Abstract
Background
Na+/K+-ATPase α1 subunit (ATP1A1) exhibits aberrant expression in various types of cancer. Moreover, its levels in specific tissues are associated with the development of cancer. Nevertheless, the mechanism and signaling pathways underlying the effects of ATP1A1 in colon cancer (CC) have not been elucidated, and its prognostic impact remains unknown.
Methods
Knockdown of ATP1A1 expression was performed in human CC cell lines HT29 and Caco2 using small interfering RNA. The roles of ATP1A1 in various biological processes of cells (i.e., proliferation, cell cycle, apoptosis, migration, and invasion) were assessed. Microarray analysis was utilized for gene expression profiling. Samples obtained from 200 patients with CC who underwent curative colectomy were analyzed through immunohistochemistry.
Results
ATP1A1 knockdown suppressed cell proliferation, migration, and invasion and induced apoptosis. The results of the microarray analysis revealed that the upregulated or downregulated gene expression in ATP1A1-depleted cells was related to the extracellular signal-regulated kinase 5 (ERK5) signaling pathway [epidermal growth factor receptor (EGFR), mitogen-activated protein kinase kinase 5 (MAP2K5), mitogen-activated protein kinase 7 (MAPK7), FOS, MYC, and BCL2 associated agonist of cell death (BAD)]. Immunohistochemical analysis demonstrated a correlation between ATP1A1 expression and pathological T stage (p = 0.0054), and multivariate analysis identified high ATP1A1 expression as an independent predictor of poor recurrence-free survival in patients with CC (p = 0.0040, hazard ratio: 2.807, 95% confidence interval 1.376–6.196).
Conclusions
ATP1A1 regulates tumor progression through the ERK5 signaling pathway. High ATP1A1 expression is associated with poor long-term outcomes in patients with CC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Colon cancer (CC) has the third highest incidence rate among cancers globally.1 In recent years, important advances have been achieved in surgery and chemotherapy. Nevertheless, tumor recurrence after curative resection is common in patients with CC, leading to a high mortality rate. Therefore, it is imperative that the molecular mechanisms involved in CC are elucidated. Also, there is an urgent need for the discovery of appropriate therapeutic targets.
The membrane-bound protein Na+/K+-ATPase (NKA) is a member of the P-type ATPase family. It is composed of four α-subunits, three β-subunits, and an isoform of FXYD protein.2 The protein generates an electrochemical gradient across the plasma membrane. This is achieved through the transportation of three sodium and two potassium ions out and into the cell, respectively; this process requires a molecule of ATP.3 NKA plays a role in multiple cellular functions, such as cell junction, adhesion, motility, apoptosis, and signal transduction.4,5,6 It is aberrantly expressed and exhibits altered activity in various types of human cancer; thus, NKA has been identified as a novel potential target for anticancer therapy.7,8 Nevertheless, the dysregulation of NKA has shown variation between different types of cancer, and the roles of these subunits in the development of cancer remain unclear.
The catalytic Na+/K+-ATPase α1 subunit (ATP1A1) contains binding sites for Mg2+, ATP, Na+, K+, and ouabain.9 The clinical significance of ATP1A1 has recently been demonstrated in numerous types of cancer (e.g., non-small-cell lung cancer,10 renal clear cell carcinoma,11 glioblastoma,12 melanoma,13 hepatocellular carcinoma,14 and esophageal squamous cell carcinoma).15 We have previously reported that ATP1A1 expression in gastric cancer cells affects cell proliferation, apoptosis, migration, and invasion, as well as prognosis.16 However, the mechanism and signaling pathways underlying the effects of ATP1A1 in CC have not been elucidated, and its prognostic impact remains unknown.17 In this study, we examined the involvement of ATP1A1 in the regulation of oncogenesis-related genes in CC and sought to determine its clinical importance.
Methods
Cell Lines and Antibodies
Human CC cell lines SW480, SW620, HT29, HCT116, RKO, Caco2, DLD1, and LoVo were utilized in this study (Riken Cell Bank, Tsukuba, Japan). These cells, except for Caco2, were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Nacalai Tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37 °C in a humidified incubator with 5% CO2. Caco2 cells were cultured in minimum essential medium (Nacalai Tesque) under the same conditions as the other cell lines. A monoclonal anti-ATP1A1 antibody (Abcam, Cambridge, UK) was used in the immunohistochemical (IHC) analysis and western blotting. A mouse monoclonal anti-actin beta (ACTB) antibody (A5441; Sigma–Aldrich, St Louis, MO, USA) was used as internal control.
Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Isolation of total RNA was carried out using an RNeasy kit (Qiagen, Valencia, CA, USA). Quantitative real-time PCR (Applied Biosystems, Foster City, CA, USA) and TaqMan Gene Expression Assays (Applied Biosystems) were used to evaluate messenger RNA (mRNA) expression. The levels of ATP1A1 (Hs00933601_m1), epidermal growth factor receptor (EGFR; Hs01076090_m1), mitogen-activated protein kinase kinase 5 (MAP2K5; Hs00177134_m1), mitogen-activated protein kinase 7 (MAPK7; Hs00611114_g1), BCL2 associated agonist of cell death (BAD; Hs00188930_m1), FOS (Hs04194186_s1), and MYC (Hs00153408_m1) (Applied Biosystems) genes were measured. For each gene, the expression levels were normalized to those of the internal control gene ACTB (Hs01060665_g1; Applied Biosystems). All experiments were performed thrice.
Western Blotting
Mammalian Protein Extraction Reagent (M-PER) lysis buffer (Pierce Biotechnology, Rockford, IL, USA) supplemented with protease inhibitors (Pierce Biotechnology) was used to harvest the cells. The concentration of proteins was determined using a modified Bradford assay (Bio-Rad, Hercules, CA, USA). Cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis to separate the proteins. Subsequently, the proteins were transferred onto polyvinylidene difluoride membranes (GE Healthcare, Piscataway, NJ, USA). This was followed by incubation with antibodies at 4 °C for 24 h. Band densities were quantified using an Amersham Imager 680 (GE Healthcare).
Transfection of Cells with Small Interfering RNA (siRNA)
HT29 and Caco2 cells were transfected with 12 nmol/l ATP1A1 siRNA (Stealth RNAi siRNA #HSS100791; Invitrogen, Carlsbad, CA, USA) using the Lipofectamine RNAiMAX reagent (Invitrogen). After 24 h, the medium was replaced with fresh medium. The control siRNA (Stealth RNAiTM siRNA Negative Control; Invitrogen) provided was used as a negative control.
Cell Proliferation Assay
HT29 and Caco2 cells were seeded in six-well plates (1.2 and 1.5 × 105 cells/well, respectively) and cultured at 37 °C in a humidified incubator with 5% CO2. The transfection of cells with siRNA was performed 24 h after seeding. Trypsin-ethylenediaminetetraacetic acid was used for cell detachment 48 and 72 h following transfection with siRNA. Finally, the cells were counted using a hemocytometer.
Cell Cycle Assay
For the detection of the cell cycle phase, fluorescence-activated cell sorting of HT29 and Caco2 cells was performed 72 h after transfection with siRNA using a BD Accuri C6 Plus Flow Cytometer (Becton–Dickinson Biosciences, San Jose, CA, USA). Trypsin-ethylenediaminetetraacetic acid and 0.2% Triton X-100 were utilized for cell detachment; the cell nuclei were stained using propidium iodide (PI) RNase staining solution (Becton–Dickinson Biosciences). Flow cytometry was performed with a BD Accuri C6 Plus Flow Cytometer (Becton–Dickinson Biosciences) to evaluate the cell cycle distribution in ≥ 1 × 104 cells.
Apoptosis Assay
At 72 h after transfection with siRNA, HT29 and Caco2 cells were harvested and stained with fluorescein isothiocyanate-conjugated annexin V and phosphatidylinositol using the ANNEXIN V-FITC Kit (Beckman Coulter, Brea, CA, USA). The proportions of early and late-stage apoptotic cells were determined using a BD Accuri C6 Plus Flow Cytometer (Becton–Dickinson Biosciences).
Wound Healing Assay
The siRNA-transfected HT29 and Caco2 cells were incubated. Subsequently, after the cells reached confluency, an incision-like line simulating a wound was performed with a pipette tip. Washes with medium were carried out for the removal of suspended cells and debris. Monitoring of wounds was conducted every 8 h, and cell-free areas were analyzed with BZ-X800 (Keyence, Osaka, Japan). All experiments were performed thrice.
Assessment of Overexpression
HT29 and Caco2 cells were transfected with either the control plasmid (V79020; Thermo Fisher Scientific, Waltham, MA, USA) or the ATP1A1 plasmid (OHu15960C; GenScript, Piscataway, NJ, USA) using the Lipofectamine 3000 reagent (Invitrogen). In the proliferation assay, HT29 and Caco2 cells were seeded in six-well plates (1.2 and 1.5 × 105 cells/well) and cultured at 37 °C in a humidified incubator with 5% CO2.
Microarray Analysis
Transfection of HT29 cells with either control or siRNA for ATP1A1 was performed. Extraction of total RNA was achieved with an RNeasy kit (Qiagen, Hilden, Germany). The RNA quality was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). For the preparation of biotinylated complementary DNA (cDNA), 100 ng of total RNA and an Affymetrix GeneChip WT PLUS Reagent Kit (Thermo Fisher Scientific, Waltham, MA, USA) were used. This was followed by fragmentation and biotinylation of 2 µg of cDNA. Thereafter, the biotinylated cDNA was hybridized to the Human Clariom S Array through an Affymetrix GeneChip Hybridization Oven 645 (Thermo Fisher Scientific). Ingenuity Pathway Analysis software (Ingenuity Systems, Inc., Redwood City, CA, USA) was used for microarray data analysis.
Patients and Primary Tissue Samples
Primary tumor samples were extracted from 200 patients with CC, who had undergone curative colectomy from 2008 to 2014 at the Division of Digestive Surgery, Department of Surgery, Kyoto Prefectural University of Medicine, Kyoto, Japan. Samples were obtained from patients treated with curative intent. All patients were stage II or III, and all underwent R0 resection. Patients with distant metastasis or synchronous/metachronous cancers, as well as those who had undergone colectomy with combined resection, or an emergency operation, were excluded from the analysis. The patients underwent colon resection and lymph node dissection according to the guidelines established by the Japanese Society for Cancer of the Colon and Rectum.18 Tumor staging was conducted using the International Union Against Cancer tumor, node, and metastasis (TNM) classification system (8th edition).19 This study was performed in accordance with the ethical standards of the Kyoto Prefectural University of Medicine and the Declaration of Helsinki. All patients provided written informed consent prior to surgery. The protocol of this study was approved by the Ethical Review Board of the Kyoto Prefectural University of Medicine (Approval Number: ERB-C-1178-1).
IHC Analysis
Paraffin-embedded tissue sections (thickness: 4 μm) were stained through the avidin-biotin-peroxidase complex method using a Vectastain ABC Elite Kit (Vector Laboratories, Burlingame, CA, USA). The prepared slides were incubated with the anti-ATP1A1 antibody (dilution: 1:5000) at 4 °C overnight. A semi-quantitative analysis of ATP1A1 expression was performed based on the staining intensity and percentage of stained cells. The intensity of cytoplasmic staining was scored as follows: 0 (no staining), 1 (weak), 2 (moderate), and 3 (strong). The proportion of stained cells was scored using a scale from 0 to 1.0 according to the stained area in the cancerous tissue. Analysis was performed by manual scoring of whole tumor cells by a pathologist and a surgeon. IHC scores were calculated as the maximum product of intensity and proportion scores (scale: 0–3.0).
Statistical Analysis
Analysis of categorical variables was carried out using chi-squared and Fisher’s exact tests. Unpaired continuous data were analyzed using Student’s t-tests. The Kaplan–Meier method was used to produce survival curves, and the log rank test was utilized for the detection of statistically significant differences. The Cox proportional hazards model was employed for the multivariate analysis of factors affecting survival. The p values < 0.05 denoted statistically significant differences. JMP software version 10 (SAS Institute, Cary, NC, USA) was utilized for all statistical analyses.
Results
ATP1A1 Expression in CC Cell Lines
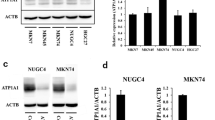
ATP1A1 expression in CC cell lines SW480, SW620, HT29, HCT116, RKO, Caco2, DLD1, and LoVo was examined using quantitative RT-PCR and western blotting (Fig. 1a, b). Among these, HT29 and Caco2 (both specifically expressing ATP1A1) were used to confirm the siRNA efficiency. HT29 and Caco2 cells transfected with the siRNA showed decreased mRNA and protein levels of ATP1A1 (Fig. 1c, d). ATP1A1 mRNA levels increased in HT29 and Caco2 cells transfected with the ATP1A1 plasmid (Supplementary Fig. 1).
ATP1A1 mRNA and protein expression in CC cells. a Quantitative RT-PCR revealed mRNA expression in CC cells. b Western blotting showed ATP1A1 expression in CC cells. c Transfection with ATP1A1 siRNA reduced the levels of ATP1A1 mRNA in HT29 and Caco2 cells. Data are presented as the mean ± SEM. n = 3. *p < 0.05 versus control siRNA. d Transfection with ATP1A1 siRNA reduced the levels of ATP1A1 protein in HT29 and Caco2 cells. ACTB, actin beta; ATP1A1, Na+/K+-ATPase α1 subunit; CC, colon cancer; Cont., control; RT-PCR, reverse transcription-polymerase chain reaction; SEM, standard error of the mean; siRNA, small interfering RNA
ATP1A1 Regulated the Proliferation and Cell Cycle of CC Cells
Knockdown of ATP1A1 was carried out in HT29 and Caco2 cells. Significantly fewer cells were transfected with ATP1A1 siRNA versus control siRNA (Fig. 2a). Moreover, in the overexpression analyses, the overexpression of ATP1A1 increased the proliferation of HT29 and Caco2 cells (Supplementary Fig. 1). According to the cell cycle analysis, following the knockdown of ATP1A1 in HT29 and Caco2 cells, the number of cells in the sub-G1 phase had increased in both cell lines (Fig. 2b).
ATP1A1 regulated the proliferation, cell cycle, survival, migration, and invasion of CC cells. a Downregulation of ATP1A1 expression reduced the proliferation of HT29 and Caco2 cells. Cells were counted at 48 and 72 h after transfection with siRNA. b Downregulation of ATP1A1 expression increased the number of cells in the sub-G1 phase in HT29 and Caco2 cells. Cells transfected with control or ATP1A1 siRNA were stained with PI and analyzed using flow cytometry. c Downregulation of ATP1A1 expression increased the proportions of early and late-stage apoptotic HT29 and Caco2 cells. Following transfection with control or ATP1A1 siRNA, the cells were stained with PI and annexin V and subjected to flow cytometric analysis. d Downregulation of ATP1A1 expression inhibited wound closure in HT29 and Caco2 cells. Data are presented as the mean ± SEM. n = 3. *p < 0.05 versus control siRNA. ATP1A1, Na+/K+-ATPase α1 subunit; CC, colon cancer, Cont., control; FITC-A, fluorescein isothiocyanate-antibody; PE-A, phycoerythrin-antibody; PI, propidium iodide; SEM, standard error of the mean; siRNA, small interfering RNA
ATP1A1 Regulated the Survival of CC Cells
The induction of apoptosis through transfection of HT29 and Caco2 cells with ATP1A1 siRNA was assessed. At 72 h after transfection, the proportions of early stage (annexin V-positive and PI-negative) and late-stage (annexin V-positive and PI-positive) apoptosis were significantly increased in ATP1A1-depleted HT29 and Caco2 cells (Fig. 2c).
ATP1A1 Controlled the Migration and Invasion of CC Cells
The effects of ATP1A1 knockdown on the migratory and invasive abilities of CC cells were also examined. The results of the wound healing assay revealed that the downregulation of ATP1A1 expression led to inhibition of wound healing in both HT29 and Caco2 cells (Fig. 2d).
Gene Expression Profiles in ATP1A1-depleted CC Cells
The results of the microarray analysis revealed that the depletion of ATP1A1 in HT29 cells resulted in > 1.7-fold alterations in the expression of 2175 genes compared with negative controls. Among those, the expression of 983 and 1192 genes was upregulated and downregulated, respectively. Table 1 includes 20 genes that exhibited the greatest upregulation or downregulation of gene expression in ATP1A1-depleted HT29 cells. The Ingenuity Pathway Analysis revealed that ‘Cancer’ and ‘Organismal Injury and Abnormalities’ were the most common ‘Disease and Disorders’ associated with the identified genes in ATP1A1-depleted HT29 cells. Notably, ‘Cell Death and Survival’ and ‘Cellular Development’ were the most commonly recorded ‘Molecular and Cellular Functions’ (Supplementary Table 1).
Signaling Pathways and Molecular Mechanisms Regulated by ATP1A1 in CC cells
According to the IPA, the ERK5 signaling pathway was among the canonical pathways most commonly affected by ATP1A1 knockdown (Supplementary Fig. 2). Figure 3a provides an illustration of the ERK5 signaling pathway. We subsequently sought to validate the results of the microarray analysis. To this end, quantitative RT-PCR was performed to further examine the expression of several genes involved in the ERK5 signaling pathway. In HT29 and Caco2 cells, the mRNA expression of EGFR, MAP2K5, MAPK7, FOS, and MYC was lower in cells with ATP1A1 knockdown versus those transfected with control siRNA. In contrast, the mRNA expression of BAD exhibited an inverse pattern (Fig. 3b). The same study was conducted using samples extracted from patients. The mRNA expression levels of EGFR, MAP2K5, MAPK7, FOS, and MYC were decreased in samples with low ATP1A1 in IHC, and the mRNA expression levels of BAD were increased compared with those with high ATP1A1 expression (Supplementary Fig. 3).
Regulation of signaling pathways by ATP1A1 in CC cells. a Signaling map of ‘ERK5 signaling’ and top-ranking canonical pathways associated with the knockdown of ATP1A1 in HT29 cells according to microarray analysis and Ingenuity Pathway Analysis. Red and green indicate genes with increased and decreased expression, respectively, versus reference levels. Blue indicates predicted inhibition. b Quantitative RT-PCR was used to verify the gene expression. Quantitative RT-PCR was used to compare the expression of six genes linked to the ERK5 signaling pathway (i.e., EGFR, MAP2K5, MAPK7, BAD, FOS, and MYC) in ATP1A1-depleted HT29 cells with that recorded in cells transfected with control siRNA (red circles in a). Data are presented as the mean ± SEM. n = 3. *p < 0.05 versus control siRNA. ACTB, actin beta; ATP1A1, Na+/K+-ATPase α1 subunit; BAD, BCL2 associated agonist of cell death; CC, colon cancer; Cont, control; CREB, cAMP responsive element binding protein; CTF1, cardiotrophin 1; EGFR, epidermal growth factor receptor; EGF, epidermal growth factor; ELK4, ETS transcription factor ELK4; ERK5, extracellular signal-regulated kinase 5; FOSL1, FOS like 1; FOXO3, forkhead box O3; GAB1, GRB2 associated binding protein 1; GNAQ, G protein subunit alpha q; GPCR, G protein-coupled receptor; H2O2, hydrogen peroxide; IL6ST, interleukin 6 cytokine family signal transducer; MAP2K5, mitogen-activated protein kinase kinase 5; MAPK7, mitogen-activated protein kinase 7; NGF, nerve growth factor; NTRK1, neurotrophic receptor tyrosine kinase 1; PTPN11, protein tyrosine phosphatase non-receptor type 11; Rsk, ribosomal protein S6 kinase; RT-PCR, reverse transcription-polymerase chain reaction; SEM, standard error of the mean; SGK1, serum/glucocorticoid regulated kinase 1; SH2D2A, SH2 domain containing 2A; siRNA, small interfering RNA; WNK1, WNK lysine deficient protein kinase 1
IHC Analysis of ATP1A1 in CC Primary Tissue
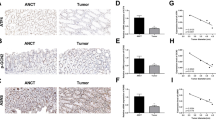
The IHC analysis of samples extracted from 200 patients indicated that ATP1A1 protein was primarily expressed in the cell membrane and cytoplasm of epithelial cells in non-cancerous areas. In cancerous areas, ATP1A1 protein was expressed on the membrane and cytoplasm of tumor cells, and the staining intensity was dependent on ATP1A1 expression levels (Fig. 4a–d). Figure 4e–h show the semi-quantitative analysis of ATP1A1 expression based on the staining intensities and proportions of positive tumor cells; the mean IHC score was 1.63 (range: 1.0–2.6). An IHC score of 1.6 was set (according to the median score) as the optimal cut-off value for the prediction of overall survival and recurrence-free survival (RFS). The classification of patients into high- and low-ATP1A1 expression groups was based on this cut-off value. Potential correlations between ATP1A1 expression and clinicopathological factors of 200 patients were subsequently investigated (Table 2). The levels of ATP1A1 were only correlated with the pathological T (pT) stage (p = 0.0054) (Supplementary Table 2). The prognostic impact of ATP1A1 expression in patients with CC following curative resection was also assessed. The 5-years overall survival rate in the high- and low-ATP1A1-expression groups was 78.93% and 86.12%, respectively (p = 0.1314) (Fig. 4i). Furthermore, the 5-years RFS rate was 69.14% and 88.88%, respectively (p = 0.0010) (Fig. 4j). The 5-years RFS rates by stage were 77.33% and 90.57% for stage II (p = 0.0554) and 58.85% and 86.49% for stage III (p = 0.0076) (Fig. 4k). As shown in Table 3, the univariate analysis identified lymphovascular invasion, pT and pathological N categories, and ATP1A1 expression after colectomy as significant prognostic factors of RFS. Similarly, the multivariate analysis identified lymphovascular invasion, the pT category, and ATP1A1 expression as independent prognostic factors (Table 3).
ATP1A1 protein expression in CC tissues and survival analysis according to the ATP1A1 levels. a HE staining of non-cancerous colonic tissue. Original magnification: ×400. b IHC analysis of primary human CC samples with an anti-ATP1A1 antibody; positivity for ATP1A1 protein was detected in the basolateral side of epithelial cells in the non-cancerous colonic tissue. Original magnification: ×400. c HE staining of CC tissue. Original magnification: ×400. d IHC analysis of primary human CC samples with an anti-ATP1A1 antibody; positivity for ATP1A1 protein was detected at the cell membrane and cytoplasm of tumor cells from CC tissue. Original magnification: ×400. e–h IHC analysis of primary CC samples with an anti-ATP1A1 antibody. Photomicrographs with examples of intensity scores 0 (e), 1 (f), 2 (g), and 3 (h) are provided. Original magnification: ×100. i Patients were classified into the low- (n = 90, blue line) or high-(n = 110, red line) ATP1A1 expression group. The 5-years overall survival rate tended to be lower in the former group versus the latter group (log-rank test, p = 0.1314). j The 5-years recurrence-free survival rate was significantly lower in the former group versus the latter group (log-rank test, p = 0.0010). k The 5-years recurrence-free survival rate by stage according to ATP1A1 expression. In stage II, the high ATP1A1expression group tended to have a worse prognosis, whereas in stage III, the high ATP1A1expression group had a significantly poorer prognosis. ATP1A1, Na+/K+-ATPase α1 subunit; CC, Colon cancer; HE, Hematoxylin and eosin; IHC, Immunohistochemical
Discussion
In this study, ATP1A1 knockdown in CC cells exerted tumor-suppressive effects. Therefore, ATP1A1 may play a critical role in the regulation of proliferation, survival, migration, and invasion of CC cells. According to the microarray analysis, knockdown of ATP1A1 suppressed the ERK5 signaling pathway in CC cells. Furthermore, IHC analysis revealed an apparent relationship between high ATP1A1 expression and poor long-term survival in patients with CC following curative resection. Based on these results, ATP1A1 may be involved in the progression of CC.
ATP1A1 is a subunit of NKA, a membrane pump with a low intracellular Na+/K+ ratio, cellular volume, and osmolarity. Apart from its involvement in numerous processes (e.g., ion transport, membrane potential generation, and nutrient uptake), NKA has been linked to signal transduction, cell junction, cell adhesion, and migration.20 Aberrant expression of ATP1A1 has been reported in various types of cancer.10,11,12,13,14,15,16 In the current analysis, knockdown of ATP1A1 led to inhibition of CC cell proliferation, survival, migration, and invasion; this is in agreement with findings reported in a previous study on gastric cancer.16 The results of in vivo experiments have also demonstrated that the tumorigenic potential of hepatocellular carcinoma was markedly decreased following transfection with ATP1A1-shRNA.14 Furthermore, it has been reported that the treatment of certain cells with specific inhibitors of NKA results in the activation of multiple signaling pathways, such as the MAPK signaling cascade and phosphatidylinositol-3 kinase (PI3K) pathway.21,22 It has been shown that the mechanisms by which ATP1A1 affects tumorigenesis and metastasis also influence K+ homeostasis. This leads to cell apoptosis or necrosis,21,22 as well as cell growth, migration, and invasion, by influencing the migration of fibroblast growth factor 2 (FGF2) from the cytosol to the extracellular space and the PI3K signaling pathway.23 Collectively, these findings demonstrate that ATP1A1 is associated with poor prognosis in cancer. This conclusion is in accordance with the data derived from a functional analysis of ATP1A1 in vitro, as well as the results of the IHC analysis, which associate high ATP1A1 expression with poor prognosis in CC.
In this investigation, the microarray analysis highlighted that the ERK5 signaling pathway is suppressed in CC cells after ATP1A1 knockdown (Fig. 3a). For the validation of these findings, quantitative RT-PCR was employed to determine the levels of genes associated with the ERK5 signaling pathway (Fig. 3b). MAPK cascades are frequently deregulated in cancer, and ERK5 is a major MAPK subfamily.24 ERK5 is activated by MAP2K5/MEK5; in turn, it induces the downstream activation of numerous transcription factors, as well as other effectors. These factors regulate multiple critical cellular events, such as proliferation, differentiation, transcriptional regulation, survival, and apoptosis.25,26 MEK5/ERK5 signaling abnormalities occur in many types of cancer, including CC.27,28 Importantly, their relationships with the induction of cell proliferation, apoptosis, tumor angiogenesis, and metastasis are attracting considerable research attention.29,30 However, the complete cellular and molecular mechanisms of the MEK5/ERK5 cascade in colorectal cancer remain to be elucidated. This is the initial evidence linking ATP1A1 to the ERK5 signaling pathway in CC. These findings provide insight into this ATP1A1-mediated signaling pathway.
This is the first investigation demonstrating the clinicopathologic and prognostic importance of ATP1A1 expression in tissue samples obtained from patients with CC. Seligson et al.11 determined the expression of ATP1A1 in 317 samples of renal clear cell carcinoma; according to their data, ATP1A1 levels were associated with the tumor grade. Also, high ATP1A1 expression significantly and independently predicted disease-specific death, the pT stage, metastasis status, and tumor grade. Nakamura et al.16 measured the levels of ATP1A1 in 192 samples of gastric cancer; their analysis showed that high ATP1A1 expression was related to venous invasion, the pT stage, and poor long-term outcomes. Consistent with previous data, in our analysis high ATP1A1 levels were associated with the pT stage of CC and may promote T stage (Supplementary Table 2). ATP1A1 expression was associated with prognosis regardless of adjuvant chemotherapy (Supplementary Fig. 4). In vitro experiments showed that ATP1A1 plays an active role in the development of colorectal cancer; the results of our IHC analysis corroborated this conclusion. As shown in Fig. 4j and Table 3, high ATP1A1 expression was identified as a significant predictor of poor prognosis in the multivariate analysis. Furthermore, in a stage-by-stage study, high ATP1A1 expression was associated with poor prognosis, and the group with high ATP1A1 expression at stage II tended to have a worse prognosis than the group with low ATP1A1 expression at stage III (Fig. 4k). These results suggest that ATP1A1 expression is associated with CC and may be used to identify high-risk stage II and low-risk stage III.
The limitations of this study should be mentioned. The investigation involved a retrospective IHC analysis, which may have been affected by selection bias. Moreover, this was a single-center study with a small cohort of patients with CC. A large prospective cohort study should be conducted to validate these findings and determine their applicability to clinical practice.
In summary, this investigation revealed that ATP1A1 is involved in the proliferation, apoptosis, migration, and invasion of CC cells. In addition, it was shown that ATP1A1 influenced the levels of genes linked to the ERK5 signaling pathway. Moreover, high ATP1A1 expression was identified as a predictor of poor prognosis in CC. Further investigation is warranted to elucidate the molecular mechanisms underlying the effects of ATP1A1. Nevertheless, the evidence indicates the potential prognostic importance of ATP1A1 and its value as a new target for the treatment of CC.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Bejček J, Spiwok V, Kmoníčková E, Rimpelová S. Na(+)/K(+)-ATPase revisited: on its mechanism of action, role in cancer, and activity modulation. Molecules. 2021;26(7):1905.
Skou JC, Esmann M. The Na, K-ATPase. J Bioenerg Biomembr. 1992;24(3):249–61.
Rajasekaran SA, Palmer LG, Quan K, et al. Na, K-ATPase beta-subunit is required for epithelial polarization, suppression of invasion, and cell motility. Mol Biol Cell. 2001;12(2):279–95.
Xiao Y, Meng C, Lin J, et al. Ouabain targets the Na(+)/K(+)-ATPase α(3) isoform to inhibit cancer cell proliferation and induce apoptosis. Oncol Lett. 2017;14(6):6678–84.
Xie Z, Askari A. Na(+)/K(+)-ATPase as a signal transducer. Eur J Biochem. 2002;269(10):2434–9.
Yang P, Cartwright C, Efuet E, et al. Cellular location and expression of Na+, K+-ATPase α subunits affect the anti-proliferative activity of oleandrin. Mol Carcinog. 2014;53(4):253–63.
Suhail M. Na, K-ATPase: ubiquitous multifunctional transmembrane protein and its relevance to various pathophysiological conditions. J Clin Med Res. 2010;2(1):1–17.
Chen JQ, Contreras RG, Wang R, et al. Sodium/potassium ATPase (Na+, K+-ATPase) and ouabain/related cardiac glycosides: a new paradigm for development of anti-breast cancer drugs? Breast Cancer Res Treat. 2006;96(1):1–15.
Mijatovic T, Roland I, Van Quaquebeke E, et al. The alpha1 subunit of the sodium pump could represent a novel target to combat non-small cell lung cancers. J Pathol. 2007;212(2):170–9.
Seligson DB, Rajasekaran SA, Yu H, et al. Na, K-adenosine triphosphatase alpha1-subunit predicts survival of renal clear cell carcinoma. J Urol. 2008;179(1):338–45.
Lefranc F, Mijatovic T, Kondo Y, et al. Targeting the alpha 1 subunit of the sodium pump to combat glioblastoma cells. Neurosurgery. 2008;62(1):211–21.
Mathieu V, Pirker C, Martin de Lassalle E, et al. The sodium pump alpha1 sub-unit: a disease progression-related target for metastatic melanoma treatment. J Cell Mol Med. 2009;13(9b):3960–72.
Zhuang L, Xu L, Wang P, et al. Na+/K+-ATPase α1 subunit, a novel therapeutic target for hepatocellular carcinoma. Oncotarget. 2015;6(29):28183–93.
Wu IC, Chen YK, Wu CC, et al. Overexpression of ATPase Na+/+ transporting alpha 1 polypeptide, ATP1A1, correlates with clinical diagnosis and progression of esophageal squamous cell carcinoma. Oncotarget. 2016;7(51):85244–58.
Nakamura K, Shiozaki A, Kosuga T, et al. The expression of the alpha1 subunit of Na(+)/K(+)-ATPase is related to tumor development and clinical outcomes in gastric cancer. Gastric Cancer. 2021;24(6):1278–92.
Sakai H, Suzuki T, Maeda M, et al. Up-regulation of Na(+), K(+)-ATPase alpha 3-isoform and down-regulation of the alpha1-isoform in human colorectal cancer. FEBS Lett. 2004;563(1–3):151–4.
Japanese Classification of Colorectal, Appendiceal, and Anal Carcinoma: the 3d English Edition [Secondary Publication]. J Anus Rectum Colon. 2019; 3(4):175–5.
Brierley JD, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. New York: Wiley; 2017.
Litan A, Langhans SA. Cancer as a channelopathy: ion channels and pumps in tumor development and progression. Front Cell Neurosci. 2015;9:86.
Pierre SV, Xie Z. The Na, K-ATPase receptor complex: its organization and membership. Cell Biochem Biophys. 2006;46(3):303–16.
Reinhard L, Tidow H, Clausen MJ, Nissen P. Na(+), K (+)-ATPase as a docking station: protein-protein complexes of the Na(+), K (+)-ATPase. Cell Mol Life Sci. 2013;70(2):205–22.
Zacherl S, La Venuta G, Müller HM, et al. A direct role for ATP1A1 in unconventional secretion of fibroblast growth factor 2. J Biol Chem. 2015;290(6):3654–65.
Paudel R, Fusi L, Schmidt M. The MEK5/ERK5 pathway in health and disease. Int J Mol Sci. 2021;22(14):7594.
Wang X, Tournier C. Regulation of cellular functions by the ERK5 signalling pathway. Cell Signal. 2006;18(6):753–60.
Cude K, Wang Y, Choi HJ, et al. Regulation of the G2-M cell cycle progression by the ERK5-NFkappaB signaling pathway. J Cell Biol. 2007;177(2):253–64.
Hu B, Ren D, Su D, et al. Expression of the phosphorylated MEK5 protein is associated with TNM staging of colorectal cancer. BMC Cancer. 2012;12:127.
Simões AE, Pereira DM, Gomes SE, et al. Aberrant MEK5/ERK5 signalling contributes to human colon cancer progression via NF-κB activation. Cell Death Dis. 2015;6(4):e1718.
Pereira DM, Simões AE, Gomes SE, et al. MEK5/ERK5 signaling inhibition increases colon cancer cell sensitivity to 5-fluorouracil through a p53-dependent mechanism. Oncotarget. 2016;7(23):34322–40.
Lochhead PA, Gilley R, Cook SJ. ERK5 and its role in tumour development. Biochem Soc Trans. 2012;40(1):251–6.
Acknowledgment
This study was supported by Grants-in-Aid for Scientific Research (C) (Grant numbers: 21K08689, 21K16456, 20K09016, 20K09084, 19K09202, and 19K09182) from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
All authors has nothing to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sumiyoshi, S., Shiozaki, A., Kosuga, T. et al. Functional Analysis and Clinical Importance of ATP1A1 in Colon Cancer. Ann Surg Oncol 30, 6898–6910 (2023). https://doi.org/10.1245/s10434-023-13779-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-023-13779-8