Abstract
Background
OBP-801 is a novel histone deacetylase inhibitor being developed as an anticancer drug. In this study, we explored genes to predict drug resistance in human cancer.
Methods
OBP-801 resistance was assessed in 37 strains of human cancer cell lines. Expression microarrays harboring 54,675 genes were used to focus on candidate genes, which were validated for both functional and clinical relevance in esophageal squamous cell carcinoma (ESCC).
Results
OBP-801 is sensitive to esophageal, gastric, and thyroid cancer, and resistant to some esophageal and colorectal cancers. We therefore used ESCC to explore genes. Comprehensive exploration focused on ΔNp63/SOX2, which were both genetically and epigenetically overexpressed in ESCC. Genomic amplifications of ΔNp63/SOX2 were tightly correlated each other (r = 0.81). Importantly, genomic amplification of ΔNp63/SOX2 in the resected tumors after neoadjuvant chemotherapy was significantly associated with histological grade of response (G1). Forced expression of either of these two genes did not induce each other, suggesting that their functional relevances were independent and showed robust drug resistance in OBP-801, as well as 5-fluorouracil. Furthermore, ΔNp63 could exert a potent oncogenic potential. RNA interference of ΔNp63 supported its oncological properties, as well as drug resistance.
Conclusion
Comprehensive exploration of genes involved in anticancer drug residence could identify critical oncogenes of ΔNp63/SOX2 that would predict chemotherapy response in ESCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
OBP-801/YM753/spiruchostatin A was discovered for a p21WAF1/Cip1-inducing agent, demonstrating tumor growth factor (TGF)-ß-like activity in the culture broth of Pseudomonas fluorescens Q71576, and proven to be a histone deacetylase (HDAC) inhibitor, giving it potential as an anticancer drug.1 Tumor-specific histone hypoacetylation was observed in solid tumors,2 and increased expression of HDAC13 HDAC23,4 and HDAC35 has been reported in human cancers. Several structural classes of HDAC inhibitors have been characterized, including short-chain fatty acids (phenylbutyrate [PB]) or hydroxamic acids (trichostatin A [TSA] and suberoylanilide hydroxamic acid [SAHA]). HDAC inhibitors affect cellular processes dysregulated in neoplastic cells,6,7 and clinical trials of these agents have been conducted.8,9,–10 Cancer is a genetic disease and is addicted to DNA aberrations, hence proposing genetic aberrations to predict treatment failure. Previous papers identified anticancer activities of OBP-801,1,2,3,4,–5,7,8,9,–10 however these abnormalities have not included genetic activities. In our current study, we investigated genetic abnormalities predicting drug resistance of OBP-801.
In our laboratory, gene expressions associated with PB sensitivity have been rigorously sought, and both ZEB1 and ASCL2 were identified as being associated with drug resistance in breast cancer and colorectal cancer, respectively.11 Histone modification is supposed to affect endogenous metabolism in human cancer, and thus HDAC inhibitors can specifically inhibit the molecular pathways modified by histone modification. Suchphysiologic inhibition is thus supposed to be less toxic than conventional chemotherapy, representing suppression of DNA synthesis. Based on this rationale, we believe that HDAC inhibitors may be a better model than toxic inhibition of DNA synthesis by chemotherapy to predict physiological pathways aberrantly activated in aggressive cancer cells. In this study, we used the novel HDAC inhibitor OBP-801 to explore genes involved in the treatment resistance of human cancers to guide treatments.
Patients and Methods
Cell Lines
We used 37 strains of human cancer cell lines, composed of 10 esophageal,12 8 gastric,13 7 colorectal,14 7 pancreatic,15 and 5 thyroid cancers. 8305c cell lines were purchased from the RIKEN BioResource Center (Ibaraki, Japan). TT, FTC-133, R082-w-1, and Nthy-ori 3-1 were obtained from the European Collection of Authenticated Cell Cultures (England, UK). 8305c, R082-w-1, and Nthy-ori 3-1 cells were maintained in RPMI 1640 medium (Gibco, Carlsbad, CA, USA), TT cells were maintained in Ham’s F-12 medium (Gibco), and FTC-133 cells were maintained in DMEM/F12 glutamax (Gibco).
OBP-801 and 5-Fluorouracil Treatments in Human Cancer Cell Lines
OBP-801 obtained from Oncolys BioPharma Inc. (Tokyo, Japan) was dissolved in DMSO (Merck KGaA, Darmstadt, Germany), and 5-fluorouracil purchased from Wako Pure Chemical Industries (Osaka, Japan) was dissolved in distilled water. On day 3 after treatment with OBP-801 or 5-fluorouracil, the cells were counted using a Countess automated cell counter (Invitrogen Life Technologies, Carlsbad, CA, USA).
Expression Microarrays
Total RNA was extracted using an RNeasy Mini Kit (Qiagen Sciences, Hilden, Germany). Gene profiles were analyzed using Affymetrix GeneChip Human Genome U133 Plus 2.0 Array (harboring 54,675 genes).
5-Aza-2′-Deoxycytidine and Trichostatin A Treatments in Esophageal Squamous Cell Carcinoma (ESCC) Cell Lines
Cells were treated once every 24 h for 4 days with 1 or 5 µM of 5-aza-2′-deoxycytidine (Merck KGaA) dissolved in 50% acetic acid (Wako Pure Chemical Industries). 300 nM of the HDAC inhibitor trichostatin A (TSA; Merck KGaA) was added to the medium for the final 24 h.
Semi-quantitative Reverse Transcriptase–Polymerase Chain Reaction
Complementary DNA (cDNA) was synthesized from RNA using SuperScript III reverse transcriptase (Invitrogen) and Oligo (dT) primers (Invitrogen). Semi-quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) was conducted using Platinum Taq DNA Polymerase (Invitrogen; primers shown in electronic supplementary Table S1).
Western Blot Analysis
Thirty micrograms of protein with 5 µL NuPAGE® LDS Sample Buffer and 2 µL NuPAGE® Sample Reducing Agent (10 ×) were separated on NuPAGE® Bis–Tris Gel (Invitrogen). Anti-Myc antibody (dilution of 1:2000; Invitrogen) and monoclonal anti-β-actin antibody (dilution of 1:5000; Sigma, St Louis, MO, USA) were used.
Plasmid and Transfection
Full-length cDNA sequences of ΔNp63 and SOX2 were made from TE10 cells into a pcDNA3.1/myc-His C expression vector (Invitrogen). Transient transfection was performed using Lipofectamine 2000 (Invitrogen) in OPTI-MEM medium (Gibco).
Anchorage-Independent Colony Formation Assay
Anchorage-independent cell growth was conducted as follows: 0.72% bottom agarose (Bacto™ Agar; Becton Dickison and Company, Franklin Lakes, NL, USA) was placed in six-well plates, and 0.36% top agarose with 1 × 105 cells was infused on bottom agarose. After 3 weeks of incubation, colonies were visualized using ethidium bromide.
Proliferation Assay
Cell proliferation and viability were assessed using a Premix WST-1 Cell Proliferation Assay System (Takara Bio, Tokyo, Japan) in 96-well plates in triplicate.
Small-Interfering RNA and Transfection
Small-interfering RNA (siRNA) targeting human ΔNp63 (SASI_Hs02_00326884, sense; GGAUGAACCGCCGUCCAAUdTdT antisense; AUUGGACGGCGGUUCAUCCdTdT) was purchased from Merck KGaA. MISSION siRNA Universal Negative Control (Merck KGaA) was used as a control. The cells were transfected with 30 nM of siRNA using Lipofectamine 2000 reagent.
ΔNp63/SOX2 Genomic Gain Status in Primary ESCC
To examine the relative copy number of the ΔNp63 and SOX2 gene, quantitative PCR (Q-PCR) and the 2−ΔΔCt method were performed. The study group included 112 esophageal squamous cell carcinoma (ESCC) patients who underwent surgical resection at Kitasato University hospital between 1996 and 2007. Genomic DNA was extracted from the formalin-fixed paraffin-embedded (FFPE) tumor tissues and corresponding non-cancerous mucosa tissues using a QIAamp DNA FFPE Kit (Qiagen).
Q-PCR was carried out in triplicate using iQ™ Supermix (Bio-Rad Laboratories, Hercules, CA, USA) and the CFX96 real-time system (Bio-Rad). The primer set of ΔNp63 and SOX2 can be seen in electronic supplementary Table S2. Genomic gain was defined as twofold in the primary tumor tissues compared with the corresponding non-cancerous mucosa tissues.
Immunohistochemistry
FFPE histological sections (3 µm thick) were immunostained, and sections were incubated at 4 °C overnight with the following antibodies: anti-p63, clone 4A4 (mouse monoclonal immunoglobulin [Ig] G2a; BioCare Medical, LLC., Pacheco, CA, USA) at an antibody concentration of 100 µg/mL, and anti-SOX2, clone GT1352 (mouse monoclonal IgG2a; Gene Tex, Inc., Irvine, CA, USA) at an antibody concentration of 20 µg/mL. SOX2 and p63 expression levels were graded according to the diagnostic criteria of American Society of Clinical Oncology/College of American Pathology 2007 guidelines.16
Statistical Analysis
Continuous variables were evaluated using Student’s t test, and categorical variables were evaluated using Fisher’s exact test or the Chi square test, as appropriate. A p value < 0.05 was considered to indicate statistical significance. All calculations were performed using JMP® 14 pro software (SAS Institute Inc., Cary, NC, USA).
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committee of Kitasato University (approval number B17-234). Written informed consent was obtained from all patients, and the study was conducted in accordance with the Declaration of Helsinki.
Results
OBP-801 Drug Resistance in Human Cancer Cells
We initially investigated OBP-801 drug resistance in 37 human cancer cell lines. The results of ESCC cells are shown in Fig. 1a, while other cells are displayed in electronic supplementary Fig. S1. The antiproliferative effect of OBP-801 is dose-dependent (5–100 nM). OBP-801 is sensitive to gastric, pancreatic, and thyroid cancers, as well as a portion of ESCCs, and is rather resistant to colorectal cancer and some portions of ESCCs. We therefore used ESCC to explore genes involved in drug resistance to OBP-801.
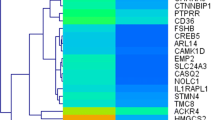
a, b OBP-801 and 5-fluorouracil drug sensitivity of ESCC cells. c–e Identification and confirmation of expression profiles of OBP-801 resistance-related genes and sensitivity-related genes. a Viable cancer cell counts after 72 h of OBP-801 administration (5–100 nM) compared with control cells. Error bar represents the standard error. Experiments were performed in triplicate. b Viable ESCC cell counts after 72 h of 5-fluorouracil administration (2.5–160 μM) compared with control cells. Experiments were performed in triplicate. c Heat map of gene expression in OBP-801-resistant ESCC strains compared with OBP-801-sensitive ESCC strains by Affymetrix expression microarray. d Representative confirmation of gene expression by RT-PCR for both OBP-801-resistant and -sensitive ESCC strains. Note that p63 primers are used for p63 common structures as indicated in electronic supplementary Fig. S2a). e Demethylation treatment robustly decreases the expression of both ΔNp63 and SOX2 in OBP-801-resistant strains. ESCC esophageal squamous cell carcinoma, RT-PCR reverse transcriptase–polymerase chain reaction
OBP-801 Drug Resistance in Human ESCC
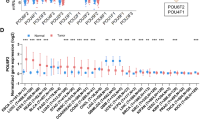
We initially selected ESCC cell lines as the most sensitive (TE1, TE4) and the most resistant (TE6, TE10, TE11) to OBP-801 (Fig. 1a). Responses to 5-fluorouracil are also shown in ESCC cell lines (Fig. 1b). Using the five cell lines, gene selection was performed using the Affymetrix expression microarray harboring 54,675 genes (Fig. 2). A heat map according to the microarray data was shown (Fig. 1c).
RT-PCR Confirmed Microarray Expression Data in the Five ESCC Cell Lines
Among the most impressive 25 genes, we initially focused on OBP-801 resistance-related genes that were increased in the OBP-801-resistant strains and decreased in expression in the OBP-801-sensitive strains. The most representative genes are shown in Fig. 1d. The resistant gene candidates, such as STEP1, C3orf58, SOX2, TP63, and IFI16, were only expressed in the resistant strains. For TP63, we designed three sets of primers, two of which were specifically targeted for TP63-specific (blue) and ΔNp63-specific (red) primers, as well as common primers (TP63, black) [electronic supplementary Fig. S2a]. PCR revealed that overexpression of TP63 in the OBP-801-resistant strains was attributed to ΔNp63.
Overexpression of both ΔNp63 and SOX2 genes in OBP-801-resistant cells is very intriguing because the genomic location of these genes is so close to each other at the 3q chromosome. Additionally, the Cancer Genome Atlas Research (CGAR) Network reported that genomic co-amplifications of both genes was recently identified as the most frequently gained region in primary ESCC17 and other SCCs.18,19 Moreover, ΔNp63/SOX2 was confirmed to represent a stem cell signature in various SCCs.20,21,–22
Concurrent 5-aza-2′-deoxycytidine and TSA was performed in the resistant strains. Both ΔNp63 and SOX2 were robustly suppressed at messenger RNA (mRNA) level by the epigenetic treatments (Fig. 1e). We also demonstrated that OBP-801 alone could not alter both ΔNp63 and SOX2 expression (electronic supplementary Fig. S2b).
Transient Transfection of ΔNp63 and SOX2 Genes, and Phenotypic Changes in OBP-801-Sensitive Strains
ΔNp63 and SOX2 expression plasmids with c-Myc tagged were constructed with sequencing confirming no mutation. Transient transfection of both genes confirmed actual expression at mRNA and protein levels (Fig. 3a). Both proteins were detected using the c-Myc antibody. At mRNA level, the ΔNp63 gene could not induce the SOX2 gene, and the SOX2 gene did not increase the ΔNp63 gene.
Functional analysis after transient transfection of both ΔNp63 and SOX2 genes to OBP-801-sensitive ESCC cell lines. Error bar represents the standard error. Experiments were performed in triplicate. a RT-PCR confirmed augmented expression of both the ΔNp63 and SOX2 genes at mRNA level in OBP-801-sensitive ESCC cell lines (TE1 and TE4) by transient transfection. Note that either of the two genes could not induce vice versa in both TE1 and TE4. Western blot analysis confirmed protein expression of the two genes (35 and 63 kDa) in both TE1 and TE4. Both proteins were detected by tagged c-Myc antibody. b OBP-801 sensitivity after transient transfection of the SOX2 and/or ΔNp63 genes. After transfections of SOX2, ΔNp63, and both genes, both TE1 and TE4 became significantly resistant to OBP-801. c 5-Fluorouracil sensitivity after transient transfection of the SOX2 and/or ΔNp63 genes. After transfections of SOX2, ΔNp63, and both genes, both TE1 and TE4 became significantly resistant to 5-fluorouracil. d Cell count assay after transient transfection of the SOX2 and/or ΔNp63 genes. There were no significant differences in cell count assay between before and after transient transfection. e The WST-1 assay after transient transfection of the SOX2 and/or ΔNp63 genes. The absorbances were significantly increased after ΔNp63 transfection in both TE1 and TE4, as well as after transfection of both genes in TE4. f Anchor-independent colony formation assay after transient transfection of the SOX2 and/or ΔNp63 genes. The number of colonies was significantly increased after ΔNp63 transfection in both TE1 and TE4, as well as after transfection in both genes in TE4. ESCC esophageal squamous cell carcinoma, RT-PCR reverse transcriptase–polymerase chain reaction, mRNA messenger RNA, NS non-significant
Transient transfection of ΔNp63 and/or SOX2 significantly increased drug resistance for OBP-801, in both TE1 and TE4 (Fig. 3b). For 5-fluorouracil, transient transfection of ΔNp63 and/or SOX2 significantly increased drug resistance in both TE1 and TE4 (Fig. 3c).
We then investigated cell proliferation by simple cell count, WST-1 assay, and soft agar colony formation assay. Differently from drug resistance, the two genes did not significantly increase cell proliferation by simple cell count in TE1 and TE4 (Fig. 3d). On the other hand, ΔNp63 is involved in augmented properties of cell viability assessed by the WST-1 assay (Fig. 3e), and increased anchorage-independent growth in the soft agar assay (Fig. 3f). These findings suggest that oncogenic potential did not necessarily represent drug resistance in ESCC cells. We could not putatively show the synergistic effects of both ΔNp63 and SOX2 due to the low efficiency of double transfection.
Knockdown of the ΔNp63 Gene in OBP-801-Resistant Cells
The ΔNp63 gene was highly expressed in all OBP-801-resistant strains (TE6, TE10, TE11), while the SOX2 gene was expressed in two of the three OBP-801-resistant cells. ΔNp63 showed stronger oncogenic properties in the OBP-801-sensitive strains. We therefore then investigated knockdown of the ΔNp63 gene in OBP-801-resistant strains of TE6, TE10, and TE11 with regard to drug resistance against both OBP-801 and 5-fluorouracil (Fig. 4a). Interestingly, ΔNp63 is essential for drug resistance in the most optimized concentration of TE6, TE10, and TE11 (Fig. 4b).
Drug resistance alteration after ΔNp63 gene knockdown experiments in OBP-801-resistant ESCC cell lines. OBP-801 and 5-fluorouracil resistance was attenuated after ΔNp63 knockdown in a specific condition. a RT-PCR confirmed suppressed expression of the ΔNp63 gene at mRNA level in OBP-801-resistant ESCC cell lines (TE6, TE10, and TE11). b In the context of ΔNp63 gene knockdown, OBP-801 drug resistance was significantly attenuated in 10 nM concentrations in TE6 and TE11, and was significantly attenuated in 20 nM concentrations in TE10. In the context of ΔNp63 gene knockdown, 5-fluorouracil drug resistance was significantly attenuated in 5 μM concentrations in TE10 and TE11, and was significantly attenuated in 10 μM concentrations in TE6 and TE10. Error bar represent the standard error. Experiments were performed in triplicate. ESCC esophageal squamous cell carcinoma, RT-PCR reverse transcriptase–polymerase chain reaction, mRNA messenger RNA, NS non-significant
Clinical Relevance of Genomic Amplification of both ΔNp63 and SOX2 Genes in the Resected Primary ESCC Tissues
Next, we investigated the clinical significance of genomic amplification of both ΔNp63 and SOX2 in the resected primary ESCC tissues (n =112). Interestingly, both genomic statuses were tightly associated with each other (p <0.0001), and the correlation coefficient was as much as 0.81, even in clinical samples (Fig. 5a).
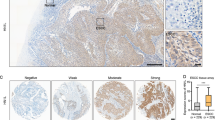
Gene amplification and immunohistochemistry of both p63 and SOX2 in ESCC. a Scatter diagram of SOX2 2−ΔΔCt and ΔNp63 2−ΔΔCt in primary ESCC tissues compared with the corresponding non-cancerous mucosa. There is a strong correlation between SOX2 2−ΔΔCt and ΔNp63 2−ΔΔCt. b IHC staining score was classified into IHC 1+ (left panels), IHC 2+ (middle panels), and IHC 3+ (right panels) for both p63 (upper panels) and SOX2 (lower panels). The amplification was 400 × magnification. c The correlation between IHC staining and genomic amplification of individual genes; gene amplification is significantly associated with strong IHC staining in both SOX2 (p =0.03) and ΔNp63 (p <0.01). ESCC esophageal squamous cell carcinoma, IHC immunohistochemical
Both gains of ΔNp63 and SOX2 were significantly associated with advanced pathological T factor (p =0.03), and cases with genomic gains therefore included those with neoadjuvant therapy (12/32, 37.5%; p =0.0003) (Table 1). Among the 32 cases with neoadjuvant therapy, both gains were significantly associated with histological grade after preoperative chemo(radio)therapy (p =0.03). The 12 cases with genomic gains of both ΔNp63/SOX2 showed histological response of G1. Conversely, seven cases with G2/G3 were all defective for genomic gains of ΔNp63/SOX2 in ESCC with neoadjuvant chemotherapy (Table 2).
Correlation Between Immunohistochemical Staining to Genomic Amplification
We investigated the immunohistochemical (IHC) staining of p63 for 15 cases with high ΔNp63 genomic gain (2−ΔΔCt ≧ 2.0) and 15 cases with low genomic gain (2−ΔΔCt < 1.0), and classified IHC 1+, 2+, and 3+. The same experiment was performed in SOX2 (Fig. 5b). Positive correlations were confirmed between genomic gains and IHC score in both p63 and SOX2 (Fig. 5c).
Discussion
We aimed to discover the genes associated with expression in OBP-801 drug resistance. Interestingly, OBP-801 drug sensitivity partially mimics that of 5-fluorouracil, suggesting that the drug resistance of OBP-801 is a good model for identifying genes to predict anticancer drugs, as in our previous reports.11,23
OBP-801 is so potent that a 10 nM concentration is enough to induce remarkable suppression of cell proliferation in all types of human cancer cells. Thus, OBP-801 could be considered a promising anticancer drug. Intriguingly though, it is very sensitive to gastric, pancreatic, and thyroid (including undifferentiated) cancers, which are well known as highly aggressive tumors, for which there are few therapeutic options at present.
Among all the genomes, ΔNp63/SOX2 were finally selected as potential candidate genes involved in OBP-801 drug resistance, and were closely positioned at the 3q chromosome. Furthermore, concurrent genomic amplification was recently reported in the CGAR Network in primary ESCC.17ΔNp63/SOX2 genomic amplification is not restricted to ESCC, which is a common characteristic of SCCs, such as head and neck SCC24 and lung SCC.25 It has also been reported that ΔNp63/SOX2 represent cancer stem cell signatures of SCC cells,18,19,20,21,–22 suggesting they may have been involved in chemotherapeutic resistant of SCC. For the first time, we demonstrated that ΔNp63/SOX2 genomic amplification status is significantly correlated with chemoresistance of neoadjuvant chemotherapy in primary ESCC patients who underwent esophagectomy.26 Our functional experiments support these results.
More intriguingly, both genes were robustly reduced in expression by the pharmacological unmasking of epigenetic treatments of concurrent 5-aza-2′-deoxycytidin with TSA that could reactivate the epigenetically suppressed genes.27,28 Importantly, these two genes are controlled at mRNA level by multiple discrete and independent mechanisms, such as genomic amplification and epigenetic regulation, indicating the important role of both genes in cancer transformation.
Genomic amplification of ΔNp63 and SOX2 have strong positive correlation, indicating that the synergistic effect of the overexpression of both genes is expected. However, our transient transfection experiments could not necessarily putatively demonstrate the synergy due to the low efficiency of the double transfection in a transient manner. In the near future, stable double transfectants could demonstrate the hypothesis in this context.
Data from in vitro experiments have provided evidence that the TA isoforms of TP63 are tumor suppressor genes, and that the ΔN isoforms of TP63 are oncogenes, which antagonize p53, TAp63 and TAp73 by inhibiting their ability to transactivate downstream target genes. The TAp63γ and TAp73β isoforms, which most closely resemble p53, are the most transcriptionally active isoforms to induce apoptosis.29 Further in vivo work has recently demonstrated that the TAp63 and TAp73 isoforms are likely to be tumor suppressors in the context of p53 research.30 All the results of our functional experiments also supported the hypothesis that ΔNp63 plays an oncogenic role in ESCC transformation, and a recent study clarified its mechanism through TGF-β.31
On the other hand, the oncogenic potential of SOX2 in our experiment is modest in contrast to ΔNp63 in ESCC, while SOX2 significantly contributed to the drug resistance of both OBP-801 and 5-fluorouracil in ESCC. SOX2 is a stem cell marker of epithelial cells, and required iPS cell induction,32 suggesting that it is involved in maintaining undifferentiation of the cells. SOX2-positive stem/progenitor cells also have tumor-inducing potential and increase the malignant stemness properties,20 suggesting that it can contribute to anticancer drug sensitivity. Among the three OBP-801-resistant cells, TE11 only expressed ΔNp63, not SOX2. Knockdown of ΔNp63 in TE11 robustly induced the weakest cell viability among the three cell lines in which the other two cells (TE6, TE10) highly expressed SOX2, indicating that SOX2 may have alternate stem cell properties to ΔNp63 with regard to drug resistance in TE6 and TE10.
Conclusions
We explored candidate genes relating to OBP-801 resistance in ESCC cell lines and discovered ΔNp63 and SOX2, which were both genetically and epigenetically overexpressed in ESCC. Forced expression of either gene showed robust drug resistance in both OBP-801 and 5-fluorouracil. Furthermore, ΔNp63 could exert a potent oncogenic potential. Genomic amplification of ΔNp63/SOX2 in the resected tumors after neoadjuvant chemotherapy was associated with histological grade of response.
According to the above results, ΔNp63/SOX2 likely have critical oncogenic properties, including anticancer drug sensitivity in ESCC.
References
Shindoh N, Mori M, Terada Y, Oda K, et al. YM753, a novel histone deacetylase inhibitor, exhibits antitumor activity with selective, sustained accumulation of acetylated histones in tumors in the WiDr xenograft model. Int J Oncol. 2008;32(3):545–55.
Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37(4):391–400.
Huang BH, Laban M, Leung CH, Lee L, et al. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differ. 2005;12(4):395–04.
Zhu P, Martin E, Mengwasser J, Schlag P, Janssen KP, Göttlicher M. Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell. 2004;5(5):455–63.
Wilson AJ, Byun DS, Popova N, Murray LB, et al. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and arederegulated in human colon cancer. J Biol Chem. 2006;281(19):13548–58.
Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov. 2002;1(4):287–99.
Minucci S, Pelicci P. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6(1):38–51.
Kelly WK, O’Connor OA, Krug LM, Chiao JH, et al. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J Clin Oncol 2005;23(17):3923–31.
O’Connor OA, Heaney ML, Schwartz L, Richardson S, et al. Clinical experience with intravenous and oral formulations of the novel histone deacetylase inhibitor suberoylanilide hydroxamic acid in patients with advanced hematologic malignancies. J Clin Oncol. 2006;24(1):166–73.
Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90.
Kikuchi M, Yamashita K, Waraya M, Minatani N et al. Epigenetic regulation of ZEB1-RAB25/ESRP1 axis plays a critical role in phenylbutyrate treatment-resistant breast cancer. Oncotarget. 2016;7(2):1741–53.
Ooki A, Yamashita K, Kikuchi S, Sakuramoto S, Katada N, Watanabe M. Phosphatase of regenerating liver-3 as a convergent therapeutic target for lymph node metastasis in esophageal squamous cell carcinoma. Int J Cancer. 2010;127(3):543–54.
Ooki A, Yamashita K, Kikuchi S, et al. Therapeutic potential of PRL-3 targeting and clinical significance of PRL-3 genomic amplification in gastric cancer. BMC Cancer. 2011;11:122.
Yokoi K, Yamashita K, Ishii S, Tanaka T, et al. Comprehensive molecular exploration identified promoter DNA methylation of the CRBP1 gene as a determinant of radiation sensitivity in rectal cancer. Br J Cancer. 2017;116(8):1046–56.
Waraya M, Yamashita K, Katoh H, et al. Cancer specific promoter CpG Islands hypermethylation of HOP homeobox (HOPX) gene and its potential tumor suppressive role in pancreatic carcinogenesis. BMC Cancer. 2012;12:397.
Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131(1):18–43.
Cancer Genome Atlas Research Network, Analysis Working Group, Asan University, BC Cancer Agency, Brigham and Women’s Hospital, Broad Institute, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–75.
Bass AJ, Watanabe H, Mermel CH, Yu S, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41(11):1238–42.
Watanabe H, Ma Q, Peng S, Adelmant G, et al. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J Clin Invest. 2014;124(4):1636–45.
Boumahdi S, Driessens G, Lapouge G, Rorive S, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511(7508):246–50.
Siegle JM, Basin A, Sastre-Perona A, Yonekubo Y, et al. SOX2 is a cancer-specific regulator of tumour initiating potential in cutaneous squamous cell carcinoma. Nat Commun. 2014;31(5):4511.
Huang H, Zhang W, Pan Y, Gao Y, et al. YAP Suppresses lung squamous cell carcinoma progression via deregulation of the DNp63-GPX2 axis and ROS accumulation. Cancer Res. 2017;77(21):5769–81.
Tanaka T, Kojima K, Yokota K, et al. Comprehensive genetic search to clarify the molecular mechanism of drug resistance identifies ASCL2-LEF1/TSPAN8 axis in colorectal cancer. Ann Surg Oncol. 2019;26(5):1401–11.
Hibi K, Trink B, Patturajan M, et al. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci USA. 2000;97(10):5462–7.
Campbell JD, Yau C, Bowlby R, Liu Y, et al. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep. 2018;23(1):194–212.
Yamashita K, Hosoda K, Moriya H, Katada C, et al. Prognostic advantage of docetaxel/cisplatin/5-fluorouracil neoadjuvant chemotherapy in clinical stage II/III esophageal squamous cell carcinoma due to excellent control of preoperative disease and postoperative lymph node recurrence. Oncology. 2017;92(4):221–8.
Yamashita K, Upadhyay S, Osada M, Hoque MO, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2(6):485–95.
Ishii S, Yamashita K, Harada H, et al. The H19-PEG10/IGF2BP3 axis promotes gastric cancer progression in patients with high lymph node ratios. Oncotarget. 2017;8(43):74567–81.
Osada M, Park H, Nagakawa Y, Yamashita K, et al. Differential recognition of response elements determines target gene specificity for p53 and p63. Mol Cell Biol. 2005;25(14):6077–89.
Tanaka T, Watanabe M, Yamashita K. Potential therapeutic targets of TP53 gene in the context of its classically canonical functions and its latest non-canonical functions in human cancer. Oncotarget. 2018;9(22):16234–47.
Abraham CG, Ludwig MP, Andrysik Z, et al. DeltaNp63alpha suppresses TGFB2 expression and RHOA activity to drive cell proliferation in squamous cell carcinomas. Cell Rep. 2018;24(12):3224–36.
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosures
This work received a research grant from Oncolys BioPharma Inc., Tokyo, Japan.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10434_2019_7795_MOESM3_ESM.pptx
Supplementary Fig. S1 OBP-801 sensitivity according to four types of cancer cells. Viable cancer cell counts s after 72 h of OBP-801 administration (5–100 nM) compared with control cells. Error bar represents the standard error. Experiments were performed for a single time only (PPTX 72 kb)
10434_2019_7795_MOESM4_ESM.pptx
Supplementary Fig. S2 Identification and confirmation of expression profiles of OBP-801 resistance-related and sensitivity-related genes. (a) Schematic representations of p63 structures and primer designs (blue: TAp63-specific; red: ΔNp63-specific; and black: common p63). Gene expression was also shown for both TAp63-specific and ΔNp63-specific PCR primers. (b) OBP-801 did not alter expressions of ΔNp63 and SOX2 in esophageal squamous cell carcinoma cell lines (PPTX 480 kb)
Rights and permissions
About this article
Cite this article
Ooizumi, Y., Kojima, K., Igarashi, K. et al. Comprehensive Exploration to Identify Predictive DNA Markers of ΔNp63/SOX2 in Drug Resistance in Human Esophageal Squamous Cell Carcinoma. Ann Surg Oncol 26, 4814–4825 (2019). https://doi.org/10.1245/s10434-019-07795-w
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-019-07795-w