and techniques used in protein material engineering. We will then discuss directed evolution, introducing common methods and concepts from the field. Finally, we discuss the application of directed evolution to protein materials, highlight current work in that direction, technical challenges, relevant developments, and look forward to potential applications.
Protein materials
Biotechnological advances in recent decades have facilitated the production of customized proteins for a variety of medical and industrial applications. Due to the rapid advance of DNA manipulation technologies, we have a high degree of control over the molecular structure of proteins, and consequently this has been exploited to produce a wide variety of synthetic protein materials. One hallmark of protein material systems is that they are composed of monomeric elements, which assemble into large-scale structures. The self-assembly is also usually hierarchical, with polymeric assemblies forming larger structures such as fibers or sheets, making up the material.
Typically, synthetic protein materials are composed of modular components containing repeated peptide sequence inspired by natural materials such as elastin, silk, or collagen. These proteins are designed in silico through DNA sequence, which is inserted onto plasmid vectors and expressed in microbial culture.[21] The resulting culture is then processed to purify and concentrate the proteins of interest. The proteins are subsequently allowed to assemble into supramolecular structures, which are then assessed for their properties or used for further applications.
There are many classes of protein materials that have found extensive use within the materials community, usually inspired by or derived from natural sources. There are several detailed reviews exploring protein materials, on aspects such as their protein sequence and structure,[22] their oligomerization,[23] their nanostructures and analysis,[24] and their ability to be combined into multicomponent materials.[25] Furthermore, for several classes of material, there are active communities of researchers and a wealth of literature. As such, we will only present a brief summary of several classes, in order to highlight sequence variation, sequence function relationships, and how these materials are designed and produced.
Silk
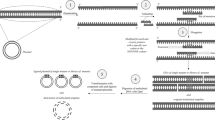
Silks are fibrous proteins with a long history of material use and extensive study, and are thus one of the best-studied protein materials. Silks are employed in many forms, including woven textiles, non-woven mats, films, or hydrogels, and have many applications due to their impressive material properties and biocompatibility.[26] Silks and silk-inspired proteins have been widely adopted as synthetic protein materials, with many established tools to engineer silk molecular structure and produce silk proteins in microbial hosts.[27] Researchers have also developed ways to spin recombinant silks into fibers with comparable physical characteristics with natural fibers[28] [Fig. 1(a)].
Examples of commonly used and recombinantly produced protein materials: (a) silk (adapted with permission from Ref. 28 copyright 2016 Nature Publishing Group), (b) elastin-like proteins (ELPs) (adapted with permission from Ref. 40 copyright 2018 American Chemical Society), (c) collagen (adapted with permission from Ref. 142 copyright 2001 Wiley Online Library), (d) curli (adapted with permission from Ref. 52 copyright 2017 American Chemical Society), (e) de novo designed protein DHF107 (adapted with permission from Ref. 60 copyright 2018 American Association for the Advancement of Science). In each case, the critical amino acid motifs are shown, with X representing variable positions, as well as electron micrographs showing high magnification images of materials assembled from the expressed proteins. Sequences encoding protein materials can be concatenated into block copolymers, to imbue the resulting proteins with complex self-assembly properties. For example, (f) Rabotyagova et al.[67] combined silk-inspired modules to produce morphogenically distinct nanoparticles (adapted with permission from Ref. 67 copyright 2009 American Chemical Society). (g) Huber et al.[68] created amphiphilic ELPs that formed capsules inside bacterial cells. Interactions between proteins can also be exploited to generate protein macromolecular assemblies (adapted with permission from Ref. 68 copyright 2015 Nature Publishing Group). Such interactions can be used to engineer (h) micrometer scale fibers (adapted with permission from Ref. 72 copyright 2017 Nature Publishing Group), (i) 2D crystalline arrays (adapted with permission from Ref. 73 copyright 2016 Nature Publishing Group), (j) covalently crosslinked hydrogels (adapted with permission from Ref. 75 copyright 2014 National Academy of Sciences).
Silk fibroin, the protein within silk materials, is an insoluble protein that assembles into fibrous polymeric structures. These proteins can be found in a range of insect and spider species, and the protein sequences found in each organism have diverged over time,[29] particularly as organisms have come to occupy different ecological niches and hunting modes.[30] In fact, single organisms are capable of producing many kinds of silk for different purposes.[31,32] The molecular organization of silk, and thus the resulting material properties, depends significantly on DNA sequence.
In general, silk fibroin proteins are composed of modular subunits with different characteristics. In the well-studied silkworm Bombyx mori for example, the proteins have a hydrophobic antiparallel beta-pleated sheet conformation, with the beta-sheet nanocrystals embedded within amorphous domains.[33] However among other arthropod species, there are a wide variety of other protein sequences and structures, with domains containing parallel beta-sheets, alpha-helices, collagen-like domains, or poly-glycine motifs.[34]
The different sequences of silk proteins give rise to distinct mechanical properties. Spiders can spin up to seven different types of silks, each with distinct functions. For the Golden Orb Weaver spider, Nephila clavipes, major ampullate dragline silk forms the main frame of the web and is composed of two proteins. At the level of amino acids, this silk is composed of crystalline poly(A) or poly(GA) domains, which repeat within segments rich in GGX motifs (where X is a variable amino acid), and these that provide strong and rigid fibers. Furthermore, the proteins contain repeats of a GPGXX motif, which form beta-spirals and confer significant elasticity to the silk.[35] Within the same spider, minor ampullate silk is used to reinforce webs, and lacks GPGXX domains, so consequently has lower elasticity.[35] On the other hand, flagelliform silk, which is used to capture prey, is composed predominantly of the GPGXX repeat, and has exceptional elasticity. Such diversity within a single class of species hints at the possible diversity of protein sequences and material properties that could be accessed with evolutionary techniques.
Elastin
Elastin-like polypeptides (ELPs), inspired by the tropoelastin proteins that provide elasticity to mammalian tissues, have also become a staple of protein material biotechnology.[36] As such, ELPs have found a range of applications, such as protein purification[37] and drug delivery.[38] ELPs are modular elements commonly composed of repeating VPGXG motifs, where the X amino acid can take a range of identities, imbuing the ELP with distinct properties. The number of repeats can also be varied, providing a further mechanism to influence properties.
ELPs are generally soluble in water at low temperatures, but undergo a reversible transition at higher temperatures to an insoluble phase, characterized by a shift in turbidity. The insoluble protein agglomerations can take a range of different architectures, dependent on peptide sequence.[39] In addition, ELPs can be combined with chemically active domains that can be crosslinked to form hydrogels, which undergo the thermoresponsive transition to form materials with complex microstructure[40] [Fig. 1(b)].
The sequence-function relations for ELPs are complex and we have only begun to understand them. Significant progress has been made by assessing many polypeptide permutations containing amino acids commonly found in ELPs.[41] This work revealed heuristics to identify ELP-like phase behavior, and showed how sequence can be changed to tune the phase transition. Li et al.[42] further showed that the directionality of an ELP sequence significantly influences its behavior. The canonical ELP sequence, poly(VPGVG), and its reverse sequence poly(VGPVG) were both studied and found to have radically different properties despite having the same atomic composition. While the canonical ELP became insoluble at higher temperatures and redissolved upon cooling, the reversed sequence remained aggregated after cooling to well below the transition temperature. Such unpredictable differences in material behavior from proteins with very similar compositions highlight the need for evolutionary techniques to fully explore the sequence space.
Collagen
Another class of protein materials is collagen, an abundant structural protein prevalent in animals where they are assembled into fibers and networks in tissues and bones. Collagens form right-handed, triple helical fibers, and their structure requires every third amino acid to be a glycine, such that they are made up of a repeating GXY motifs.[43] The amino acids in the X and Y positions are variable, but are most often prolines and hydroxyprolines. This freedom in sequence lends collagen proteins a diverse set of properties, with some forming fibrils and others traversing cell membranes.[44] The prediction of collagen structure presents a challenge, however some progress has been made to understand how different amino acids impact collagen stability.[45]
Collagen proteins can also be found in bacteria,[46] which also form the characteristic triple-helical structures despite lacking hydroxyproline amino acids.[47] Since hydroxyproline is formed through post-translational modification by machinery not found in bacteria, bacterial collagens can be produced readily by bacteria, and have begun to find a variety of applications.[48]
Curli
A further biological material that has emerged in recent years is the bacterial amyloid protein curli. Curli is a bacterial amyloid protein that assembles into fibers, and is produced by bacteria in biofilms[49] [Fig. 1(d)]. The primary component of curli fibers, CsgA, is composed of five repeating subunits, with each repeat assembling into a strand-loop-strand motif, with the entire protein taking on a beta-sheet conformation.
Curli fibers are remarkably robust, resisting degradation from proteases, detergents, or boiling.[50] Curli fibrillation is also tolerant of a variety of translational fusions[51] and variants can be efficiently produced from bacterial culture.[52] As such it is an attractive platform for a variety of applications, such as bioremediation to sequester toxins from the environment,[53] or for in vivo therapeutic use.[54] The mechanical properties of curli fibers have not been extensively studied in isolation; however, they can provide stiffness to natural biofilms,[55] and when added to alginate gels.[56]
Bacterial amyloid proteins are present throughout the bacterial kingdom, with a wide variety of sequence compositions.[57] While the best characterized curli proteins from Escherichia coli or Salmonella enteritidis have five repeats of the canonical curli sequence, some homologs, such as those in the Shewanella genus, have up to 22 repeats. However, the functional properties of these protein variants are unexplored.
Unnatural protein materials
Advances in the understanding of protein structure have facilitated the de novo design of proteins that are not based on natural templates. In the recent years, there have been several such successful designs that can undergo macromolecular assembly and form structures and materials. King et al.[58] for example, designed 24-subunit protein cages composed of two components using the Rosetta software suite[59] to design and model the protein structure. Using a similar computational approach, Shen et al.[60] took a further step and designed de novo proteins that self-assembled into micrometer scale helical filaments [Fig. 1(e)]. Through this computational model, they were able to carefully tune the helix geometry within the filaments, such as the helix diameter through variation of subunit repeats.
Another strategy for the design of de novo proteins is the use of coiled-coil helical assemblies, which have well-characterized properties and oligomerization states.[61] As such, proteins composed of coiled-coil domains have been engineered to self-assemble into 100 nm spheres,[62]- as well as alpha-helical barrels.[63] These domains are increasingly being used as modular building blocks that can be used to accurately design the nanostructure of the resulting assembles.[64]
The variation found in natural protein materials highlights the vast capabilities of proteins to exhibit a range of useful material properties. These properties have evolved over time, a process that could serve as a model for the production of novel materials. Furthermore, the creation of de novo protein materials further underscores that there may be interesting regions of protein sequence space untapped even by nature.
Block copolymers
One strategy commonly employed to create novel protein materials is the use of block copolymer constructions.[65] This technique relies on the concatenation of modular protein material domains into longer sequences. This feature is common in natural materials. Looking again to silks, we see their protein sequences composed of modular domains arranged together in single-polypeptide chains.[66]
The modular subunits of a block copolymer can have different properties, and the combination of functionally distinct domains can generate emergent properties in the resulting protein that are not exhibited by the domains in isolation. However, as with most emergent phenomena, these properties can be difficult to predict.
This concatenation strategy has been exploited to design synthetic protein materials with novel properties. Rabotyagova et al.[67] for example, designed amphiphilic silk-like protein clock copolymers with both hydrophobic (B) and hydrophobic (A) domains [Fig. 1(f)]. The resulting proteins self-assembled into nanoparticles whose morphologies depended on the sequence of the copolymer, with the BA peptide organizing into rod-like structures and BAA forming spherical structures.
ELPs are also commonly employed in block copolymer assemblies, and can similarly be programed to perform self-assembly. Huber et al.[68] combined hydrophilic or hydrophobic ELP domains into a single amphiphilic molecule that formed spherical protein compartments inside E. coli bacterial cells [Fig. 1(g)]. The amphiphilic protein was fused to GFP, encapsulating the fluorescent protein within replicating E. coli cells.
Many material hybrids can be made, combining different sequences derived from silk, ELP, and collagen, or other proteins into single polypeptides with complex properties.[69] Other functional domains can also be fused to material domains, for example, Chen et al.[70] fused metal binding domains to curli fibers to template the assembly of gold nanoparticles, resulting in conductive curli wires. The scope for fusing functional protein domains to material domains is nearly endless, facilitating a vast range of function as well as control over the spatial distribution of function.[71]
Engineering interactions
Another strategy to design protein materials is to control the oligomerization between proteins, directing the assembly of supramolecular structures such as fibers, sheets, or matrices.
For example, Garcia-Seisdedos et al.[72] engineered spontaneous fibril formation by introducing hydrophobic interactions in bacterial proteins. The researchers took symmetric homomeric protein complexes present in E. coli, and introduced point mutations designed to produce surface hydrophobicity. The resulting proteins were expressed in yeast, and many formed non-amyloid aggregates, including micrometer-scale fibers [Fig. 1(h)].
Suzuki et al.[73] created two-dimensional (2D) crystalline protein materials, by introducing disulfide bonds and metalcoordination interactions between proteins. Using the square-shaped homotetrameric protein RhuA, researchers inserted either cystine or dihistidine amino acids into the corners of the homotetramer square. The cystine-mediated disulfide bonds, or the histidine-mediated metal binding interactions, facilitated the assembly of some homotetramers into micrometer scale 2D lattices with highly ordered geometries [Fig. 1(i)]. Furthermore, these crystalline complexes had interesting properties, being capable of undergoing deformation without a loss of crystallinity.
A further way to engineer interactions between proteins is through covalent bond formation between subunits, a feature that stabilizes naturally evolved proteins. SpyTag and SpyCatcher are protein domains that spontaneously form an isopeptide bond between them, even when expressed as separate peptides.[74] Sun et al.[75] introduced these binding domains fused to ELP sequences in such a way as to form a covalently linked network [Fig. 1(j)]. These networks formed hydrogels that were further engineered to be powerful scaffolds for tissue culture, regulating the states of the cells growing within the hydrogel.
The examples listed above show that the sequence space of proteins spans many interesting material properties. Furthermore, block copolymers have been constructed, and these have been shown to possess novel properties not found in single subunits. De novo designed proteins can also undergo self-assembly and have emergent material properties. There is a rich space of protein sequences than span many material properties, much of which we are only beginning to explore. Indeed, the sequence-function space is so vast that it may be impossible to explore fully using rational design alone. The process of directed evolution in the laboratory has been used extensively to explore the functionality of protein sequences in high-throughput, and therefore offers an attractive approach to develop protein materials.
Directed evolution
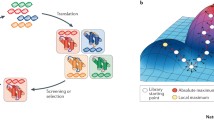
Directed evolution of proteins is a process of mutation and selection, whereby the encoding DNA is mutated, and the resulting variants are screened and selected for specific functionality (Fig. 2). This process can overcome our ignorance of the many molecular mechanisms and functions within the vast expanse of protein sequence space. There is a wealth of literature on directed evolution, as well as many excellent reviews, covering the history and future of directed evolution[76]; methods in the field[77,78]; and more theoretical concepts.[79,80] As such we will only cover these methods briefly to introduce what is possible for mutagenesis and selection.
General schematic of a protein directed evolution experiment. Starting from the top right, DNA encoding for a particular function of interest is mutagenized, producing a library of mutants. The DNA library is transformed into microbes, and made to express proteins. The resulting microbial variants are screened, and the best performing variants are selected for further evolution.
Mutagenesis methods
Directed evolution relies on the selection of particular proteins from pools of variants, and thus the first step of many directed evolution experiments is the creation of a pool of DNA encoding variants. There is an extensive range of mutational methods available, each with their own properties and applicability.
Some of the more common and established mutagenesis strategies are polymerase chain reaction (PCR)-based methods, which introduce point mutations with error prone DNA polymerases [Fig. 3(a)], or through assembly PCR [Fig. 3(b)] incorporating DNA oligomers containing variable nucleotide sequences. The resulting DNA amplicons are then cloned into plasmids and expressed in microbial hosts.
Schematics of common in vitro DNA mutagenesis techniques. (a) Error prone PCR introduces mutations from a single template, producing a library of mutants. (b) For assembly PCR, DNA oligos with random segments can be inserted into specific locations to generate diversity. (c) DNA shuttling begins with homologous sequences for a particular function, which are digested by endonuclease and reassembled into chimeras. (d) Golden gate shuffling can create chimeras by randomly assembling defined modular components. (e) Incremental truncation for the creation of hybrid enzymes (ITCHY) uses exonucleasesto degrade the DNA and concatenate the resulting fragments.
Another class of mutagenesis methods is DNA shuf-fling,[81,82] where experiments generally start with many similar DNA sequences that encode for the same functional protein. The sequences are then digested with DNAse I endonuclease enzymes to create many short fragments, which are then reassembled randomly into chimeras containing sequences from several original source sequences [Fig. 3(c)]. The initial pool of variants is usually derived from homologs of genes from many species, encompassing a rich library of functional mutants. This diversity can lead to the screening of many relevant mutations, which can significantly speed the evolutionary process.[83]
There are many other enzymatic methods to manipulate DNA. Type-II restriction enzymes[84] can be used to digest and subsequently specifically reassemble modular DNA sequences from many sources in one pot, facilitating the creation of chimeric sequences [Fig. 3(d)]. Other methods, such as ITCHY[85] [Fig. 3(e)], use exonucleases to truncate DNA sequences, which are subsequently ligated to create chimeras without requiring homology. Furthermore, DNA recombinases can be used to randomly rearrange DNA sequences in vivo,[86] shuffling many genes within bacterial cells.
The mutagenesis methods covered so far target relatively short DNA pieces, often on plasmids; however, a further class of methods generates mutations across the genome a target organism. These methods are usually employed in order to enhance an existing function within the target organism that relies on a number of native processes. The simplest of these methods utilizes chemical mutagens, which generate mutations throughout the genome in an uncontrolled manner, creating a library of mutants for subsequent screening.[87]
A more sophisticated technique for genomic manipulation is Multiplex Genome Engineering or MAGE.[88] MAGE relies on viral proteins to incorporate DNA oligos throughout the genome, at locations specified by the DNA oligo homology. These oligos can tolerate non-homologous segments, allowing for mutations, insertions or deletions throughout a genome, producing targeted modification to many genes simultaneously. This technique can be employed to optimize a particular multigene pathway within a host organism in order to maximize the yield of a desired product. Several other genome-editing tools have also been developed, including those that exploit the capacity of the CRISPR Cas9 enzyme to target specific loci on the genome for mutations.[89]
An evolutionary process can produce desired changes in a system without requiring much underlying systemic knowledge. However, some knowledge of a protein structure allows mutations to be designed in important regions. In such a way, rational design can be combined with directed evolution to create small and functionally rich pools of variants that can be screened more efficiently.[90] One prominent way of identifying effective mutations is to use protein structure as a guide, by designing mutations at locations predicted to effect properties such as the stability of the folded complex.[91]
Screening and selection
There is an extensive suite of DNA manipulation methods, and given their relative ease, the primary challenge of most directed evolution experiments is the selection of variants from the pool of mutants.[92] The process of selection involves measuring a particular property of interest and choosing variants that survive to further stages of the experiment. As such, directed evolution can only be performed for functions that can be readily detected, using methods amenable to measuring many samples to screen large libraries of mutants.
There are several classes of screening and selection methods. One of the simplest couples the function of the protein of interest to cell growth and survival. A classic example here is antibiotic resistance genes, which evolve both naturally and in the laboratory through the strong selective pressure of the antibiotic. Although this method is effective, few experiments can be designed in this way, as the proteins of interest are rarely directly responsible for any essential functions in the cell. More often, the expression of the proteins of interest imposes a metabolic burden on the cells. However, ingenious experimental design can couple growth to a biomolecule of interest, through the use of genetic circuits that sense a particular metabolite. In the work of Raman et al.,[93] such genetic sensor circuits were used to produce a selectable reporter, inhibiting growth when the metabolite of interest was present at low levels. In this way, high-producing mutants within a variant pool can be directly selected and enriched through growth.
A more common screening method is optical measurement, which requires visible markers for the processes of interest.
Such methods typically measure the absorbance or fluorescence properties of microbial cells, colonies, or cultures, on agar plates, in microplates or through flow cytometry. Many fluorescent proteins have been developed in this way, for example the commonly used superfolder green fluorescent protein (GFP). Wild-type GFP was found to misfold and malfunction when translationally fused to some proteins, therefore Pendelacq et al.[94] created a library of GFP mutants that were fused to a protein that inhibited proper folding of the wild-type. The variants were expressed in E. coli, and bright cells were selected by fluorescence-assisted flow cytometry (FACS). The resulting superfolder GFP variant had significantly improved stability and folding kinetics, and was a much more favorable partner for fusion experiments.
However, most evolution targets do not provide visible output; therefore visible reporters need to be crafted into the experimental design. Often, this requires the use of exogenously added fluorescent substances that interact with the process of interest. Another example is the use of fluorescent biosensor proteins that have been engineered to sense a particular metabolite, providing a fluorescent signal when high levels of the metabolite are present.[95] These biosensors can therefore provide an intracellular measurement of metabolite concentration, allowing for the selection of high-producing mutants.
Genotype–phenotype linkage
In order to successfully extract desired variants, it is critical to link genotype to phenotype in such a way that the desired genotype can be readily isolated. Most commonly, this is carried out by exploiting the inherent encapsulation of the variant DNA molecule within a host microbe. Once a specific mutant is selected, the microbe can be grown to amplify the DNA for extraction and any further processing.
However, while proteins are produced and mostly contained intracellularly, their functions cannot always be screened within the cell. For example, enzymes can synthesize metabolites that can pass through cell membranes, leading to serious difficulties in extracting desired variants from a pool of mutants. One prominent solution to this problem is the use of microfluidics, whereby single cells are encapsulated and grown within aqueous picoliter scale droplets in an emulsion within an oil phase.[96] This extreme miniaturization allows for the isolation of single variants within the droplets, which can be screened for the subsequent selection of desired variants.
Another practical challenge for genotype-phenotype linkage is the evolution of proteins that are designed to bind to an extracelhilar target, such as antibodies.[97] This challenge has been overcome by displaying proteins on the surface of bacteriophages, which are viruses that infect bacteria. Phages such as fd or M13 naturally infect bacteria, reproducing inside them and creating protein shells that encapsulate their own DNA. The proteins making up the shells can be fused to targets of interest, leading to protein capsules decorated with proteins that encapsulate the relevant DNA. The phages can then be selected with affinity chromatography, whereby they pass through a column containing the desired binding target, isolating variants that bind well.[98] The phages can then be extracted from the column and used to infect bacteria, recovering the DNA of successful variants.
Phages can also be used in experiments where the evolution is continuous. In such experiments, mutation and selection are happening constantly within the reaction flask, rapidly generating many rounds of evolution. In one prominent example, Esvelt et al.[99] modified the phage lifecycle to evolve proteins within constantly growing bacterial populations. Phage infection was dependent on the evolution of the target protein, so phages encoding for high functionality were replicated more, outcompeting poorer variants.
Several other innovative genotype-phenotype linkage strategies have also been developed in recent years. For example, one technique covalently attaches proteins to their encoding mRNA, allowing for the recovery of the coding sequences from selected proteins.[100]
High-throughput methods
Researchers within synthetic biology have been increasingly making use of technologies that allow for the screening of large libraries of samples. This high-throughput approach is largely driven by the difficulty in predicting the outcomes of changes to biological systems, leading to the need to empirically evaluate many variants. This approach has facilitated new experiments that synthesize and evaluate huge numbers of DNA variants, providing rich datasets and functional insights. For example, Kosuri et al.[101] studied the combination of simple bacterial regulatory DNA elements, screening over 12,000 constructs in one experiment, providing insight into the complex behavior of the genetic elements when combined together.
One of the more prominent technologies that has enabled high-throughput experiments for directed evolution is microfluidics.[102] Aside from coupling genotype to phenotype, the miniaturization of experiments in this way allows for a huge number of samples to be screened and sorted. This technique is capable of screening droplets at a rate of 107 samples per hour.[103] The high sample throughput facilitates the robust isolation and selection of rare variants from very large pools of mutants. For example, Colin et al.[104] screened libraries of 1,250,000 enzyme variants isolated from environmental bacteria, selecting 14 that best chemically modified a dye molecule. Similarly, Agresti et al.[103] mutated and screened around 107 enzyme variants in a single round of their evolution experiment, identifying 100 for use in subsequent rounds.
Droplets in microfluidic experiments are typically screened in custom microscopes for fluorescence [Fig. 4(a)], however absorbance measurements are also possible.[105] More complex optical droplet assessments can also be performed, such as measuring the morphological characteristics of cells encapsulated within the droplets.[106] However, the computational load of the image processing typically slows the screening rate significantly. There are a variety of techniques to actively control the movement of droplets within the microfluidic chips, allowing for the rapid sorting of droplets with desirable optical properties.[107] Furthermore, if droplets are made to be water-in-oil-in-water emulsions, screening and selection can be performed with FACS,[108] resulting in rapid and sensitive fluorescent selection.
Examples of microfluidic screening systems. (a) Picolitre-scale droplets encapsulating cells expressing a particular enzyme variant that are lysed and screened for fluorescence using a reporter for the enzymatic process.[119] Microfluidic systems can also sort eukaryotic cells by their mechanical and electrical properties, for example (b) their stiffness (adapted with permission from Ref. 129 copyright 2017 Nature Publishing Group), (c) electrical conductivity (adapted with permission from Ref. 131 copyright 2009 American Chemical Society), or (d) compressibility (adapted with permission from Ref. 128 copyright 2014 National Academy of Sciences).
Fitness landscapes
One concept critical to the understanding of evolutionary processes is fitness landscapes. Such landscapes are defined by a sequence space, which encompasses the genotype sequence, and describes the functional performance of any given coordinate within the space. Directed evolution of proteins can then be thought of an exploration of this landscape.[109] Under a particular screening paradigm, each point in sequence space for a protein has an associated fitness, which describes the function of the protein as evaluated by the screen. Thus, we can imagine a multidimensional sequence landscape, with points representing proteins with a particular sequence. The points within sequence space can move about the landscape through mutation, entering peaks or troughs of a desired function.
Directed evolution therefore explores the fitness landscape by generating many variants throughout the sequence space, and selecting for protein sequences with the desired properties. Many mutations are neutral with respect to the function of interest, however they can open pathways to other regions of the fitness landscape.[110]
The properties of the fitness landscape have significant implications on the design of an evolutionary process, as highlighted in work on evolutionary algorithms.[111] By visualizing the fitness landscape, we can tailor the mutational exploration of the space to maximize the chance of finding new peaks. For example, fitness maxima may be hidden behind the regions of poor fitness, so are not reachable by short jumps through the landscape, requiring large shifts in sequence space.
Now that we have covered, albeit briefly, protein materials and directed evolution, we will discuss the evolution of protein materials specifically.
Directed evolution of materials
Current examples
While directed evolution has found much success improving many individual proteins, metabolic pathways, and other biological processes, it has not yet found significant use in the field of protein materials. Unlike with traditional evolution experiments, the properties critical to the function of protein materials are the mechanical properties and macromolecular assembly. While there are many examples of selection for biochemical properties, protein stability or ligand binding, there are relatively few examples of selection for macromolecular assembly.
One such example is the creation of protein cages, which act as synthetic capsules containing other molecules such as DNA, RNA, or other biomolecules. These protein assemblies are inspired by viruses in nature, which are often strands of DNA or RNA encapsulated in a protein cage, and have a constantly evolving and changing capsule structure. Such cages can be used in a range of therapeutic applications, for drug delivery or as artificial vaccines.[112]
Wörsdörfer et al.[113] evolved protein containers initially formed from the Lumazine Synthase from Aquifex aeolicus (AaLS), a protein known to form icosahedral assemblages. The researchers produced these protein capsids intracellularly in E. coli, which also expressed a toxic protease protein that was engineered to bind to the inner surface of the capsid. In this way, mutants that better sequestered the toxic protein grew more efficiently and were thus selected for. As a result, the experiments had identified a protein container with a 5–10 fold higher loading capacity than wild-type, coinciding with seven point mutations.
A further development in protein containers was the de novo design of proteins that assembled into polyhedral structures. Butterfield et al.[114] used two proteins that were designed to form 120-subunit icosahedra,[115] which were further designed to bind and encapsulate RNA molecules encoding their sequence. The researchers created a deep combinatorial library of mutant DNA encoding for an amino acid mutation at all positions along the polypeptide. These synthetic nucleocapsids were then expressed intracellularly in E. coli, liberated from the bacteria and challenged with RNAse enzymes, heat, blood, and in vivo environments inside mice. Synthetic nucleocapsids that survived the challenges were then analyzed. Consequently, the researchers discovered variants with over 133-fold improved RNA packing, outcompeting even some natural viruses.
Both of these examples demonstrate how directed evolution can be used to select for and thus improve macromolecular assembly, a critical aspect of protein materials.
Generating protein material variants
A directed evolution experiment typically begins with many variants that can be screened for functionality. As we have seen, there are many techniques to obtain variable libraries of DNA molecules; however, for protein materials there are several considerations to select an appropriate strategy.
Many protein materials are composed of relatively short repeating modules, and as such, the encoding DNA would contain highly repetitive sequences. The repetition poses significant issues for synthetic DNA library construction, hindering PCR and DNA assembly, due to incorrect hybridization. Additionally, many mutagenesis methods rely on homology, and the repetitive sequences could lead to recombination resulting in undesired truncated sequences. This issue can be mitigated in several ways. Tang and Chilkoti[116] for example, seeking to synthesize repetitive proteins, developed a codon-scrambling algorithm that minimized repetition and generated sequences that were robustly assembled. Additionally, several mutagenesis methods either do not rely on homology, or rely on the homology of only short single stranded overhang regions between double stranded modular elements.[84] In order to avoid the inherent issues associated with the repetitive nature of many protein materials, such techniques need to be employed to synthesize or mutate the encoding DNA for expression and screening.
Proteins are generally produced intracellularly, and in many cases, the intracellular accumulation of proteins is toxic to microbial cells due to the formation of aggregates that hinder essential cellular processes.[117] These aggregates are typically composed of misfolded proteins in an amyloid-like conformation. However, the natural curli material system is composed of functional amyloid proteins, and offers an insight into overcoming toxicity. Since the curli fiber monomers can form toxic aggregates inside bacterial cells, the curli operon contains a chaperone protein, CsgC, which inhibits amyloid formation intracellularly.[118] In this way, curli toxicity is reduced, allowing the bacteria to maintain high-intracellular CsgA concentrations that are subsequently secreted into the extracellular space, where the amyloid-like curli fibers are formed.
The inherent intracellularity of protein translation presents further challenges, as many protein materials assemble into structures many times the size of a single-microbial cell. The protein materials can thus be misfolded inside cells, and incapable of adopting conformations with significant material properties. As such, the biopolymer has to be not only produced but also removed from the cell for assessment. Established methods for the microbial production of protein materials in a laboratory setting invariably involve lysis, purification, and concentration steps. These sample processing procedures are usually performed manually, with large sample volumes, limiting the number of samples that can be processed simultaneously. While cell lysis can be performed in microfluidic chips[119] at high throughput, samples are not purified or concentrated, as there is sufficient optical signal directly from microbial culture within the droplet. In order to fit protein materials into this screening paradigm, sufficient material needs to be produced to induce a noticeable change in culture properties.
In addition to the challenges of typical directed evolution experiments, the evolution of protein materials requires additional considerations. Repetition within protein material sequences, and the issues of intracellular expression need to be addressed by potential experimenters to effectively generate libraries of variants for further screening and selection.
Screening protein materials
Screening protein materials for directed evolution poses unique practical challenges. Traditional characterization methods for materials are relatively low throughput, and present a bottleneck to directed evolution experiments. In pioneering protein block copolymer research, Cappello et al.[120] produced materials through the random concatenation of DNA encoding silk or ELP domains. While in principle the concatenation could produce a large number of variants, the material characterization was performed with x-ray diffraction for only four variants. While more sophisticated methods have been developed for producing such block copolymers,[121] material characterization techniques have not yet been developed to screen large libraries of variants. Common techniques such as electron microscopy, x-ray, spectroscopic methods, or AFM require cumbersome sample preparation protocols, and are currently unfeasible to employ on large libraries where genotype-phenotype linkages are retained.
However, new measurement paradigms are emerging that show potential for successful integration with the directed evolution techniques described above. Novel and relevant technologies are being developed to measure the mechanical properties of eukaryotic cells, in order to understand cell mechanics and determine phenotypes for diagnostic applications. Many such techniques are compatible with high-throughput approaches, and a recent review by Darling and Di Carlo[122] provides detailed explanations of these methods. In short, there are several promising microfluidic techniques that can measure and sort cells based on their mechanical properties.
High-throughput measurements can be made of cells by optical means, rapidly applying image-processing algorithms to sort cell types by their visual appearance.[123] Furthermore, cells can be forced through constrictions, or deformed by hydrodynamic stretching, and the resulting shapes of the cells can be analyzed, providing mechanical data at rates of up to 20,000 cells per second.[124] More generally, the rapid quantification of particle deformation can elucidate the stiffness of the particles.[125] Further optical techniques have arisen in recent years capable of measuring rheology by tracking the trajectories of sub-micrometer-sized fluorescent beads injected into living cells,[126] or with fluorescent probes that respond to local viscosity.[127]
Sorting can also be performed based on the mechanical properties of cells, and these techniques have primarily been developed to characterize eukaryotic cells. One such technique used acoustic standing waves in microfluidic chambers to apply forces dependent on the relative compressibility and density of cells[128] [Fig. 4(d)], isolating cells by these properties. Another recent example sorted cells by their stiffness by forcing cells through a series of constrictions diagonal to the flow of the cells. In this way, deformable cells passed through, whereas stiffer cells were deflected and isolated[129] [Fig. 4(b)]. A further example used a device with a series of micrometer-scale gaps to filter cells by their capacity to deform and pass through the increasingly smaller gaps.[130]
Microfluidic sorting systems have been made to sort particles and cells by their electrical conductivity.[131] Here, particles or cells were made to flow through a chamber containing a transverse conductivity gradient, and electrodes dielectrophoretically deflected cells depending on their conductivities [Fig. 4(c)]. This technique has been used to screen a library of Saccharomyces cerevisiae genome deletion strains, identifying genes that contributed to the electrical properties of the yeast.[132] Sorting with magnetic fields has also been performed, for example, Tay et al.[133] used such a microfluidic setup to select Magnetospirillum magneticum cells that produced higher levels of magnetic nanoparticles.
Microbial cultures inside microfluidic droplets can in some ways be viewed as crude eukaryotic cells, acting as organelles that produce proteins. If the microbes are secreting protein materials, or they are lysed to release them, these proteins can then assemble into larger-scale structures. These can therefore act analogously to structural proteins within eukaryotic cells, and will consequently influence the mechanical properties of the droplet. This strategy, combined with the tools described above, seems like a promising avenue for screening and sorting protein materials in high-throughput.
Theoretical aspects
It is important to consider the fitness landscape of a protein designed for material self-assembly, in order to appropriately design experiments to effectively explore the space of the emergent material properties. Protein-protein interactions are critical to self-assembly, and so such proteins necessarily have many regions of interaction with other proteins. Self-assembling proteins therefore have much of their sequence involved in interaction, and thus many mutations would be deleterious to the assembly process.[134] As such, the evolution of multimeric proteins can be slow, and be dependent on the evolutionary trajectories of any other proteins they interact with. Also, their functional fitness landscapes are rugged, with regions of fitness between large regions where the protein has poor function.
Furthermore, highly expressed proteins in natural systems evolve slowly, as there is an extra selection pressure against mistranslated proteins.[135] The aggregates that can accumulate inside cells and harmfully impact an organism’s fitness, would act as further selection pressure that may reduce access to certain regions of sequence space.
Therefore, since macromolecular assembly can be easily disrupted by mutation, and the added selective pressure inherent in high expression, successful variants may be rare. Taken together, these aspects highlight the need for screening many samples in order to adequately explore sequence space and discover interesting variants.
Conclusions
As we have discussed in this work, there are many examples throughout nature of protein sequences with powerful material properties. Recent developments in de novo protein design also reveal that there are protein sequences that exhibit material qualities that are unused by nature. Furthermore, there are many protein engineering strategies to combine material properties, or introduce interactions between components. While the landscape of protein materials is vast and largely unexplored, it contains immense potential, and there are many avenues for the design of such material systems.
The field of synthetic biology is constantly contributing novel tools to robustly manipulate and engineer biological systems. These new techniques can generate mutants for directed evolution experiments, allowing for greater control of DNA libraries. Similarly, new strategies for protein material biosynthesis are emerging. The secretion of recombinant materials for example, is emerging as a technique to produce materials directly with living systems, as cells export proteins into the extracellular space.[136] Another possibility is the production of materials with cell-free systems,[137] where the inner workings of cells are extracted and used in vitro to produce proteins. Both of these strategies overcome the need to process material samples inherent in traditional protein production methods, by removing the requirement to lyse and purify samples. Such methods appear attractive to simplify the material characterization of protein materials, opening a path for highthroughput approaches necessary for effective directed evolution.
Screening and characterization methods are the primary technical challenge for the high-throughput assessment of material variants. However, there are many promising technologies emerging that present attractive options. Microfluidic technologies in particular offer many attractive qualities, from genotype-phenotype linkage to a growing array of tools to select variants. Additionally, microfluidic systems are capable performing many laboratory processes in miniature, such as PCR[138] or centrifugation,[139] creating options for high-throughput sample processing. New sorting paradigms, based on mechanical or electrical properties, also offer approaches to evolve proteins for material properties directly.
Directed evolution is a powerful method to produce new and improved biological systems. Indeed, evolutionary techniques have been adopted across many disciplines, for example inspiring genetic algorithms in computer science. In one relevant example, researchers simulated artificial moving robots composed of several materials.[140] These material subunits served different functions, some generating forces by contracting and expanding, while others were static, and were either soft or hard. The algorithm rearranged various subunits to optimize the movement of the robot, finding configurations that could walk efficiently. This work underscores the power of evolution, and shows in silico the potential of evolving material properties for emergent function.
Although this review has focused on materials that are made of a limited number of monomeric subunit types, many natural materials are made up of large numbers of diverse components. Materials such as sporopollenin, the tough coating of plant pollen grains, have a poorly understood composition consisting of many polymers.[141] Sporopollenin is remarkably tough, allowing pollen to survive in harsh environments, in some cases remaining intact for millennia. Despite our ignorance of such complex systems and the molecular mechanisms underpinning their properties, directed evolution would allow us to evolve complex multi-component materials. While the controlled manipulation of many elements within such a complex system currently presents significant technical challenges, the future scope of material directed evolution offers many opportunities.
Directed evolution is a powerful mechanism, and appears essential to explore the possibilities of protein materials. Such experiments are technically challenging, requiring ideas and techniques from a range of disciplines. However, overcoming these challenges will allow evolution to design and fabricate the next generation of biological materials.
References
A.A. Cheng and T.K. Lu: Synthetic biology: an emerging engineering discipline. Annu. Rev. Biomed. Eng. 14, 155–178 (2012).
Y.-J. Chen. P. Liu, A.A.K. Nielsen, J.A.N. Brophy, K. Clancy, T. Peterson, and C.A. Voigt: Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods 10, 659–664 (2013).
A.A.K. Nielsen, B.S. Der, J. Shin, P. Vaidyanathan, V. Paralanov, E.A. Strychalski, D. Ross, D. Densmore, and C.A. Voigt: Genetic circuit design automation. Science 352, aac7341 (2016).
K.M. Esvelt and H.H. Wang: Genome-scale engineering for systems and synthetic biology. Mol. Syst. Biol. 9, 641 (2013).
M.T. Mee, J.J. Collins, G.M. Church, and H.H. Wang: Syntrophic exchange in synthetic microbial communities. Proc. Natl. Acad. Sci. USA 111, E2149–E2156 (2014).
M. Elowitz and W.A. Lim: Build life to understand it. Nature 468, 889–890 (2010).
C.J. Paddon, P.J. Westfall, D.J. Pitera, K. Benjamin, K. Fisher, D. McPhee, M.D. Leavell, A. Tai, A. Main, D. Eng, D.R. Polichuk, K.H. Teoh, D.W. Reed, T. Treynor, J. Lenihan, H. Jiang, M. Fleck, S. Bajad, G. Dang, D. Dengrove, D. Diola, G. Dorin, K.W. Ellens, S. Fickes, J. Galazzo, S.P. Gaucher, T. Geistlinger, R. Henry, M. Hepp, T. Horning, T. Iqbal, L. Kizer, B. Lieu, D. Melis, N. Moss, R. Regentin, S. Secrest, H. Tsuruta, R. Vazquez, L.F. Westblade, L. Xu, M. Yu, Y. Zhang, L. Zhao, J. Lievense, P.S. Covello, J.D. Keasling, K.K. Reiling, N.S. Renninger, and J.D. Newman: High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532 (2013).
P. Ball: Synthetic biology—Engineering nature to make materials. MRS Bull. 43, 477–484 (2018).
R.A. Le Feuvre and N.S. Scrutton: A living foundry for synthetic biological materials: a synthetic biology roadmap to new advanced materials. Synth. Syst. Biotechnol. 3, 105–112 (2018).
M.K. Rice and W.C. Ruder: Creating biological nanomaterials using synthetic biology. Sci. Technol. Adv. Mater. 15, 014401 (2013).
S.R. MacEwan and A. Chilkoti: Applications of elastin-like polypeptides in drug delivery. J. Controlled Release 190, 314–330 (2014).
K.G. DeFrates, R. Moore, J. Borgesi, G. Lin, T. Mulderig, V. Beachley, and X. Hu: Protein-based fiber materials in medicine: a review. Nanomaterials (Basel, Switz.) 8, 457 (2018).
N.H. Romano, D. Sengupta, C. Chung, and S.C. Heilshorn: Protein-engineered biomaterials: nanoscale mimics of the extracellular matrix. Biochim. Biophys. Acta 1810, 339–349 (2011).
G. Chan and D.J. Mooney: New materials for tissue engineering: towards greater control over the biological response. Trends Biotechnol. 26, 382–392 (2008).
J.M. Caves, V.A. Kumar, A.W. Martinez, J. Kim, C.M. Ripberger, C.A. Haller, and E.L. Chaikof: The use of microfiber composites of elastin-like protein matrix reinforced with synthetic collagen in the design of vascular grafts. Biomaterials 31, 7175–7182 (2010).
C. Gilbert and T. Ellis: Biological engineered living materials: growing functional materials with genetically programmable properties. ACS Synth. Biol. 8, 1–15 (2019).
P.Q. Nguyen, N.-M.D. Courchesne, A. Duraj-Thatte, P. Praveschotinunt, and N.S. Joshi: Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials. Adv. Mater. 30, 1704847 (2018).
S.E. Naleway, M.M. Porter, J. McKittrick, and M.A. Meyers: Structural design elements in biological materials: application to bioinspiration. Adv. Mater. 27, 5455–5476 (2015).
K.A. Dill and J.L. MacCallum: The protein-folding problem, 50 years on. Science 338, 1042–1046 (2012).
R.E. Cobb, N. Sun, and H. Zhao: Directed evolution as a powerful synthetic biology tool. Methods 60, 81–90 (2013).
J.C.M. van Hest and D.A. Tirrell: Protein-based materials, toward a new level of structural control. Chem. Commun. 1897–1904 (2001).
Y.J. Yang, A.L. Holmberg, and B.D. Olsen: Artificially engineered protein polymers. Annu. Rev. Chem. Biomol. Eng. 8, 549–575 (2017).
L. Yang, A. Liu, S. Cao, R.M. Putri, P. Jonkheijm, and J.J.L.M. Cornelissen: Self-assembly of proteins: towards supramolecular materials. Chem.–Eur. J. 22, 15570–15582 (2016).
M.S. Ekiz, G. Cinar, M.A. Khalily, and M.O. Guler: Self-assembled peptide nanostructures for functional materials. Nanotechnology 27, 402002 (2016).
B.O. Okesola and A. Mata: Multicomponent self-assembly as a tool to harness new properties from peptides and proteins in material design. Chem. Soc. Rev. 47, 3721–3736 (2018).
C. Vepari and D.L. Kaplan: Silk as a biomaterial. Prog. Polym. Sci. 32, 991–1007 (2007).
O. Tokareva, V.A. Michalczechen-Lacerda, E.L. Rech, and D.L. Kaplan: Recombinant DNA production of spider silk proteins. Microb. Biotechnol. 6, 651–663 (2013).
Q. Peng, Y. Zhang, L. Lu, H. Shao, K. Qin, X. Hu, and X. Xia: Recombinant spider silk from aqueous solutions via a bio-inspired microfluidic chip. Sci. Rep. 6, 36473 (2016).
C.L. Craig: Evolution of arthropod silks. Annu. Rev. Entomol. 42, 231–267 (1997).
F. Vollrath and P. Selden: The role of behavior in the evolution of spiders, silks, and webs. Annu. Rev. Ecol. Evol. Syst. 38, 819–846 (2007).
X. Hu, K. Vasanthavada, K. Kohler, S. McNary, A.M.F. Moore, and C.A. Vierra: Molecular mechanisms of spider silk. Cell. Mol. Life Sci. 63, 1986–1999 (2006).
L. Römer and T. Scheibel: The elaborate structure of spider silk. Prion 2, 154–161 (2008).
L.P. Gage and R.F. Manning: Internal structure of the silk fibroin gene of Bombyx mori. I. The fibroin gene consists of a homogeneous alternating array of repetitious crystalline and amorphous coding sequences. J. Biol. Chem. 255, 9444–9450 (1980).
K.M. Rudall and W. Kenchington: Arthropod silks: the problem of fibrous proteins in animal tissues. Annu. Rev. Entomol. 16, 73–96 (1971).
A. Rising, H. Nimmervoll, S. Grip, A. Fernandez-Arias, E. Storckenfeldt, D.P. Knight, F. Vollrath, and W. Engström: Spider silk proteins— mechanical property and gene sequence. Zool. Sci. 22, 273–281 (2005).
T. Kowalczyk, K. Hnatuszko-Konka, A. Gerszberg, and A.K. Kononowicz: Elastin-like polypeptides as a promising family of genetically-engineered protein based polymers. World J. Microbiol. Biotechnol. 30, 2141–2152 (2014).
W. Hassouneh, T. Christensen, and A. Chilkoti: Elastin-like Polypeptides as a Purification Tag for Recombinant Proteins. Curr. Protoc. Protein Sci. Editor. Board John E Coligan Al CHAPTER, Unit–6.11 (2010).
R. Saxena and M.J. Nanjan: Elastin-like polypeptides and their applications in anticancer drug delivery systems: a review. Drug Deliv. 22, 156–167 (2015).
K.E. Inostroza-Brito, E. Collin, O. Siton-Mendelson, K.H. Smith, A. Monge-Marcet, D.S. Ferreira, R.P. Rodríguez, M. Alonso, J.C. Rodríguez-Cabello, R.L. Reis, F. Sagués, L. Botto, R. Bitton, H.S. Azevedo, and A. Mata: Co-assembly, spatiotemporal control and morphogenesis of a hybrid protein–peptide system. Nat. Chem. 7, 897 (2015).
H. Wang, A. Paul, D. Nguyen, A. Enejder, and S.C. Heilshorn: Tunable control of hydrogel microstructure by kinetic competition between selfassembly and crosslinking of elastin-like proteins. ACS Appl. Mater. Interfaces 10, 21808–21815 (2018).
F.G. Quiroz and A. Chilkoti: Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater. 14, 1164 (2015).
N.K. Li, S. Roberts, F.G. Quiroz, A. Chilkoti, and Y.G. Yingling: Sequence directionality dramatically affects LCST behavior of elastin-like polypeptides. Biomacromolecules 19, 2496–2505 (2018).
M.D. Shoulders and R.T. Raines: Collagen structure and stability. Annu. Rev. Biochem. 78, 929–958 (2009).
K. Gelse, E. Pöschl, and T. Aigner: Collagens—structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 55, 1531–1546 (2003).
A.V. Persikov, J.A.M. Ramshaw, and B. Brodsky: Prediction of collagen stability from amino acid sequence. J. Biol. Chem. 280, 19343–19349 (2005).
S. Lukomski, K. Nakashima, I. Abdi, V.J. Cipriano, R.M. Ireland, S.D. Reid, G.G. Adams, and J.M. Musser: Identification and characterization of the scl gene encoding a group a streptococcus extracellular protein virulence factor with similarity to human collagen. Infect. Immun. 68, 6542–6553 (2000).
Z. Yu, B. An, J.A.M. Ramshaw, and B. Brodsky: Bacterial collagen-like proteins that form triple-helical structures. J. Struct. Biol. 186, 451–461 (2014).
E. Cosgriff-Hernandez, M.S. Hahn, B. Russell, T. Wilems, D. Munoz-Pinto, M.B. Browning, J. Rivera, and M. Höök: Bioactive hydrogels based on designer collagens. Acta Biomater. 6, 3969–3977 (2010).
M.M. Barnhart and M.R. Chapman: Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147 (2006).
S.K. Collinson, L. Emödy, K.H. Müller, T.J. Trust, and W.W. Kay: Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173, 4773–4781 (1991).
P.Q. Nguyen, Z. Botyanszki, P.K.R. Tay, and N.S. Joshi: Programmable biofilm-based materials from engineered curli nanofibres. Nat. Commun. 5, 4945 (2014).
N.-M. Dorval Courchesne, A. Duraj-Thatte, P.K.R. Tay, P.Q. Nguyen, and N.S. Joshi: Scalable production of genetically engineered nanofibrous macroscopic materials via Filtration. ACS Biomater. Sci. Eng. 3, 733–741 (2017).
P.K.R. Tay, P.Q. Nguyen, and N.S. Joshi: A synthetic circuit for mercury bioremediation using self-assembling functional amyloids. ACS Synth. Biol. 6, 1841 (2017).
A.M. Duraj-Thatte, P. Praveschotinunt, T.R. Nash, F.R. Ward, and N.S. Joshi: Modulating bacterial and gut mucosal interactions with engineered biofilm matrix proteins. Sci. Rep. 8, 3475 (2018).
G. Zeng, B.S. Vad, M.S. Dueholm, G. Christiansen, M. Nilsson, T. Tolker-Nielsen, P.H. Nielsen, R.L. Meyer, and D.E. Otzen: Functional bacterial amyloid increases Pseudomonas biofilm hydrophobicity and stiffness. Front. Microbiol. 6, 1099 (2015).
E. Axpe, A. Duraj-Thatte, Y. Chang, D.-M. Kaimaki, A. Sanchez-Sanchez, H.B. Caliskan, N.-M. Dorval Courchesne, and N.S. Joshi: Fabrication of amyloid curli fibers–alginate nanocomposite hydrogels with enhanced stiffness. ACS Biomater. Sci. Eng. 4, 2100–2105 (2018).
M.S. Dueholm, M. Albertsen, D. Otzen, and P.H. Nielsen: Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS ONE 7, e51274 (2012).
N.P. King, J.B. Bale, W. Sheffler, D.E. McNamara, S. Gonen, T. Gonen, T. O. Yeates, and D. Baker: Accurate design of co-assembling multicomponent protein nanomaterials. Nature 510, 103–108 (2014).
A. Leaver-Fay, M. Tyka, S.M. Lewis, O.F. Lange, J. Thompson, R. Jacak, K. Kaufman, P.D. Renfrew, C.A. Smith, W. Sheffler, I.W. Davis, S. Cooper, A. Treuille, D.J. Mandell, F. Richter, Y.-E. A. Ban, S.J. Fleishman, J.E. Corn, D.E. Kim, S. Lyskov, M. Berrondo, S. Mentzer, Z. Popović, J.J. Havranek, J. Karanicolas, R. Das, J. Meiler, T. Kortemme, J.J. Gray, B. Kuhlman, D. Baker, and P. Bradley: Rosetta3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574 (2011).
H. Shen, J.A. Fallas, E. Lynch, W. Sheffler, B. Parry, N. Jannetty, J. Decarreau, M. Wagenbach, J.J. Vicente, J. Chen, L. Wang, Q. Dowling, G. Oberdorfer, L. Stewart, L. Wordeman, J.D. Yoreo, C. Jacobs-Wagner, J. Kollman, and D. Baker: De novo design of selfassembling helical protein filaments. Science 362, 705–709 (2018).
A.S. Cristie-David, A. Sciore, S. Badieyan, J.D. Escheweiler, P. Koldewey, J.C.A. Bardwell, B.T. Ruotolo, and E.N.G. Marsh: Evaluation of de novodesigned coiled coils as off-the-shelf components for protein assembly. Mol. Syst. Des. Eng. 2, 140–148 (2017).
J.M. Fletcher, R.L. Harniman, F.R.H. Barnes, A.L. Boyle, A. Collins, J. Mantell, T.H. Sharp, M. Antognozzi, P.J. Booth, N. Linden, M.J. Miles, R.B. Sessions, P. Verkade, and D.N. Woolfson: Self-assembling cages from coiled-coil peptide modules. Science 340, 595–599 (2013).
F. Thomas, W.M. Dawson, E.J.M. Lang, A.J. Burton, G.J. Bartlett, G.G. Rhys, A.J. Mulholland, and D.N. Woolfson: De novo-designed α-helical barrels as receptors for small molecules. ACS Synth. Biol. 7, 1808 (2018).
A. Ljubetič, F. Lapenta, H. Gradišar, I. Drobnak, J. Aupič, Ž. Strmšek, D. Lainšček, I. Hafner-Bratkovič, A. Majerle, N. Krivec, M. Benčina, T. Pisanski, T. Ć. Veličković, A. Round, J.M. Carazo, R. Melero, and R. Jerala: Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo. Nat. Biotechnol. 35, 1094 (2017).
O.S. Rabotyagova, P. Cebe, and D.L. Kaplan: Protein-based block copolymers. Biomacromolecules 12, 269–289 (2011).
R. Valluzzi, S. Winkler, D. Wilson, and D.L. Kaplan: Silk: molecular organization and control of assembly. Philos. Trans. R. Soc. B: Biol. Sci. 357, 165–167 (2002).
O.S. Rabotyagova, P. Cebe, and D.L. Kaplan: Self-assembly of genetically engineered spider silk block copolymers. Biomacromolecules 10, 229–236 (2009).
M.C. Huber, A. Schreiber, P. von Olshausen, B.R. Varga, O. Kretz, B. Joch, S. Barnert, R. Schubert, S. Eimer, P. Kele, and S.M. Schiller: Designer amphiphilic proteins as building blocks for the intracellular formation of organelle-like compartments. Nat. Mater. 14, 125 (2015).
N. Dinjaski and D.L. Kaplan: Recombinant protein blends: silk beyond natural design. Curr. Opin. Biotechnol. 39, 1–7 (2016).
A.Y. Chen, Z. Deng, A.N. Billings, U.O.S. Seker, M.Y. Lu, R.J. Citorik, B. Zakeri, and T.K. Lu: Synthesis and patterning of tunable multiscale materials with engineered cells. Nat. Mater. 13, 515–523 (2014).
J.K. Polka, S.G. Hays, and P.A. Silver: Building spatial synthetic biology with compartments, scaffolds, and communities. Cold Spring Harb. Perspect. Biol. 8, a024018 (2016).
H. Garcia-Seisdedos, C. Empereur-Mot, N. Elad, and E.D. Levy: Proteins evolve on the edge of supramolecular self-assembly. Nature 548, 244 (2017).
Y. Suzuki, G. Cardone, D. Restrepo, P.D. Zavattieri, T.S. Baker, and F.A. Tezcan: Self-assembly of coherently dynamic, auxetic, two-dimensional protein crystals. Nature 533, 369–373 (2016).
B. Zakeri, J.O. Fierer, E. Celik, E.C. Chittock, U. Schwarz-Linek, V.T. Moy, and M. Howarth: Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 109, E690–E697 (2012).
F. Sun, W.-B. Zhang, A. Mahdavi, F.H. Arnold, and D.A. Tirrell: Synthesis of bioactive protein hydrogels by genetically encoded SpyTag-SpyCatcher chemistry. Proc. Natl. Acad. Sci. USA 111, 11269–11274 (2014).
R.E. Cobb, R. Chao and H. Zhao: Directed evolution: past, present and future. AIChE J. Am. Inst. Chem. Eng. 59, 1432–1440 (2013).
K.L. Tee and T.S. Wong: Polishing the craft of genetic diversity creation in directed evolution. Biotechnol. Adv. 31, 1707–1721 (2013).
M.S. Packer and D.R. Liu: Methods for the directed evolution of proteins. Nat. Rev. Genet. 16, 379–394 (2015).
F.H. Arnold: Design by directed evolution. Acc. Chem. Res. 31, 125–131 (1998).
A. Currin, N. Swainston, P.J. Day, and D.B. Kell: Synthetic biology for the directed evolution of protein biocatalysts: navigating sequence space intelligently. Chem. Soc. Rev. 44, 1172–1239 (2015).
W.P.C. Stemmer: Rapid evolution of a protein in vitro by DNA shuffling. Nature 370, 389–391 (1994).
H. Zhao and F.H. Arnold: Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res. 25, 1307–1308 (1997).
A. Crameri, S.-A. Raillard, E. Bermudez, and W.P.C. Stemmer: DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391, 288–291 (1998).
C. Engler, R. Gruetzner, R. Kandzia, and S. Marillonnet: Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE 4, e5553 (2009).
M. Ostermeier, J.H. Shim, and S.J. Benkovic: A combinatorial approach to hybrid enzymes independent of DNA homology. Nat. Biotechnol. 17, 1205 (1999).
D. Bikard, S. Julié-Galau, G. Cambray, and D. Mazel: The synthetic integron: an in vivo genetic shuffling device. Nucleic Acids Res. 38, e153–e153 (2010).
P.L. Foster: In vivo mutagenesis. Methods Enzymol. 204, 114–125 (1991).
H.H. Wang, F.J. Isaacs, P.A. Carr, Z.Z. Sun, G. Xu, C.R. Forest, and G.M. Church: Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009).
S.O. Halperin, C.J. Tou, E.B. Wong, C. Modavi, D.V. Schaffer, and J.E. Dueber: CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 560, 248 (2018).
S. Lutz: Beyond directed evolution—semi-rational protein engineering and design. Curr. Opin. Biotechnol. 21, 734–743 (2010).
P. Heinzelman, C.D. Snow, I. Wu, C. Nguyen, A. Villalobos, S. Govindarajan, J. Minshull, and F.H. Arnold: A family of thermostable fungal cellulases created by structure-guided recombination. Proc. Natl. Acad. Sci. USA 106, 5610–5615 (2009).
P.A.G. Tizei, E. Csibra, L. Torres, and V.B. Pinheiro: Selection platforms for directed evolution in synthetic biology. Biochem. Soc. Trans. 44, 1165–1175 (2016).
S. Raman, J.K. Rogers, N.D. Taylor, and G.M. Church: Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci. USA 111, 17803–17808 (2014).
J.-D. Pédelacq, S. Cabantous, T. Tran, T.C. Terwilliger, and G.S. Waldo: Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006).
S.-A. Morgan, D.C. Nadler, R. Yokoo, and D.F. Savage: Biofuel metabolic engineering with biosensors. Curr. Opin. Chem. Biol. 35, 150–158 (2016).
S.R.A. Devenish, M. Kaltenbach, M. Fischlechner, and F. Hollfelder: Droplets as Reaction Compartments for Protein Nanotechnology. In Protein Nanotechnology: Protocols, Instrumentation, and Applications, 2nd ed., edited by J A. Gerrard (Humana Press, 2013), pp. 269–286.
K.T. O’Neil and R.H. Hoess: Phage display: protein engineering by directed evolution. Curr. Opin. Struct. Biol. 5, 443–449 (1995).
A. Fernandez-Gacio, M. Uguen, and J. Fastrez: Phage display as a tool for the directed evolution of enzymes. Trends Biotechnol. 21, 408–414 (2003).
K.M. Esvelt, J.C. Carlson, and D.R. Liu: A system for the continuous directed evolution of biomolecules. Nature 472, 499 (2011).
H. Leemhuis, V. Stein, A.D. Griffiths, and F. Hollfelder: New genotype–phenotype linkages for directed evolution of functional proteins. Curr. Opin. Struct. Biol. 15, 472–478 (2005).
S. Kosuri, D.B. Goodman, G. Cambray, V.K. Mutalik, Y. Gao, A.P. Arkin, D. Endy, and G.M. Church: Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl. Acad. Sci. USA. 110, 14024 (2013).
A. Zinchenko, S.R.A. Devenish, B. Kintses, P.-Y. Colin, M. Fischlechner, and F. Hollfelder: One in a million: flow cytometric sorting of single celllysate assays in monodisperse picolitre double emulsion droplets for directed evolution. Anal. Chem. 86, 2526–2533 (2014).
J.J. Agresti, E. Antipov, A.R. Abate, K. Ahn, A.C. Rowat, J.-C. Baret, M. Marquez, A.M. Klibanov, A.D. Griffiths, and D.A. Weitz: Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. USA 107, 4004–4009 (2010).
P.-Y. Colin, B. Kintses, F. Gielen, C.M. Miton, G. Fischer, M.F. Mohamed, M. Hyvönen, D.P. Morgavi, D.B. Janssen, and F. Hollfelder: Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics. Nat. Commun. 6, 10008 (2015).
F. Gielen, R. Hours, S. Emond, M. Fischlechner, U. Schell, and F. Hollfelder: Ultrahigh-throughput–directed enzyme evolution by absorbance-activated droplet sorting (AADS). Proc. Natl. Acad. Sci. USA 113, E7383–E7389 (2016).
M. Girault, H. Kim, H. Arakawa, K. Matsuura, M. Odaka, A. Hattori, H. Terazono, and K. Yasuda: An on-chip imaging droplet-sorting system: a real-time shape recognition method to screen target cells in droplets with single cell resolution. Sci. Rep. 7, 40072 (2017).
H.-D. Xi, H. Zheng, W. Guo, A.M. Gañán-Calvo, Y. Ai, C.-W. Tsao, J. Zhou, W. Li, Y. Huang, N.-T. Nguyen, and S.H. Tan: Active droplet sorting in microfluidics: a review. Lab Chip 17, 751–771 (2017).
S.S. Terekhov, I.V. Smirnov, A.V. Stepanova, T.V. Bobik, Y.A. Mokrushina, N.A. Ponomarenko, A.A. Belogurov, M.P. Rubtsova, O.V. Kartseva, M.O. Gomzikova, A.A. Moskovtsev, A.S. Bukatin, M.V. Dubina, E.S. Kostryukova, V.V. Babenko, M.T. Vakhitova, A.I. Manolov, M.V. Malakhova, M.A. Kornienko, A.V. Tyakht, A.A. Vanyushkina, E.N. Ilina, P. Masson, A.G. Gabibov, and S. Altman: Microfluidic droplet platform for ultrahigh-throughput single-cell screening of biodiversity. Proc. Natl. Acad. Sci. USA 114, 2550–2555 (2017).
P.A. Romero and F.H. Arnold: Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 10, 866–876 (2009).
J.D. Bloom and F.H. Arnold: In the light of directed evolution: pathways of adaptive protein evolution. Proc. Natl. Acad. Sci. USA 106, 9995–10000 (2009).
S. Kauffman and S. Levin: Towards a general theory of adaptive walks on rugged landscapes. J. Theor. Biol. 128, 11–45 (1987).
J.G. Heddle, S. Chakraborti, and K. Iwasaki: Natural and artificial protein cages: design, structure and therapeutic applications. Curr. Opin. Struct. Biol. 43, 148–155 (2017).
B. Wörsdörfer, K.J. Woycechowsky, and D. Hilvert: Directed evolution of a protein container. Science 331, 589–592 (2011).
G.L. Butterfield, M.J. Lajoie, H.H. Gustafson, D.L. Sellers, U. Nattermann, D. Ellis, J.B. Bale, S. Ke, G.H. Lenz, A. Yehdego, R. Ravichandran, S.H. Pun, N.P. King, and D. Baker: Evolution of a designed protein assembly encapsulating its own RNA genome. Nature 552, 415–420 (2017).
J.B. Bale, S. Gonen, Y. Liu, W. Sheffler, D. Ellis, C. Thomas, D. Cascio, T. O. Yeates, T. Gonen, N.P. King, and D. Baker: Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 353, 389–394 (2016).
N.C. Tang and A. Chilkoti: Combinatorial codon scrambling enables scalable gene synthesis and amplification of repetitive proteins. Nat. Mater. 15, 419 (2016).
N.G. Bednarska, J. Schymkowitz, F. Rousseau, and J. Van Eldere: Protein aggregation in bacteria: the thin boundary between functionality and toxicity. Microbiology 159, 1795–1806 (2013).
M.L. Evans, E. Chorell, J.D. Taylor, J. Åden, A. Götheson, F. Li, M. Koch, L. Sefer, S.J. Matthews, P. Wittung-Stafshede, F. Almqvist, and M.R. Chapman: The bacterial curli system possesses a potent and selective inhibitor of amyloid formation. Mol. Cell 57, 445–455 (2015).
B. Kintses, C. Hein, M.F. Mohamed, M. Fischlechner, F. Courtois, C. Lainé, and F. Hollfelder: Picoliter cell lysate assays in microfluidic droplet compartments for directed enzyme evolution. Chem. Biol. 19, 1001–1009 (2012).
Cappello Joseph, Crissman John, Dorman Mary, Mikolajczak Marcia, Textor Garret, Marquet Magda, and Ferrari Franco: Genetic engineering of structural protein polymers. Biotechnol. Prog. 6, 198–202 (1990).
M.C. Huber, A. Schreiber, W. Wild, K. Benz, and S.M. Schiller: Introducing a combinatorial DNA-toolbox platform constituting defined protein-based biohybrid-materials. Biomaterials 35, 8767–8779 (2014).
E.M. Darling and D. Di Carlo: High-throughput assessment of cellular mechanical properties. Annu. Rev. Biomed. Eng. 17, 35–62 (2015).
N. Nitta, T. Sugimura, A. Isozaki, H. Mikami, K. Hiraki, S. Sakuma, T. Iino, F. Arai, T. Endo, Y. Fujiwaki, H. Fukuzawa, M. Hase, T. Hayakawa, K. Hiramatsu, Y. Hoshino, M. Inaba, T. Ito, H. Karakawa, Y. Kasai, K. Koizumi, S. Lee, C. Lei, M. Li, T. Maeno, S. Matsusaka, D. Murakami, A. Nakagawa, Y. Oguchi, M. Oikawa, T. Ota, K. Shiba, H. Shintaku, Y. Shirasaki, K. Suga, Y. Suzuki, N. Suzuki, Y. Tanaka, H. Tezuka, C. Toyokawa, Y. Yalikun, M. Yamada, M. Yamagishi, T. Yamano, A. Yasumoto, Y. Yatomi, M. Yazawa, D. Di Carlo, Y. Hosokawa, S. Uemura, Y. Ozeki, and K. Goda: Intelligent image-activated cell sorting. Cell, 175, 266–276.e13 (2018).
J.S. Dudani, D.R. Gossett, H.T.K. Tse, and D.D. Carlo: Pinched-flow hydrodynamic stretching of single-cells. Lab Chip 13, 3728–3734 (2013).
M.Y. Hwang, S.G. Kim, H.S. Lee, and S.J. Muller: Elastic particle deformation in rectangular channel flow as a measure of particle stiffness. Soft Matter 14, 216 (2017). doi: 10.1039/C7SM01829K.
P.-H. Wu, C.M. Hale, W.-C. Chen, J.S.H. Lee, Y. Tseng, and D. Wirtz: High-throughput ballistic injection nanorheology to measure cell mechanics. Nat. Protoc. 7, 155 (2012).
T. Liu, X. Liu, D.R. Spring, X. Qian, J. Cui, and Z. Xu: Quantitatively mapping cellular viscosity with detailed organelle information via a designed PET fluorescent probe. Sci. Rep. 4, 5418 (2014).
X. Ding, Z. Peng, S.-C. S. Lin, M. Geri, S. Li, P. Li, Y. Chen, M. Dao, S. Suresh, and T.J. Huang: Cell separation using tilted-angle standing surface acoustic waves. Proc. Natl. Acad. Sci. USA 111, 12992–12997 (2014).
M. Islam, H. Brink, S. Blanche, C. DiPrete, T. Bongiorno, N. Stone, A. Liu, A. Philip, G. Wang, W. Lam, A. Alexeev, E.K. Waller, and T. Sulchek: Microfluidic sorting of cells by viability based on differences in cell stiffness. Sci. Rep. 7, 1997 (2017).
N.K. Gill, C. Ly, K.D. Nyberg, L. Lee, D. Qi, B. Tofig, M. Reis-Sobreiro, O. Dorigo, J. Rao, R. Wiedemeyer, B. Karlan, K. Lawrenson, M.R. Freeman, R. Damoiseaux, and A.C. Rowat: A scalable filtration method for high throughput screening based on cell deformability. Lab Chip 19, 343–357 (2019).
M.D. Vahey and J. Voldman: High-throughput cell and particle characterization using ISO-dielectric separation. Anal. Chem. 81, 2446–2455 (2009).
M.D. Vahey, L.Q. Pesudo, J.P. Svensson, L.D. Samson, and J. Voldman: Microfluidic genome-wide profiling of intrinsic electrical properties in Saccharomyces cerevisiae. Lab Chip 13, 2754–2763 (2013).
A. Tay, C. Murray, and D. Di Carlo: Phenotypic selection of Magnetospirillum magneticum (AMB-1) overproducers using magnetic ratcheting. Adv. Funct. Mater. 27, 1703106 (2017).
H.B. Fraser, A.E. Hirsh, L.M. Steinmetz, C. Scharfe, and M.W. Feldman: Evolutionary rate in the protein interaction network. Science 296, 750–752 (2002).
D.A. Drummond, J.D. Bloom, C. Adami, C.O. Wilke, and F.H. Arnold: Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. USA 102, 14338–14343 (2005).
P.Q. Nguyen. Synthetic biology engineering of biofilms as nanomaterials factories. Biochem. Soc. Trans. 45, 585–597 (2017).
J. Shin and V. Noireaux: An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth. Biol. 1, 29–41 (2012).
Y. Schaerli and F. Hollfelder: The potential of microfluidic water-in-oil droplets in experimental biology. Mol. Biosyst. 5, 1392–1404 (2009).
A.J. Mach, J.H. Kim, A. Arshi, S.C. Hur, and D.D. Carlo: Automated cellular sample preparation using a centrifuge-on-a-chip. Lab Chip 11, 2827–2834 (2011).
N. Cheney, R. MacCurdy, J. Clune, and H. Lipson: Unshackling Evolution: Evolving Soft Robots with Multiple Materials and a Powerful Generative Encoding. In Proceedings of the 15th Annual Conference on Genetic and Evolutionary Computation (ACM, New York, NY, USA, 2013), pp. 167–174.
G. Mackenzie, A.N. Boa, A. Diego-Taboada, S.L. Atkin, and T. Sathyapalan: Sporopollenin, The least known yet toughest natural biopolymer. Front. Mater. 2, 66 (2015).
M. Nokelainen, H. Tu, A. Vuorela, H. Notbohm, K.I. Kivirikko, and J. Myllyharju: High-level production of human type I collagen in the yeast Pichia pastoris. Yeast 18, 797–806 (2001).
Acknowledgments
This work was supported by NSF Grant 1410751 (Division of Materials Research), the National Institutes of Health (1R01DK11077001A1), and the Wyss Institute for Biologically Inspired Engineering. The authors would also like to thank the reviewers for their time and insightful contributions to the article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kan, A., Joshi, N.S. Towards the directed evolution of protein materials. MRS Communications 9, 441–455 (2019). https://doi.org/10.1557/mrc.2019.28
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1557/mrc.2019.28