Abstract
Background
Few studies have reported the function and activation mechanism of ANO9 in esophageal squamous cell carcinoma (ESCC). The current study aimed to investigate the role of ANO9 in the regulation of tumor progression.
Methods
Knockdown experiments with human ESCC cell lines were performed using ANO9 siRNA, and the effects on cell proliferation, the cell cycle, apoptosis, and cellular movement were analyzed. Immunohistochemistry (IHC) analysis was performed on 57 primary tumor samples obtained from ESCC patients.
Results
In an in vitro study, depletion of ANO9 reduced cell proliferation, invasion, and migration in KYSE150 and KYSE 790 cells. In the cell cycle analysis, depletion of ANO9 increased the number of cells in G0/G1 arrest. In addition, the knockdown of ANO9 increased apoptosis. The results of the microarray analysis indicated that various centrosome-related genes such as CEP120, CNTRL, and SPAST were up- or downregulated in ANO9-depleted KYSE150 cells. The IHC results showed that high expression of ANO9 was associated with poor prognosis.
Conclusions
The results of the current study suggest that ANO9 regulates the cell cycle via centrosome-related genes in ESCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The anoctamin family consists of transmembrane proteins in 10 isoforms, and anoctamins (ANOs) are broadly expressed in epithelial and nonepithelial tissues.1 Findings show that ANOs mediate various functions such as ion transport and phospholipid scramblase across the membrane.1,2,3,4,5,6,7,–8 The cited reports demonstrate that each subtype has either ion channel activity, scramblase activity, or both activities, but details on these functions are unknown.
The best-known anoctamin gene is anoctamin 1 (ANO1), a Cl− channel activated by Ca2+ (CaCC) and expressed in various tissues, including secretory epithelia and smooth muscles.5,9,10 As reported, ANO1 is related to prognosis in gastric cancer, esophageal cancer, breast cancer, lung cancer, prostate cancer, and head and neck cancer.11,12,13,14,15,16,17,–18 Anoctamin 6 (ANO6) works as a phospholipid scramblase that exposes phosphatidylserine (PS) to the cell surface.19 Reports show that ANO6 is related to the metastatic potential of breast cancer.20
Recent studies show that anoctamin 9 (ANO9) plays important roles in various cancers such as colorectal cancer 21 and pancreatic cancer.22 However, the roles of ANO9 in tumor progression for patients with esophageal squamous cell carcinoma (ESCC) and the clinical significance of its expression remain unclear.
We previously reported that several ion channels were expressed and played important roles in human ESCC. For example, Na+/K+/2Cl− cotransporter 1 (NKCC1) affects the G2/M checkpoint,23 K+–Cl− cotransporter 3 (KCC3) regulates cellular invasion,24 anion exchanger 1 (AE1) regulates tumor progression,25 anion exchanger 2 (AE2) suppresses cellular movement,26 Na+/H+ exchanger 1 (NHE1) plays a suppressive role in cancer progression, and aquaporin 5 (AQP5) increases cell proliferation and survival.27 The current study aimed to investigate the roles of ANO9 in the regulation of cancer growth and its clinicopathologic significance in ESCC.
Materials and Methods
Cell Lines and Antibodies
Human ESCC cell lines KYSE150 and KYSE790 were purchased from Kyoto University (Kyoto, Japan). The immortalized fibroblast cell line WI-38 was obtained from the Japanese Cancer Research Resource Bank (JCRB) Cell Bank Center (Osaka, Japan), and the immortalized esophageal squamous cell line Het-1A as well as the immortalized mesothelial cell line MeT-5A were purchased from ATCC (Rockville, MD, USA). These cells were cultured in RPMI-1640 (Nacalai Tesque, Kyoto, Japan) supplemented with 100 μg/ml of streptomycin, 100 U/ml of penicillin, and 10% fetal bovine serum (FBS). The cells were cultured at 37 °C in a 5% carbon dioxide (CO2) incubator. The rabbit polyclonal anti-ANO9 antibody used in the immunohistochemical (IHC) analysis and Western blotting was purchased from Abcam (ab140087; Cambridge, MA, UK). The mouse monoclonal anti-β-actin (ACTB) antibody was purchased from Sigma-Aldrich (St. Louis, MO, USA). Horseradish peroxidase (HRP)-conjugated anti-rabbit and mouse secondary antibodies were obtained from Cell Signaling Technology (Beverly, MA, USA).
Western Blotting
The cells were washed two times with ice-cold phosphate-buffered saline (PBS) and lysed in M-PER buffer (Pierce, Rockford, IL, USA) with protease inhibitors (Pierce). Protein concentrations were measured with a modified Bradford assay (Bio-Rad, Hercules, CA, USA). Equal amounts of protein (10 mg/lane) were subjected to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to polyvinylidene difluoride (PVDF) membranes (GE Healthcare, Piscataway, NJ, USA). These membranes were incubated with antibodies for 24 h at 4 °C. An Amersham Imager 680 (GE Healthcare) was used to analyze band densities.
Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
From cancer cells, RNA was extracted using a RNeasy kit (Qiagen, Valencia, CA, USA). Real-time quantitative PCR analysis was performed using the Step One plus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and TaqMan Gene Expression Assays (Applied Biosystems). The PCR thermal cycle conditions were as follows: initial step at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and at 60 °C for 1 min. We measured the expression levels of the following genes: ANO9 (Hs00947743_g1), CEP120 (Hs00537880_m1), CNTRL (Hs00949316_m1), and SPAST(Hs01057020_m1) (Applied Biosystems). Expression was normalized for each gene to ACTB (Hs01060665_g1; Applied Biosystems). Assays were performed in triplicate.
Immunofluorescence Staining
Both KYSE150 and KYSE790 cells were cultured on glass coverslips and fixed with 4% paraformaldehyde. After fixation, the cells were rinsed twice with PBS and permeabilized with 0.1% Triton X-100. Then the cells were blocked with 1% bovine serum albumin and stained with the anti-ANO9 antibody and rhodamine phalloidin. Fluorescence microscopy (BZ-X810; Keyence, Osaka, Japan) was used to examine the distribution of ANO9.
siRNA Transfection
Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA, USA) was used for all small interfering RNA (siRNA) reverse transfection procedures at a final siRNA concentration of 24 nmol/l according to the manufacturer’s instructions. Both ANO9 siRNA (Stealth RNAiTM siRNA #HSS179461) and control siRNA (Stealth RNAiTM siRNA Negative Control) were purchased from Invitrogen.
Cell Proliferation Assay
On 6-well plates, KYSE150 and KYSE790 were seeded at a density of 1.2 × 105 cells/well and incubated at 37 °C in a 5% CO2 incubator. At the same time as the seeding, siRNA was transfected. These cells were detached from the plates with trypsin–EDTA and counted with a hemocytometer.
Cell Cycle Assay
The phase of the cell cycle was analyzed 48 h after siRNA transfection using flow cytometry with BD Accuri C6 (BD Biosciences, Bedfold, MA, USA). Cells were detached from the plates by trypsin–EDTA, treated with 0.2% Triton X-100, and stained with PI RNase staining buffer (Becton–Dickinson Biosciences, San Jose, CA, USA). The cells were analyzed using flow cytometry. At least 20,000 cells were analyzed.
Apoptosis Assay
The cells were evaluated 48 h after transfection and stained with the ANNEXIN V-FITC Kit (Beckman Coulter, Brea, CA, USA). The proportion of early or late apoptotic cells was measured by flow cytometry with BD Accuri C6. At least 20,000 cells were analyzed.
Migration and Invasion Assay
A migration assay was performed using a Cell Culture Insert with a pore size of 8 μm (BD Biosciences, Bedfold, MA, USA) and 24-well plates, and an invasion assay was performed using a Biocoat Matrigel (BD Biosciences, Bedfold, MA, USA). Cells (4.0 × 105 cells/well in KYSE150 and 3.0 × 105 cells/well in KYSE790) were seeded in the upper chamber in RPMI-1640 (serum-free) 48 h after siRNA transfection. The lower chamber contained RPMI-1640 with 10% FBS. After 48 h of incubation, the Matrigel and cells remaining in the upper chamber were removed. Migrated or invaded cells were stained with Diff-Quick staining reagents (Sysmex, Kobe, Japan) and counted in four independent fields of view. Each assay was performed in triplicate.
Low-Chloride Stimulation and Measurement of Fluorescence Intensity
We previously showed that the culture in the chloride-replaced medium (replacement of Cl− by NO3−) reduced the intracellular chloride concentration ([Cl−]i) using an MQAE reagent that was a chloride-sensitive fluorescence probe.28 We prepared a low-chloride medium supplemented with 10% FBS in chloride-free RPMI-1640 (replacement of Cl− by NO3−) (Nacalai Tesque, Kyoto, Japan). On 24-well plates, KYSE150 and KYSE790 cells were seeded at a density of 4.0 × 104 cells/well and incubated in the normal medium at 37 °C with 5% CO2. At 48 h after seeding, MQAE reagent dissolved in normal medium or low-chloride medium was applied, and the plates were incubated at 37 °C in a CO2 incubator for 12 h. The plates were washed five times with PBS, and the fluorescence intensity of MQAE was measured by fluorescence microscopy (BZ-X810; Keyence, Osaka, Japan).
Intracellular calcium concentration ([Ca2+]i) levels were evaluated by [Ca2+]i staining chemical dye with a Fluo-8 medium removal calcium assay kit (Abcam, Cambridge, MA, USA). At 48 h after seeding, Fluo-8 reagent was applied and incubated at 37 °C in a CO2 incubator for 60 min. The fluorescence intensity of Fluo-8 was measured by fluorescence microscopy (BZ-X810; Keyence, Osaka, Japan).
Microarray Analysis
The KYSE150 cells were transfected with ANO9 siRNA, and RNA was extracted using an RNeasy kit (Qiagen). Cyanine 3 (Cy3) labeled cRNA was prepared from 0.1 μg total RNA with the Low Input Quick Amp Labeling Kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions, followed by RNeasy column purification (Qiagen). Dye incorporation and cRNA yield were analyzed with the NanoDrop ND-2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and 0.6 μg of Cy3-labeled cRNA was fragmented at 60 °C for 30 min in a reaction volume of 25 μl containing 1× Agilent fragmentation buffer and 2× Agilent blocking agent following the manufacturer’s instructions.
At completion of the fragmentation reaction, 25 μl of 2× Agilent hybridization buffer was added to the fragmentation mixture and hybridized to SurePrint G3 Human GE 8 × 60 K Microarray ver3.0 (Agilent Technologies, Santa Clara, CA, USA) for 17 h at 65 °C in a rotating Agilent hybridization oven. After hybridization, microarrays were washed 1 min at room temperature with GE Wash Buffer 1 (Agilent Technologies, Santa Clara, CA, USA) and 1 min with 37 °C GE Wash buffer 2 (Agilent Technologies, Santa Clara, CA, USA). Slides were scanned on the Agilent SureScan Microarray Scanner (G2600D) using one color scan setting for 8 × 60k array slides. Microarray data were analyzed using Ingenuity Pathway Analysis software (Ingenuity Systems, Inc, Redwood City, CA, USA).
Patients and Primary Tissue Samples
This study enrolled 57 patients with ESCC who underwent esophagectomy between 1999 and 2009 at Kyoto Prefectural University of Medicine. All the patients provided informed consent before enrollment. The study excluded patients who underwent non-curative resection or preoperative chemotherapy and/or radiation therapy. Recurrence for 18 patients (31.6%) resulted in death, and 23 patients (40.4%) had recurrence within 5 years after surgery. Lymphatic metastasis occurred for 15 patients (26.3%), and 11 patients had hematogenous metastasis (19.3%).
The median follow-up period for the patients was 5.39 years (range, 0.38–16.70 years). The patients were staged using the International Union Against Cancer (UICC)/tumor-node-metastasis (TNM) Classification of Malignant Tumors (8th edition).29 This study was approved by the Institutional Review Board at the Kyoto Prefectural University of Medicine (ERB-C-1187).
Immunohistochemistry
The IHC staining was performed with a Vectastain ABC Elite Kit (Vector Laboratories, Burlingame, CA, USA) using the avidin-biotinylated peroxidase complex (ABC) method. The sections were deparaffinized in xylene and rehydrated in an ethanol series. Endogenous peroxidase activity was destroyed by incubation in 0.3% H2O2 for 30 min. An Avidin/Biotin Blocking Kit (Vector Laboratories) was used to block endogenous biotin, biotin receptors, and avidin binding sites. Then the slides were incubated with ANO9 antibody diluted 1:100 at room temperature for 1 h at 4 °C overnight. Visualization was performed by a standard ABC method. Counterstaining was performed with hematoxylin. Human breast cancer tissue was used as a positive control, and human esophageal cancer tissue without ANO9 antibody was used as a negative control.
The ANO9 score was graded semi-quantitatively by considering the staining intensity and proportion of stained cancer cells. Staining intensity was scored as 3 (strong), 2 (moderate), 1 (weak), or 0 (no staining). The proportion of stained cells was evaluated as a percentage of the stained area in the cancer area and scored from 0 to 1. We calculated the ANO9 score as the maximum multiplied product of the intensity and proportion scores (0–3).
Statistical Analysis
Categorical data were analyzed using Fisher’s exact test. The survival rates were estimated using the Kaplan–Meier method, and statistical analysis of the survival curves was performed using the log-rank test for equality. Prognostic factors for the multivariate analysis were determined using the Cox’s proportional hazard model. The statistical software JMP Ver12 (SAS Institute Inc., Cary, NC, USA) was used for these analyses. Differences were considered significant when the p value was lower than 0.05.
Results
Protein Expression of ANO9 in ESCC Cells
To evaluate ANO9 expression in ESCC, we performed Western blotting for nine esophageal cancer cell lines. As a result, ANO9 was expressed in various cells in ESCC (Fig. 1a). Immunofluorescence staining of ANO9 indicated that ANO9 was distributed in the cytoplasm and cell membranes of ESCC cells (Fig. 1b). We compared the mRNA level of ANO9 in cancerous and non-cancerous tissues of ESCC patients (Fig. S1A), and we compared the mRNA level of ANO9 in ESCC cell lines and other cells (Fig. S1B). The findings showed that ANO9 expression was higher in cancerous than in non-cancerous cells.
Expression of anoctamin 9 (ANO9) in esophageal squamous cell carcinoma (ESCC) cells. a Western blotting showed that ANO9 was expressed in various cell lines in ESCC. b Immunofluorescence staining with the ANO9 antibody demonstrated that ANO9 was mainly distributed in the cell membranes and cytoplasm of KYSE790 cells. Magnification ×1000. c Western blotting showed that ANO9 siRNA effectively reduced ANO9 protein levels in KYSE150 and KYSE790 cells. d ANO9 siRNA effectively reduced ANO9 mRNA levels in KYSE150 and KYSE790 cells. Mean ± SEM; n = 3. *p < 0.05 (significantly different from control siRNA)
We performed knockdown experiments using ANO9 siRNA in KYSE150 and KYSE790 cells that highly expressed ANO9 and investigated the effects of ANO9 depletion on tumor progression. Knockdown of ANO9 using siRNA reduced ANO9 protein levels (Fig. 1c) and ANO9 mRNA levels (Fig. 1d) in KYSE150 and KYSE790.
ANO9-Regulated Cell Proliferation and the Cell Cycle in ESCC Cells
In the KYSE150 cells, the cell number transfected ANO9 siRNA was significantly lower than the transfected control siRNA after transfection. In the KYSE790 cells, the number of cells 72 h after transfection was significantly lower in ANO9 siRNA (Fig. 2a). In the cell cycle analysis, the depletion of ANO9 increased the number of cells in the G0/G1 arrest in KYSE150 and KYSE790 cells (Fig. 2b). These results indicated that ANO9 functioned in regulating the proliferation and cell cycle of ESCC cells.
Anoctamin 9 (ANO9) controlled proliferation and the cell cycle in esophageal squamous cell carcinoma (ESCC) cells. a The downregulation of ANO9 inhibited the proliferation of KYSE150 and KYSE790 cells. The number of cells was counted 48 h and 72 h after siRNA transfection. Mean ± SEM; n = 3. *p < 0.05 (significantly different from control siRNA). b The downregulation of ANO9 increased the number of cells in the G0/G1 phase in KYSE150 and KYSE790 cells. Cells transfected with control or ANO9 siRNA were stained with propidium iodide (PI) and analyzed by flow cytometry. Mean ± SEM; n = 3. *p < 0.05 (significantly different from control siRNA)
ANO9-Regulated Apoptosis in ESCC Cells
To investigate the role of ANO9 in cell survival, apoptosis assays in the KYSE150 and KYSE790 cells was performed. The depletion of ANO9 significantly increased early apoptosis (annexin V-positive and PI-negative) and late apoptosis (annexin V-positive and PI-positive) in the KYSE150 and KYSE790 cells 48 h after siRNA transfection (Fig. 3a).
Anoctamin 9 (ANO9) controlled apoptosis, cell migration, and invasion in esophageal squamous cell carcinoma (ESCC) cells. a The downregulation of ANO9 increased the early and late apoptosis phases in KYSE150 and KYSE790 cells. Cells transfected with control or ANO9 siRNA were stained with PI and annexin V, then analyzed by flow cytometry. Mean ± SEM; n = 3. *p < 0.05 (significantly different from control siRNA). b The downregulation of ANO9 significantly reduced the migration and invasion of KYSE150 and KYSE790 cells. Cell migration and invasion were assessed by a Boyden chamber assay. Magnification ×40. Mean ± SEM; n = 3. *p < 0.05 (significantly different from control siRNA)
Depletion of ANO9-Reduced Cell Migration and Invasion in ESCC Cells
We analyzed the effects of ANO9 knockdown on cell migration and invasion in the KYSE150 and KYSE790 cells using a Boyden chamber assay. Depletion of ANO9 significantly decreased the number of migration and invasion cells in the KYSE150 and KYSE790 cells, with significant differences (Fig. 3b).
Gene Expression Profiling in ANO9 siRNA-Transfected ESCC Cells
We determined the gene expression profiles of ANO9-depleted KYSE150 cells by microarray analysis. The results of microarray analysis showed that 4290 genes displayed in-fold changes of more than 2.0 in ANO9-depleted KYSE150 cells. Of these, 3124 were upregulated, and 1166 were down-regulated. The most strongly up- or downregulated genes in ANO9-depleted KYSE150 cells are listed in Table 1. An Ingenuity Pathway Analysis showed that “inflammatory response” and “cancer” were two of the top-ranked diseases and disorders and that “cell-to-cell signaling and interaction” and “cellular movement” were two of the top-ranked molecular and cellular functions (Table S1). Furthermore, “acute phase response signaling” and “role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis” were the top two canonical pathways when ANO9 was depleted in the KYSE150 cells (Table S1).
ANO9 Regulated the Cell Cycle Network in ESCC Cells
Several types of network were found in ANO9-depleted KYSE150 cells. Of these, we focused on the “cell cycle, cellular assembly and organization, cellular function, and maintenance” network of ANO9 functions because the network could explain the results of the in vitro experiment (Table S1, Fig. 4a). The CEP120, and CNTRL genes are centrosomal proteins, and SPAST is a microtube regulator.30,31,–32 To verify the gene expression profiling data, CEP120, CNTRL, and SPAST were examined by RT-PCR. The expression levels of CEP120 and CNTRL mRNA were increased, and SPAST mRNA was decreased by ANO9 siRNA-transfection in KYSE150 and KYSE790 cells (Fig. 4b).
Network analyses by ingenuity pathway analysis. a The cell cycle network was one of the top-ranked networks, and several genes related to this pathway were upregulated in anoctamin 9 (ANO9)-depleted KYSE150 cells. b To verify the gene expression profiling data, CEP120, CNTRL, and SPAST were examined further by quantitative reverse transcription-polymerase chain reaction (RT-PCR). The expression levels of CEP120 and CNTRL mRNA were increased, and SPAST mRNA was decreased by ANO9 siRNA-transfection in KYSE150 and KYSE790 cells. Gene expression levels were normalized to the level of ACTB. Mean ± SEM; n = 3. *p < 0.05 (significantly different from control siRNA)
Next, we measured the fluorescence intensity of MQAE, a chloride-sensitive fluorescence probe, and Fluo-8, a calcium-sensitive fluorescence probe to evaluate the intracellular ion concentration. As a result, the fluorescence intensity of MQAE increased by knockdown of ANO9 in KYSE150 and KYSE790 cells (Fig. S2A). However, the fluorescence intensity of Fluo-8 was not affected by knockdown of ANO9 (Fig. S2B). To clarify the cell cycle-regulating mechanism, ANO9, CEP120, and CNTRL were examined by quantitative RT-PCR using low-chloride stimulation. The expression levels of ANO9, CEP120, and CNTRL mRNA were increased by low-chloride stimulation in KYSE150 and KYSE790 cells (Fig. S3). These results indicated that ANO9 regulated the cell cycle via centrosome-related genes by controlling intracellular chloride.
Immunohistochemical Analysis of ANO9 in ESCC Tissues
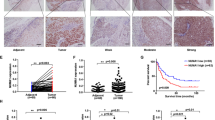
Immunohistochemistry for ANO9 indicated that ANO9 was expressed in the middle layers of the non-cancerous esophageal epithelia, and that vascular smooth muscle cells are stained by ANO9 in the submucosal layer (Fig. 5a). In ESCC tissues, the ANO9 was expressed mainly in the cell membranes and cytoplasm (Fig. 5b). The median score of ANO9 expression was 1.0 (range 0–2.7), and the mean score of ANO9 expression was 1.2 ± 0.8. The cutoff value of ANO9 was determined so that the p value for comparing the 5-year survival (OS) rate was the smallest.
Anoctamin 9 (ANO9) protein expression in esophageal squamous cell carcinoma (ESCC) cells. a Immunohistochemistry (IHC) staining of non-cancerous esophageal epithelia with the ANO9 antibody. Magnification ×100. b IHC staining of primary human ESCC samples with the ANO9 antibody. Magnification ×400. The IHC results are shown with an example intensity of 0. c Intensity 1. d Intensity 2 and e intensity 3. f Magnification ×400. g All patients were classified into two groups: a low ANO9 expression group (< 1.3, n = 31, solid line) and a high ANO9 expression group (≥ 1.3, n = 26, dotted line)
The 5-year OS rates with each cutoff value are shown in Table S2. According to the cutoff value, the patients were divided into low ANO9 expression (ANO9 scores < 1.3, n = 31) and high ANO9 expression (ANO9 scores ≥ 1.3, n = 26) groups (Fig. 5g). In the analysis of clinicopathologic features, ANO9 expression correlated with tumor length, venous invasion, pathologic T stage, and pathologic N stage (Table S3).
The pattern of recurrence was compared according to ANO9 expression (Table S4). The group with high ANO9 expression showed significantly more lymphatic metastasis (p = 0.035) and hematogenous metastasis (p = 0.039).
The study evaluated the prognostic significance of ANO9 after surgery by comparing the following 10 variables: gender, age, tumor location, tumor length, histological differentiation, lymphatic invasion, venous invasion, pathological T stage, pathological N stage and ANO9 expression. Univariate analysis showed that prognosis significantly correlated with the tumor length (p = 0.016), venous invasion (p = 0.039), pathologic T stage (p = 0.023), pathologic N stage (p = 0.024), and ANO9 expression (p = 0.0001) categories. In the multivariate analysis, high ANO9 expression (≥ 1.3) was an independent prognostic factor in ESCC patients (p = 0.025) (Table 2). The 5-year survival rate of the high ANO9 expression (49.5%) was significantly lower than that of the low ANO9 expression (82.9%).
Discussion
The knockdown of ANO9 using siRNA showed tumor suppressive effects in ESCC cells. Microarray analysis suggested that cell cycle was controlled via centrosome-related genes. We also investigated the expression of ANO9 in ESCC tissue and analyzed its relationships with the clinicopathologic factors and prognoses of patients with ESCC. As a result, ANO9 was found to be an independent poor prognostic factor for ESCC patients.
Known as transmembrane protein 16J (TMEM16J), ANO9 is expressed in the human nasal and colonic epithelium, as well as in the respiratory, digestive, skeletal, and integumentary systems during development.33 In addition, ANO9 was found to be expressed in human colorectal, lung, and breast cancers in silico.34 However, its biologic functions and clinical relevance in cancer are unknown. Previous studies reported that ANO9 regulated tumor progression via the epidermal growth factor receptor (EGFR)-signaling pathway in pancreatic cancer.22 Regarding the other isoforms of ANOs, it was reported that ANO1 regulated PI3K/Akt signaling in ovarian cancer,35 tumor necrosis factor (TNF)-alpha signaling in prostate cancer,36 and EGFR signaling in head and neck cancer.17 In addition, we newly found that ANO9 regulates the cell cycle via centrosome-related genes such as CEP120 and CNTRL in ESCC.
Betleja et al.37 reported that CEP120 played an important inhibitory role at the daughter centriole and that overexpression of CEP120 disrupted centriolar satellite localization and downregulated pericentriolar matrix 1 (PCM-1). Downregulation of PCM-1 is known to induce G0/G1 arrest.38 Gromley et al.39 reported that CNTRL-regulated cytokinesis activity and overexpression of CNTRL inhibition induced cytokinesis failure and G0/G1 arrest. These reports support our hypothesis that depletion of ANO9 induced G0/G1 arrest via centrosome-related genes.
To investigate the mechanism of regulating cell migration and invasion, we extracted mesenchymal markers from microarray data. Downregulation of mesenchymal markers such as MMP12, EST1, ACTA2, SOX10, and VIM were observed (Table S5). These results indicated that these genes regulated invasion and migration related to ANO9.
Single nucleotide polymorphisms (SNPs) in the single immunoglobulin interleukin 1 receptor gene region (SIGIRR), including the adjacent genes PKP3 and ANO9, were reported to be associated with immune pathways and adaptive immune responses, and to act as a negative regulator of Toll-like-receptor (TLR) signaling, which is important in mycobacterium tuberculosis pathogenesis.40,41 Our microarray analysis also showed depletion of ANO9-upregulated immune-related pathways such as the iCOS-iCOSL-signaling pathway and the Th1 pathway. These results indicate that ANO9 regulates immune-related functions.
Previous studies reported that high expression of ANO1 in IHC was a biomarker for predicting unfavorable outcome for ESCC patients,15 but ANO9 expression in ESCC was unclear. To clarify the clinical implications of ANO9 expression, we analyzed the survival rate for 57 patients who underwent curative resection for primary ESCC. Our IHC results showed that high expression of ANO9 was a poor prognostic factor in ESCC patients. Jun et al.22 also reported that ANO9 was a poor prognostic factor in pancreatic cancer. In contrast, Li et al.21 reported that high expression of ANO9 was correlated with good prognostic outcomes in colorectal cancer.
The reasons for the different effects of ANO9 on esophageal, pancreatic, and colorectal cancer are unclear. In pancreatic cancer, ANO9 has been reported to regulate cancer growth via the EGFR pathway, but our microarray data did not correlate with the EGFR pathway. Therefore, ANO9 is thought to be involved in the regulation of cancer growth by different mechanisms depending on the cancer type. This is the first report on the relationship between ANO9 expression and prognosis in ESCC patients, and further studies may clarify additional important roles of ANO9 in oncogenesis.
We further evaluated the recurrence-free survival (RFS) rate for patients with ESCC according to various clinicopathologic parameters (Table S6). The group with high ANO9 expression had a significantly lower RFS rate than the group with low ANO9 expression (37.4% vs. 75.2%; p = 0.002), and lymph node metastasis was an independent prognostic factor in 5-year RFS. We hypothesized that OS and RFS depended on different factors because ANO9 affected chemotherapy resistance after recurrence. Further research is needed to clarify these in the future.
Some articles reported that ANO9 had calcium-dependent chloride release channel activity.42,43 However, some articles reported that ANO9 did not have chloride channel activity.2,19,22 Whether ANO9 has chloride channel activity or not is a controversial issue. In the current study, depletion of ANO9 increased the fluorescence intensity of MQAE. On the other hand, it did not affect the fluorescence intensity of Fluo-8 in ESCC cells, suggesting that ANO9 regulated the intracellular chloride environment. These results indicate that ANO9 regulates the cell cycle via centrosome-related genes such as CEP120 and CNTRL by controlling intracellular chloride.
In summary, we showed that ANO9 played the important role in the cell cycle and progression of ESCC cells. The microarray analysis indicated that ANO9 strongly affected the expression of centrosome-related genes. The IHC analysis also suggested that the high expression of ANO9 was a poor prognostic factor in ESCC patients. The results of the current study indicate that ANO9 has potential as a biomarker for cancer growth and as a therapeutic target for ESCC such as inhibitor or RNA interference of ANO9.
References
Schreiber R, Uliyakina I, Kongsuphol P, et al. Expression and function of epithelial anoctamins. J Biol Chem. 2010;285:7838–45.
Brunner JD, Schenck S, Dutzler R. Structural basis for phospholipid scrambling in the TMEM16 family. Curr Opin Struct Biol. 2016;39:61–70.
Kunzelmann K. TMEM16, LRRC8A, bestrophin: chloride channels controlled by Ca(2+) and cell volume. Trends Biochem Sci. 2015;40:535–43.
Wanitchakool P, Wolf L, Koehl GE, et al. Role of anoctamins in cancer and apoptosis. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130096.
Pedemonte N, Galietta LJ. Structure and function of TMEM16 proteins (anoctamins). Physiol Rev. 2014;94:419–59.
Kunzelmann K, Tian Y, Martins JR, et al. Anoctamins. Pflugers Arch. 2011;462:195–208.
Kunzelmann K, Kongsuphol P, Chootip K, et al. Role of the Ca2+-activated Cl– channels bestrophin and anoctamin in epithelial cells. Biol Chem. 2011;392:125–34.
Kunzelmann K, Kongsuphol P, Aldehni F, et al. Bestrophin and TMEM16-Ca(2+) activated Cl(−) channels with different functions. Cell Calcium. 2009;46:233–41.
Duran C, Hartzell HC. Physiological roles and diseases of Tmem16/Anoctamin proteins: are they all chloride channels? Acta Pharmacol Sin. 2011;32:685–92.
Hartzell HC, Yu K, Xiao Q, Chien LT, Qu Z. Anoctamin/TMEM16 family members are Ca2+-activated Cl− channels. J Physiol. 2009;587(Pt 10):2127–39.
Lu G, Shi W, Zheng H. Inhibition of STAT6/anoctamin-1 activation suppresses proliferation and invasion of gastric cancer cells. Cancer Biother Radiopharm. 2018;33:3–7.
Wu H, Wang H, Guan S, et al. Cell-specific regulation of proliferation by ANO1/TMEM16A in breast cancer with different ER, PR, and HER2 status. Oncotarget. 2017;8:84996–5013.
Seo Y, Ryu K, Park J, et al. Inhibition of ANO1 by luteolin and its cytotoxicity in human prostate cancer PC-3 cells. PLoS ONE. 2017;12:e0174935.
Fujimoto M, Inoue T, Kito H, et al. Transcriptional repression of HER2 by ANO1 Cl(−) channel inhibition in human breast cancer cells with resistance to trastuzumab. Biochem Biophys Res Commun. 2017;482:188–94.
Shang L, Hao JJ, Zhao XK, et al. ANO1 protein as a potential biomarker for esophageal cancer prognosis and precancerous lesion development prediction. Oncotarget. 2016;7:24374–82.
Jia L, Liu W, Guan L, Lu M, Wang K. Inhibition of calcium-activated chloride channel ANO1/TMEM16A suppresses tumor growth and invasion in human lung cancer. PLoS ONE. 2015;10:e0136584.
Bill A, Gutierrez A, Kulkarni S, et al. ANO1/TMEM16A interacts with EGFR and correlates with sensitivity to EGFR-targeting therapy in head and neck cancer. Oncotarget. 2015;6:9173–88.
Britschgi A, Bill A, Brinkhaus H, et al. Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc Natl Acad Sci USA. 2013;110:E1026–34.
Suzuki J, Fujii T, Imao T, Ishihara K, Kuba H, Nagata S. Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J Biol Chem. 2013;288:13305–16.
Dutertre M, Lacroix-Triki M, Driouch K, et al. Exon-based clustering of murine breast tumor transcriptomes reveals alternative exons whose expression is associated with metastasis. Cancer Res. 2010;70:896–905.
Li C, Cai S, Wang X, Jiang Z. Identification and characterization of ANO9 in stage II and III colorectal carcinoma. Oncotarget. 2015;6:29324–34.
Jun I, Park HS, Piao H, et al. ANO9/TMEM16J promotes tumourigenesis via EGFR and is a novel therapeutic target for pancreatic cancer. Br J Cancer. 2017;117:1798–809.
Shiozaki A, Nako Y, Ichikawa D, et al. Role of the Na(+)/K(+)/2Cl(−) cotransporter NKCC1 in cell cycle progression in human esophageal squamous cell carcinoma. World J Gastroenterol. 2014;20:6844–59.
Shiozaki A, Takemoto K, Ichikawa D, et al. The K-Cl cotransporter KCC3 as an independent prognostic factor in human esophageal squamous cell carcinoma. Biomed Res Int. 2014;2014:936401.
Shiozaki A, Kudou M, Ichikawa D, et al. Expression and role of anion exchanger 1 in esophageal squamous cell carcinoma. Oncotarget. 2017;8:17921–35.
Shiozaki A, Hikami S, Ichikawa D, et al. Anion exchanger 2 suppresses cellular movement and has prognostic significance in esophageal squamous cell carcinoma. Oncotarget. 2018;9:25993–6006.
Shimizu H, Shiozaki A, Ichikawa D, et al. The expression and role of Aquaporin 5 in esophageal squamous cell carcinoma. J Gastroenterol. 2014;49:655–66.
Miyazaki H, Shiozaki A, Niisato N, Marunaka Y. Physiological significance of hypotonicity-induced regulatory volume decrease: reduction in intracellular Cl– concentration acting as an intracellular signaling. Am J Physiol Renal Physiol. 2007;292:F1411–7.
James DB, Mary KG, Christian W. International Union Against Cancer (UICC) TNM Classification of Malignant Tumors. 8th ed. Wiley, New York. 2017.
Kuo YW, Trottier O, Mahamdeh M, Howard J. Spastin is a dual-function enzyme that severs microtubules and promotes their regrowth to increase the number and mass of microtubules. Proc Natl Acad Sci USA. 2019;116:5533–41.
Kumari A, Panda D. Regulation of microtubule stability by centrosomal proteins. IUBMB Life. 2018;70:602–11.
Kumar A, Rajendran V, Sethumadhavan R, Purohit R. CEP proteins: the knights of centrosome dynasty. Protoplasma. 2013;250:965–83.
Rock JR, Harfe BD. Expression of TMEM16 paralogs during murine embryogenesis. Dev Dyn. 2008;237:2566–74.
Katoh M, Katoh M. Identification and characterization of human TP53I5 and mouse Tp53i5 genes in silico. Int J Oncol. 2004;25:225–30.
Liu Z, Zhang S, Hou F, Zhang C, Gao J, Wang K. Inhibition of Ca(2+)-activated chloride channel ANO1 suppresses ovarian cancer through inactivating PI3K/Akt signaling. Int J Cancer. 2019;144:2215–26.
Song Y, Gao J, Guan L, Chen X, Gao J, Wang K. Inhibition of ANO1/TMEM16A induces apoptosis in human prostate carcinoma cells by activating TNF-alpha signaling. Cell Death Dis. 2018;9:703.
Betleja E, Nanjundappa R, Cheng T, Mahjoub MR. A novel Cep120-dependent mechanism inhibits centriole maturation in quiescent cells. Elife. 2018;7:e35439.
Srsen V, Gnadt N, Dammermann A, Merdes A. Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J Cell Biol. 2006;174:625–30.
Gromley A, Jurczyk A, Sillibourne J, et al. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J Cell Biol. 2003;161:535–45.
O’Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP. The immune response in tuberculosis. Annu Rev Immunol. 2013;31:475–527.
Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39:1003–18.
Kunzelmann K, Ousingsawat J, Benedetto R, Cabrita I, Schreiber R. Contribution of anoctamins to cell survival and cell death. Cancers (Basel). 2019;11(3):382.
Wanitchakool P, Ousingsawat J, Sirianant L, MacAulay N, Schreiber R, Kunzelmann K. Cl(−) channels in apoptosis. Eur Biophys J. 2016;45:599–610.
Acknowledgment
This work was supported by Grants-in-Aid for Scientific Research (C) (17K10602, 17K10710, 18K08628, 18K08689, 19K09202) from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig.
1. ANO9 expression of normal cells compared with ESCC cells. (A) The expression levels of ANO9 mRNA were higher in ESCC cells than other cells. Gene expression levels were normalized to the level of ACTB. Mean ± SEM. n = 3. (B) The expression levels of ANO9 mRNA were higher in cancerous tissue than non-cancerous tissue in ESCC patients (n = 5). Gene expression levels were normalized to the level of ACTB. *p < 0.05 (significantly different from cancerous tissue) (TIFF 28567 kb)
Supplementary Fig.
2. ANO9 regulated the intracellular ion environment. (A) Fluorescence intensity of MQAE increased by knockdown of ANO9 in KYSE150 and KYSE790 cells. (B) Fluorescence intensity of Fluo-8 was not affected by knockdown of ANO9 in KYSE150 and KYSE790 cells (TIFF 28567 kb)
Supplementary Fig.
3. Change in gene expression by low chloride stimulation. ANO9, CEP120 and CNTRL were examined by quantitative RT-PCR. The expression levels of ANO9, CEP120 and CNTRL mRNA were increased by low chloride stimulation in KYSE150 and KYSE790 cells. Gene expression levels were normalized to the level of ACTB. Mean ± SEM. n = 3. *p < 0.05 (significantly different from normal medium) (TIFF 28567 kb)
Rights and permissions
About this article
Cite this article
Katsurahara, K., Shiozaki, A., Kosuga, T. et al. ANO9 Regulated Cell Cycle in Human Esophageal Squamous Cell Carcinoma. Ann Surg Oncol 27, 3218–3230 (2020). https://doi.org/10.1245/s10434-020-08368-y
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-020-08368-y