Abstract
Background
Colorectal cancer (CRC) is a heterogeneous disease caused by genetic and epigenetic alterations. This study aimed to describe the mutation frequency of 12 genes in different CRC phenotypes.
Methods
Patients who underwent surgery at the Taipei Veterans General Hospital during 2000–2010 for CRC (n = 1249) were enrolled. The endpoint was overall survival. The prognostic value was determined with the log-rank test and Cox regression analysis.
Results
We found 1836 mutations of 12 genes in 997 (79.8 %) tumors. Mutations were most frequently in KRAS (485, 38.8 %), TP53 (373, 29.9 %), APC (363, 29.0 %), and PIK3CA (179, 14.3 %); 137 (11.0 %) cancers had high microsatellite instability (MSI). Women had significantly higher high MSI (14.3 %) and BRAF mutation (6.3 %) frequencies. The abnormal MSI (21.7 %) and KRAS (44.6 %), BRAF (8.6 %), PIK3CA (19.4 %), AKT1 (2.2 %), and TGF - βR (9.6 %) mutation frequencies were significantly higher in proximal colon cancer. The high MSI (35.6 %) and BRAF (20.3 %), TGF - βR (18.6 %), PTEN (5.1 %), and AKT1 (3.4 %) mutation frequencies were significantly higher in 59 (4.7 %) poorly differentiated tumors. The high MSI (21.3 %) and KRAS (51.9 %), BRAF (8.3 %), PIK3CA (25.0 %), AKT1 (4.6 %), and SMAD4 (8.3 %) mutation frequencies were significantly higher in 108 mucinous tumors. TNM stage, lymphovascular invasion, and mucinous histology were significantly associated with patient outcomes in univariate and multivariate analyses. Only NRAS mutation (hazard ratio 1.59, 95 % confidence interval 1.06–2.38) affected patient survival.
Conclusions
Mutational spectra differ significantly between CRC subtypes, implying diverse carcinogenetic pathways. The NRAS mutation is important, despite its low frequency.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Colorectal cancer (CRC) is the most common malignancy and the third leading cause of death in Taiwan.1 Several elegant models revealed that CRC is a biologically heterogeneous disease that develops from the progressive accumulation of genetic and epigenetic alterations, leading to the transformation of the normal colonic epithelium into colon adenocarcinoma.2–4 One of the hallmarks of colorectal cancer is chromosomal instability, caused by alterations in tumor suppressor genes and oncogenes, including APC, TP53, and KRAS.3,4 In addition, 10–15 % of all cases of CRC show microsatellite instability (MSI), which is caused by a germline mutation in the mismatch repair system, or somatic hypermethylation of the MLH1 gene promoter.5,6
With the recent progress in DNA sequencing technologies, several genome-wide analyses of CRCs have identified acquired somatic mutations in several hundred genes, with an average of 80 mutations in any single patient with CRC.7–11 However, only a few of these mutations are able to initiate or promote tumorigenesis, acting as “driver” mutators.7–9 The remaining thousands of mutations are considered to occur coincidentally and do not confer a selective growth advantage during the large number of cell divisions associated with the neoplastic process.10 Based on the findings derived from these studies, alterations in WNT, mitogen-activated protein kinase (MAPK), phosphoinositol-3-kinase (PI3K), transforming growth factor beta (TGF-β), TP53, and MSI pathways have been considered to be important contributors to CRC development.7,12–15 Analyzing the association between tumor phenotypes, including different clinicopathological features and molecular alterations, could increase our knowledge of CRC carcinogenesis and has the potential to improve the prognostic evaluation of CRC patients.12,13 Therefore, in this study, we determined the MSI and mutation status of 12 genes within the WNT, MAPK, PI3K, and TGF-β pathways.
Materials and Methods
Clinical Data
We enrolled 1249 colorectal cancer patients who underwent surgery at the Taipei Veterans General Hospital from 2000 to 2010. Exclusion criteria were preoperative radiochemotherapy, emergency operations, or death within 30 days of surgery. The clinical information that was prospectively obtained and stored in a database included age, sex, personal and family medical history, location of tumor, TNM stage, differentiation, pathological prognostic features, and follow-up conditions. The proximal colon was defined as from the cecum to the splenic flexure colon. The distal colon was from the splenic flexure to the rectosigmoid colon. The rectum was within 15 cm of the anal verge.
Following surgery, patients were monitored every 3 months for the first 2 years and semiannually thereafter. The follow-up protocol included physical examination, digital rectal examination, carcinoembryonic antigen analysis, chest radiography, abdominal sonogram, and computerized tomography, if needed. Proton emission tomography or magnetic resonance imaging was arranged for patients with an elevated carcinoembryonic antigen level but an uncertain site of tumor recurrence.
Tumor Tissues
Before sample collection, written, informed consent for tissue collection was obtained from all patients. Samples were meticulously dissected and collected from different quadrants of the tumors, immediately frozen in liquid nitrogen, and stored in the Taipei Veterans General Hospital Biobank.
DNA Isolation and Quantification
After approval by the Institutional Review Board of the Taipei Veterans General Hospital (Number 2013-04-042B), the samples for this study were obtained from the Biobank. DNA was extracted using the QIAamp DNA Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s recommendations. The quality and quantity of DNA were confirmed using the Nanodrop 1000 Spectrophotometer (Thermo Scientific).
Mas ARRAY-Based Mutation Characterization
The MassDetect CRC panel (v2.0), enabling the identification of 139 mutations in 12 genes (Supplementary Table 1), was selected from hotspots found in previous studies and the COSMIC database.15,16 The polymerase chain reaction (PCR) and extension primers for the mutations were designed using the MassArray Assay Design 3.1 software (Sequenom, San Diego, CA). The details of the MassARRAY-based mutation detection methods are described in Supplementary Table 1. The PCR products of the multiplexed reactions were spotted onto SpectroCHIP II arrays, and the DNA fragments were resolved on the MassARRAY Analyzer 4 System (Sequenom). Each spectrum was then analyzed using the Typer 4.0 software (Sequenom) to identify mutations. According to a previous study, the sensitivity of mutation detection is 1–10 % for the Sequenom assays, depending on the specific mutation.17 In this study, we defined a 5 % abnormal signal as a putative mutation. Putative mutations were then filtered by manual review. Any detected mutations in BRAF, PTEN, NRAS, and KRAS were confirmed by Sanger sequencing. The concordance of these two methods was 99.1 %.
MSI Analysis
Five reference microsatellite markers were used according to the international criteria for determination of MSI: D5S345, D2S123, BAT25, BAT26, and D17S250. The primer sequences were obtained from GenBank (www.gdb.org). The detection of MSI was performed as previously described.18 Samples with ≥2 MSI markers were defined as having MSI, and those with 0–1 MSI markers were classified as microsatellite stable.
Statistical Analysis
The statistical endpoint was overall survival (OS) measured from the date of surgery. Patients not known to have died were censored at the date of last follow-up. Kaplan–Meier survival curves were plotted and compared using the log-rank test. The impact of clinicopathological features and genotype mutations on OS were assessed using Cox regression univariate and multivariate analyses. The Chi square test and two-tailed Fisher’s exact procedure were used to compare the genotype frequency by clinicopathological features. The numerical values were compared using the Student’s t test. Data were expressed as mean ± standard deviation. Statistical significance was defined as p < 0.05. Statistical analyses were performed using SPSS for Windows (version 16.0).
Results
The patient population was composed of 822 men (65.8 %) and 427 women (34.2 %). The demographic features and mutational status are shown in Tables 1 and 2. The mean age at the time of tumor resection was 70.3 ± 11.4 years (range 27–108 years; median 72 years). There were 314 (25.1 %) proximal colon, 461 (36.9 %) distal colon, and 474 (38.0 %) rectal cancers. The histology showed poorly differentiated cancer, mucinous type, and lymphovascular invasion in 59 (4.7 %), 108 (8.6 %), and 175 (14.0 %) patients, respectively. The distribution of tumor staging (TNM) was stage I in 212 patients (17 %), stage II in 564 (45.2 %), and stage III in 473 (37.9 %).
We found 1836 mutations in 12 genes in 997 (79.8 %) of the 1249 tumors analyzed. The number of mutations per tumor ranged between 0 and 7. A total of 437 cases (35 %) had one mutation. The occurrence of two or three simultaneous mutations were found in 348 (27.9 %) and 164 (13.1 %) patients, respectively. Detailed information is shown in Supplementary Table 1. The most frequently mutated gene was KRAS (mutated in 485 cases, 38.8 %), followed by TP53 (in 373, 29.9 %), APC (in 363, 29.0 %), and PIK3CA (in 179, 14.3 %), and 137 (11.0 %) patients had cancers with high MSI. The detailed information on the mutations in individual genes is described in Supplementary Table 1.
Correlation Between Molecular Alterations and the Clinical Features
According to the World Health Organization classification, age 65 years is the cutoff between the old-age and the young-age groups (Table 1). The distributions of molecular alterations in different genes were similar between the old-age and the young-age patients. Women had a significantly higher frequency of high MSI (14.3 %) and BRAF mutations (6.3 %) than men (9.2 %, p = 0.009; 2.9 %, p = 0.006). The frequency of NRAS mutations was higher in men (5.2 %) than in women (2.6 %, p = 0.041). The patterns of molecular alterations were significantly different according to the tumor location. Alterations in MSI (21.7 %) and mutations in KRAS (44.6 %), BRAF (8.6 %), PIK3CA (19.4 %), AKT1 (2.2 %), and TGF-βR (9.6 %) were significantly more frequent in proximal colon cancer than in distal colon and rectal cancer. The frequency of TP53 mutations was significantly higher in distal (30.6 %) and rectal cancer (32.1 %) than in proximal colon cancer (25.5 %, p = 0.049).
Correlation Between Molecular Alterations and Pathological Features
There was a linear association between TP53 mutation frequency and tumor stages. TP53 was mutated in 23.6 % of cases in stage I disease, and the rate significantly increased to 30.3 and 32.1 % in stage II and III disease, respectively (p = 0.036). The distribution of other mutations was similar in all disease stages (Table 2).
In 59 (4.7 %) poorly differentiated tumors, there was a significantly higher frequency of high MSI (35.6 %) and mutations in BRAF (20.3 %), TGF -βR (18.6 %), PTEN (5.1 %), and AKT1 (3.4 %) than in the well to moderately differentiated ones (3.3, 3.0, 0.8, and 0.8 %, respectively). The frequency of APC mutation was significantly lower in poorly differentiated tumors (11.9 %) than in well to moderately differentiated ones (29.9 %, p = 0.005).
The frequency of high MSI (21.3 %) and of mutations in KRAS (51.9 %), BRAF (8.3 %), PIK3CA (25.0 %), AKT1 (4.6 %), and SMAD4 (8.3 %) was significantly higher in the 108 tumors with mucinous histology than in those without a mucinous component (10, 37.6, 3.7, 13.3, 0.6, and 3.6 %, respectively). The frequency of APC mutations was significantly lower in tumors with mucinous histology (16.7 %) than in those without mucinous histology (30.2 %, p = 0.004).
Metastasis and Outcome
During a follow-up period of 6–160 (median 62) months, 442 patients died of the disease or from other causes; 236 patients had tumor recurrence or metastases, including 95 liver, 96 lung, 15 bone, 45 peritoneal metastases, 51 among others. In the patients with lung metastasis, there was a significantly higher frequency of NRAS mutation (11.6 %) than in those without lung metastasis (3.7 %, p = 0.002). Mutation of other genes was not associated with metastasis sites.
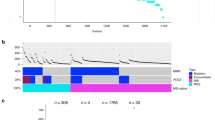
Table 3 shows the final analysis of OS in the 1249 CRC patients after a stepwise elimination process: TNM stage, status of lymphovascular invasion, and mucinous histology were significantly associated with patient outcomes in the univariate and multivariate analyses. Beside NRAS (hazard ratio 1.59, 95 % confidence interval 1.06–2.38; Fig. 1), the mutation status of individual genes was not associated with patient survival. Patients with tumors with high MSI or a mutation in TGF -βR had better outcomes than those of patients with microsatellite stable tumors or tumors without a TGF -βR mutation; however, the difference was not statistically significant (Supplementary Table 2).
Discussion
This large-scale molecular analysis, including 1249 CRC patients, provided three major contributions to the present knowledge. First, the mutation patterns were significantly different between tumors arising at different locations. Second, mutations in specific genes were associated with tumor differentiation. Third, TP53 and NRAS mutations were associated with tumor stage and prognosis.
Similar to a previous study, our results showed mutation patterns were significantly different between MSS and MSI-high tumors.15 T P53 (31 vs. 20.4 %, p = 0.01) was frequently mutated in MSS tumors but less so in MSI-high tumors. BRAF PTEN AKT1, and TGF -BR were recurrently mutated in the MSI-high cancers but not in the MSS samples. BRAF V600E, associated with CIMP, could be an initiator of the MSI pathway by inducing MLH1 hypermethylation. The majority of mutations in PTEN AKT1, and TGF -BR are located in tandem repeated sequences that are easily mutated in MMR deficiency tumors.
As other studies previously described, our series showed that right-sided colon cancer had a higher frequency of high MSI and mutations in the PI3K, MAPK, and TGF-β pathways, but lower mutation frequencies in the APC/P53 pathway.19–22 Left-sided colon cancer and rectum cancer shared a similar distribution of molecular alterations. We considered the possibility that the mutation differences by tumor locations were due to MSI. As shown in supplementary Table 3, in MSS tumors, the frequency of KRAS and PIK3CA in the proximal colon was significantly higher than in the distal colon and rectum. Even in MSI-high tumors, TGF-βR and BRAF had significantly higher mutation rates in the proximal colon. These results implied that both MSI (acquired or hereditary) and the microenvironment, such as microbiota differences, between colon and rectum might trigger a different carcinogenesis process.23–25
Molecular alterations in PI3K, MAPK, TGF-β, or MSI were associated with progression and prognosis of colorectal cancer, especially with the level of differentiation. Our series showed that 70.7 % of poorly differentiated tumors and 78.7 % of tumors with mucinous histology had either high MSI or at least one mutation in KRAS, BRAF, PIK3CA, PTEN, AKT1, and TGF -βR. On the other hand, the frequency of APC/p53 mutations was low in poorly differentiated or mucinous tumors. An in vitro study showed that a constitutively active PI3K as a consequence of a PIK3CA mutation could induce mucinous differentiation of colon cancer independent of aberrant WNT signaling.26 In an in vivo model, inactivation of TGF-β signaling together with loss of PTEN could induce colon cancer, and the majority of such tumors were mucinous adenocarcinomas with transmural effacement by neoplastic glands and large lakes of tumor-produced mucin.27 The inducible transgenic mice model suggested that the activation of the BRAF/MAPK signaling axis can affect cellular hierarchies and thus modulate cellular differentiation during intestinal tumor initiation and progression.28 After elimination of the MSI effect on tumor differentiation (Supplementary Table 3), we observed that MSS tumors with poor differentiation had higher mutation rates in BRAF and PTEN but lower APC mutation rates. In addition, MSS tumors with mucinous histology had recurrent mutations in KRAS, PIK3CA, AKT1, and SMAD4. These results suggest that PI3K, MAPK, or TGF-β alterations play a significant role in tumor behaviors without MSI influence.
In this large-scale molecular study, only NRAS affected patient outcomes significantly in univariate and multivariate analysis. The association between NRAS mutations and tumor staging was not very significant, because it occurred in a small percentage of patients (<5 %). It is of note that NRAS mutations seem to occur quite late in colon cancer progression, similar to mutations in TP53.4 We also noted that only TP53 had a linear association with tumor stage, varying from 23.6 % in stage I disease to 32.1 % in stage III disease. Our results suggest that mutated NRAS and KRAS may have different impacts on tumor behavior even if they are highly related oncogenes. The functional difference could be dissected by a gene-targeting mice model showing that activated KRAS mutation affects proliferation and differentiation, whereas activated NRAS mutation suppresses apoptosis.29
The prognostic impact of these molecular alterations described in our series had some inconsistency compared with other studies, which could be explained by the heterogeneity of colon cancer, the complex interactions between these alterations, and how many variables were analyzed.30–34 Furthermore, complex molecular subtypes, for example those defined by MSI and BRAF, but not alterations in single genes, can predict patient outcomes more reliably.14,35
Because we selected only the most common mutation sites, coverage was limited especially in tumor suppressor genes. Compared with the COSMIC database (as shown in the Supplementary Figs. S1, S2), the coverage in our study was approximately 96–99 % in oncogenes, including KRAS and BRAF. For APC and TP53, the coverage was 70–80 %. The mutation spectra of PTEN are widely scattered, and our coverage was only 25.3 %. Compared with other studies using methods similar to ours, our series had a slightly lower detection rate of mutations than sequencing methods.36,37
In this study, we demonstrated a rapid tumor genotyping system to detect MSI and mutations in several genes. Mutational spectra between proximal colon cancer and rectal cancer were significantly different and might have originated from these tumors arising from different microenvironments. Mutations or alterations in MAPK, PI3K, TGF-β signaling, and MSI might affect the differentiation of colorectal cancer. The importance of NRAS needs to be further addressed.
References
The Department of Health. Healthy statistics: Cancer Registry Annual Report in Taiwan Area. Taiwan, R.O.C.: The Department of Health, the Executive Yuan, 2007.
Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135(4):1079–99.
Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–67.
Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319(9):525–32.
Cunningham JM, Kim CY, Christensen ER, et al. The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet. 2001;69(4):780–90.
Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361(25):2449–60.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58.
Garraway LA, Lander ES. Lessons from the Cancer Genome. Cell. 2013;153(1):17–37.
Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–24.
Tomasetti C, Vogelstein B, Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc Natl Acad Sci USA. 2013;110(6):1999–2004.
Casey G, Conti D, Haile R, Duggan D. Next generation sequencing and a new era of medicine. Gut. 2013;62(6):920–32.
Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50(1):113–30.
Sinicrope FA, Sargent DJ. Molecular pathways: microsatellite instability in colorectal cancer: prognostic, predictive, and therapeutic implications. Clin Cancer Res. 2012;18(6):1506–12.
Lochhead P, Kuchiba A, Imamura Y, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105(15):1151–6.
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7.
Lin JK, Lin PC, Lin CH, et al. Clinical relevance of alterations in quantity and quality of plasma DNA in colorectal cancer patients: based on the mutation spectra detected in primary tumors. Ann Surg Oncol. 2014;21(Suppl 4):680–6.
Arcila M, Lau C, Nafa K, Ladanyi M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011;13(1):64–73.
Chang SC, Lin JK, Yang SH, Wang HS, Li AF, Chi CW. Relationship between genetic alterations and prognosis in sporadic colorectal cancer. Int J Cancer. 2006;118(7):1721–7.
Missiaglia E, Jacobs B, D’Ario G, et al. Distal and proximal colon cancers differ in terms of molecular, pathological, and clinical features. Ann Oncol. 2014;25(10):1995–2001.
Shen Y, Wang J, Han X, et al. Effectors of epidermal growth factor receptor pathway: the genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PLoS One. 2013;8(12):e81628.
Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61(6):847–54.
Albuquerque C, Baltazar C, Filipe B, et al. Colorectal cancers show distinct mutation spectra in members of the canonical WNT signaling pathway according to their anatomical location and type of genetic instability. Genes Chromosomes Cancer. 2010;49(8):746–59.
Dejea CM, Wick EC, Hechenbleikner EM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci USA. 2014;111(51):18321–6.
Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota dysbiosis is associated with colorectal cancer. Front Microbiol. 2015;6:20.
Tahara T, Yamamoto E, Suzuki H, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74(5):1311–8.
Leystra AA, Deming DA, Zahm CD, et al. Mice expressing activated PI3K rapidly develop advanced colon cancer. Cancer Res. 2012;72(12):2931–6.
Yu M, Trobridge P, Wang Y, et al. Inactivation of TGF-beta signaling and loss of PTEN cooperate to induce colon cancer in vivo. Oncogene. 2014;33(12):1538–47.
Riemer P, Sreekumar A, Reinke S, et al. Transgenic expression of oncogenic BRAF induces loss of stem cells in the mouse intestine, which is antagonized by Beta-catenin activity. Oncogene. 2015;34(24):3164–75.
Haigis KM, Kendall KR, Wang Y, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40(5):600–8.
Ogino S, Liao X, Imamura Y, et al. Predictive and prognostic analysis of PIK3CA mutation in stage III colon cancer intergroup trial. J Natl Cancer Inst. 2013;105(23):1789–98.
Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367(17):1596–606.
Mouradov D, Domingo E, Gibbs P, et al. Survival in stage II/III colorectal cancer is independently predicted by chromosomal and microsatellite instability, but not by specific driver mutations. Am J Gastroenterol. 2013;108(11):1785–93.
Warren RS, Atreya CE, Niedzwiecki D, et al. Association of TP53 mutational status and gender with survival after adjuvant treatment for stage III colon cancer: results of CALGB 89803. Clin Cancer Res. 2013;19(20):5777–87.
Yu J, Wu WK, Li X, et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut. 2015;64(4):636–45.
Phipps AI, Limburg PJ, Baron JA, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148(1):77–87.e2.
Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2(5):146–58.
Russo AL, Borger DR, Szymonifka J, et al. Mutational analysis and clinical correlation of metastatic colorectal cancer. Cancer. 2014;120(10):1482–90.
Acknowledgment
This research was funded by grants from the Taipei Veterans General Hospital (V101E2-005) and the Ministry of Science and Technology, Taiwan (103-2314-B-075-041). Department of Healthy, Taipei City Government (10301-62-058; 10401-62-031).
Author information
Authors and Affiliations
Corresponding author
Additional information
Shih-Ching Chang and Pei-Ching Lin have contributed equally to the manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chang, SC., Lin, PC., Lin, JK. et al. Mutation Spectra of Common Cancer-Associated Genes in Different Phenotypes of Colorectal Carcinoma Without Distant Metastasis. Ann Surg Oncol 23, 849–855 (2016). https://doi.org/10.1245/s10434-015-4899-z
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-015-4899-z