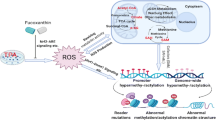
Abstract
Overexposure to ultraviolet radiation and environmental carcinogens drive skin cancer development through redox imbalance and gene mutation. Antioxidants such as triterpenoids have exhibited anti-oxidative and anti-inflammatory potentials to alleviate skin carcinogenesis. This study investigated the methylome and transcriptome altered by tumor promoter 12-O-tetradecanoylphorbol-13-acetate (TPA) or TPA with 2-cyano 2,3-dioxoolean-1,9-dien-28-oic acid (CDDO). The results show that CDDO blocks TPA-induced transformation dose dependently. Several differential expressed genes (DEGs) involved in skin cell transformation, while counteracted by CDDO, were revealed by differential expression analysis including Lyl1, Lad1, and Dennd2d. In CpG methylomic profiles, the differentially methylated regions (DMRs) in the promoter region altered by TPA while showing the opposite methylation status in the CDDO treatment group were identified. The correlation between DNA methylation and RNA expression has been established and DMRs showing inverse correlation were further studied as potential therapeutic targets. From the CpG methylome and transcriptome results, CDDO significantly restored gene expression of NAD(P)H:quinone oxidoreductase 1 (Nqo1) inhibited by TPA by decreasing their promoter CpG methylation. Ingenuity Pathways Analysis (IPA) shows that CDDO neutralized the effect of TPA through modulating cell cycles, cell migration, and inflammatory and immune response regulatory pathways. Notably, Tumor Necrosis Factor Receptor 2 (TNFR2) signaling was significantly downregulated by CDDO potentially contributing to prevention of TPA-induced cell transformation. Overall, incorporating the transcriptome, CpG methylome, and signaling pathway network, we reveal potential therapeutic targets and pathways by which CDDO could reverse TPA-induced carcinogenesis. The results could be useful for future human study and targets development for skin cancer.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Non-melanoma skin cancer (NMSC), comprising basal cell carcinoma, cutaneous squamous cell carcinoma, and Merkel cell carcinoma, is the most prevalent cancer in the USA, and its new case number surpasses the combination of all the other human malignancies (1). The most recent estimated annual new cases of NMSC increased up to 5.4 million in the USA (2). The increased incidence rate of NMSC worldwide is highly relevant to cumulative exposure to UV irradiations and chemical carcinogens (3,4,5). Chemical-induced skin tumorigenesis in mouse skin or mouse skin cells has been widely employed to understand the mechanisms of skin carcinogenesis and evaluate potential interventions (6). 12-O-tetradecanoylphorbol-13-acetate (TPA) is a frequently used skin tumor promoter that exerts its effect by enhancing the production of reactive oxygen species (ROS) and pro-inflammatory cytokines both in vitro and in vivo (6, 7).
Over the last few decades, epigenetic reprogramming has been considered a hallmark of cancer. Growing evidence suggest that DNA methylation leading to the addition of a methyl group to cytosine nucleotide adjacent to guanine base in CpG island is one of the best-described epigenetic modifications in various cancers including melanoma and NMSC (8, 9). Naturally abundant phytochemicals have received considerable attention due to their chemopreventive ability through the alteration of diverse epigenetic processes such as DNA CpG methylation (10). Triterpenoids are naturally occurring compounds synthesized in plants, fungi, or marine organisms through cyclization of squalene and possess antioxidant, anti-inflammatory, and anticancer activities (11, 12). Our previous studies showed that various triterpenoids including corosolic acid and ursolic acid exert skin cancer protective effects through epigenetic regulations (13,14,15).
CDDO (2-cyano 2,3-dioxoolean-1,9-dien-28-oic acid, also known as bardoxolone), a synthetic derivative of oleanolic acid, was found to be one of the most potent anti-inflammatory and anti-carcinogenic triterpenoids with activities found at concentrations below 1 nM (16). A previous review has shown that CDDO and its C28 modified derivative, CDDO-Me (2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid methyl ester), exert their anti-cancer effect through multifunctional pathways in various cancers, including melanoma, lymphoma, lung, breast, prostate, and colorectal cancers (11). We further illustrated the chemoprevention signaling and epigenetic mechanism of CDDO derivatives and other triterpenoids (17). In vitro and in vivo studies have shown that CDDO and CDDO derivatives presented differentiating, antiproliferative, anti-inflammatory, and tumor suppression activities (18,19,20,21,22). In addition, CDDO activates transcription factor, nuclear factor-erythroid factor 2-related factor 2 (Nrf2), and its downstream phase 2 cytoprotective enzymes including NAD(P)H quinone dehydrogenase 1 (NQO1), heme oxygenase 1 (HO1, HMOX1), superoxide dismutase (SOD), thioredoxin, and catalase to alleviate the electrophilic and oxidative stress in various cancer models (23,24,25,26). Several studies investigated CDDO’s epigenetic/epigenomic modifications in cancers, as summarized in our review (17). A previous study reported that CDDO-Me inhibited the expression of epigenetic markers DNMT1 and DNMT3a to regulate CpG methylation in the hTERT promoter in pancreatic cancer (27). Moreover, CDDO reversed the human hepatocarcinogen aflatoxin B1–induced epigenetic changes to block the early-stage carcinogen in rats (28). However, the epigenetic regulation by CDDO to protect NMSC has not been studied yet.
To fill the gap, this study characterized differentially methylated regions (DMRs) and gene expression changes after CDDO treatment using methyl-seq and RNA-seq. The comparison of DMRs with the corresponding gene expression revealed genes and regulatory regions of interest for skin cancer chemoprevention.
Materials and Methods
Materials
Minimum Essential Medium (MEM), fetal bovine serum (FBS), penicillin–streptomycin (10,000 U/mL), puromycin, versene, and trypsin-EDTA were supplied by Gibco (Grand Island, NY, USA). Dimethyl sulfoxide (DMSO) was purchased from Sigma-Aldrich (St. Louis, MO, USA). TPA was obtained from Alexis Biochemicals (San Diego, CA, USA). CDDO compound is a generous gift from Dr. Michael Sporn (Dartmouth).
Cell Culture and Treatment
Mouse epidermal JB6 P+ cell line was purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in MEM with 5% FBS in a humidified 5% CO2 atmosphere as instructed by ATCC. The cells were first seeded and grown in plates for 24 h. Then, the cells were treated with various concentrations of CDDO in 1% FBS medium every other day for 1, 3, or 5 days, and 0.1% DMSO was used as vehicle control.
Cell Proliferation Assay (MTS Assay)
JB6 P+ cells were seeded into a 96-well plate at a density of 5 × 102 cells per well and grown for 24 h. Then, the cells were incubated in different concentrations of CDDO (12.5, 25, 50, 100, 200, 400, 800, or 1600 nM) or 0.1% DMSO (vehicle control) for 1, 3, or 5 days. The complete solubility of CDDO was maintained in DMSO over the concentration ranges from 12.5 to 1600 nM with no precipitation observed. The treatment with CDDO in the cell culture medium was renewed every other day. Cell viability at different concentrations for different durations was quantified with the CellTiter 96 AQueous One Solution Cell Proliferation Assay System (Promega, Madison, WI, USA) at 490 nm absorbance according to the manufacturer.
Anchorage-Independent Cell Transformation Assay
TPA-induced JB6 cell neoplastic transformation was established in our previous publication (29). JB6 P+ cells (8 × 103) were seeded in 6-well plates for 24 h and then treated with different concentrations of CDDO (12.5, 25, 50, or 100 nM) or 0.1% DMSO for 5 days. The cell culture medium with CDDO was renewed every other day. Then, pretreated cells were subjected to an anchorage-independent cell transformation assay in 6-well plates with 0.3% BME (Basal Medium Eagle) agar as the upper agar and 0.5% BME agar as the bottom agar. The 0.3% and 0.5% BME agar were made by mixing 10% FBS-containing BME medium with 0.6% or 1% bacteriological agar (Sigma-Aldrich) in a 1:1 proportion. The cells pretreated with CDDO or 0.1% DMSO were maintained in the upper agar, containing 10 ng/mL TPA or 0.1% DMSO (vehicle control). After 14 days of incubation, we captured the images of the cell colonies in the soft agar by a microscope camera using Nikon ACT-1 software (version 2.20; LEAD Technologies, Charlotte, NC, USA). The colonies were quantified with the ImageJ program (version 1.52a; NIH, Bethesda, MD, USA).
Nucleic Acid Isolation and Next-Generation Sequencing
Total RNA and DNA were isolated using an AllPrep DNA/RNA Mini Kit (QIAGEN, Germantown, MD, USA). The concentration and quality of extracted nucleic acids were measured using a NanoDrop spectrophotometer and an Agilent 2100 Bioanalyzer. RNA samples from all treatment groups with one replicate were subjected to RNA-seq with library preparation and sequencing performed by RUCDR Infinite Biologics (Piscataway, NJ, USA). First, the RNA library was prepared using an Illumina TruSeq RNA preparation kit (Illumina, San Diego, CA, USA) based on the manufacturer’s manual. Next, the RNA library was sequenced on an Illumina NextSeq 500 instrument (75 bp, single end), generating 30–40 million reads per sample. On the other hand, DNA samples from all treatment groups with one replicate were subjected SureSelect Methyl-seq, and library preparation and sequencing were conducted by RUCDR Infinite Biologics. First, the DNA samples were processed using an Agilent Mouse SureSelect Methyl-seq Target Enrichment System (Agilent Technologies, Santa Clara, CA, USA). Then, EZ DNA Methylation-Gold Kit (Zymo Research, USA) was applied to perform bisulfite conversion as instructed by the manufacturer. Finally, the DNA library was sequenced on an Illumina NextSeq 500 instrument (75 bp, single end), generating 30–40 million reads per sample.
RNA-Seq Analyses
The RNA-seq reads were preprocessed by Cutadapt for Illumina Universal Adapter sequence recognition and removal (30). The reads were then aligned to the mouse genome (mm10) with HISAT2 (31), and PCR duplicates were removed. Next, we quantified the reads overlapping predetermined genomic features in different treatment groups using the FeatureCounts program (version 1.5.1) (32). Finally, the reads were analyzed for differential expression with DEGSeq (version 1.36.0) in R (version 3.4.0), and the genes with the total reads across samples less than 10 were removed (33). The raw counts in FASTQ format and FeatureCounts results were uploaded to Gene Expression Omnibus (GEO) repository (GSE206168).
DNA SureSelect Methyl-Seq Analyses
The DNA reads were aligned to the bisulfite-converted mouse genome (mm10) with the Bismark (version 0.15.0) alignment algorithm (34). After alignment, DMRfinder (version 0.1) was used to extract CpG counts and cluster CpG sites into DMRs (35). Each DMR contained at least three CpG sites. Methylation differences greater than 0.10 with p values less than 0.05 were considered significant. Genomic annotation was performed with ChIPseeker (version 1.10.3) in R (version 3.4.0) (36). The raw data in FASTQ format and CpG methylation ratio result files were uploaded to GEO repository (GSE206168).
Correlation Study between DNA Methylation and RNA Expression
DNA methylation on cytosine bases at C5 in gene promoters was linked to gene repression (37, 38). This study compared DMRs with corresponding gene expression in TPA versus control and CDDO with TPA versus TPA comparisons to understand the correlation between methylome and transcriptome. We obtained a list of 88062 and 85613 DMRs with their corresponding gene expression in both comparisons. We then filtered the results by applying the thresholds of 10% changes in DNA methylation ratio and log2 2-fold changes for gene expression and visualized the correlations between DMRs and DEGs in a starburst plot (Fig. 5a). With the thresholds, the DMRs with an inverse correlation between the change of CpG methylation ratio and gene expression in the promoter region were labeled by their gene names.
Canonical Pathway Analysis and Pathway Network Analysis by Ingenuity Pathway Analysis Software
Canonical pathway analysis was performed using the differential expression results from RNA-seq. The Comparison Analysis program of IPA was applied to determine the most significant biological pathways and upstream regulators involved in skin cell transformation and CDDO treatment using expression patterns (Fig. 5b). In addition, the Path Explorer tool of IPA was applied to build a pathway network (Fig. 6) based on the direct and indirect interactions among gene of interest with an inversed correlation between gene expression and CpG methylation (Table I). The analyses were generated using QIAGEN IPA (QIAGEN Inc., https://digitalinsights.qiagen.com/IPA).
Statistical Analysis
The quantitative results of MTS assay, cell transformation assay, and qPCR are expressed as mean ± SEM from at least three independent experiments. Welch’s t-test was applied to evaluate the significance of statistical analysis with * indicating p <0.05 and ** indicating p <0.01, compared to the control group. In addition, # symbolizes p <0.05, and ## symbolizes p <0.01, compared to TPA-alone group. The cutoffs of p <0.05 and absolute value of log2 fold change ≥2 were used for RNA-seq analysis and visualization including the results presented in MA plots, Venn diagrams, and a heatmap (Fig. 3) with Benjamini–Hochberg procedure applied to control the false discovery rate. The threshold of p <0.05 and 10% DNA methylation ratio difference were used for Methyl-seq analysis. The threshold of p <0.05 was used for IPA canonical pathway analysis.
Results
CDDO Reduced Cell Viability in a Dose- and Time-Dependent Manner
Determination of optimal treatment duration and concentration involves a trade-off between toxicity and efficacy. The concentration and treatment duration were determined by cytotoxicity of CDDO assessed with an MTS cell proliferation assay as elaborated in the “Materials and Methods” section. CDDO inhibited cell viability in a time- and dose-dependent manner (Fig. 1b). The cell viabilities after 1, 3, and 5 days of 50 nM CDDO treatments are 99.67%, 92.63%, and 87.26%, respectively. The effect of CDDO 5-day treatment at concentrations around 50 nM was further tested in the subsequent efficacy and mechanistic studies.
a Chemical structure of CDDO. b Effect of CDDO on the viability of JB6 P + cells. JB6 P + cells were treated with various concentrations of CDDO for 1, 3, or 5 days as described in the “Materials and Methods” section. Cell viability was determined with an MTS cell proliferation assay and is presented as the mean ± SEM from three independent experiments
CDDO Prevented TPA-Mediated JB6 P + Cell Transformation
The cells which showed anchorage‐independent growth ability in the soft agar colony formation assay reflect their neoplastic transformation. TPA, a well-established promoter of skin carcinogenesis both in vitro and in vivo (39, 40), was applied to induce transformation in JB6 P + cells and was shown to effectively increase colony formation by 14.6-fold compared to the vehicle control (Fig. 2). CDDO treatment significantly attenuated the anchorage‐independent growth by 41.0%, 54.2%, 57.5%, and 81.1% at the concentration of 12.5, 25, 50, and 100 nM, respectively (Fig. 2). To conclude, the above result indicates that CDDO can significantly inhibit the TPA-induced anchorage‐independent growth of JB6 P + cells from 12.5 to 100 nM in a dose-dependent manner.
Inhibitory effects of CDDO on TPA-induced JB6 P + cell transformation. JB6 P + cells (3 × 10.5/10-cm dish) pretreated with 12.5, 25, 50, and 100 nM of CDDO for 5 days were transferred to soft agar containing TPA and CDDO for another 14 days. Then, the colonies showing anchorage-independent growth on soft agar were measured under a microscope and analyzed using ImageJ software. a Representative images of transformed JB6 colonies. b The quantities of colonies presented as the mean ± SEM from at least three independent studies. Significant differences were evaluated by Welch’s t-test (*p < 0.05; **p < 0.01)
CDDO Reversed Part of Transcriptome Dysregulated during TPA-Induced Neoplastic Transformation
Differential expression analysis by DEGSeq was employed to probe into the gene expression profiles of TPA and CDDO treatments. The p value was adjusted to q value by the Benjamini–Hochberg methods, considering the false discovery rate (41). The log2 fold changes between samples were used to describe the gene expression differences. As shown in the MA plot (Fig. 3a), q value less than 0.05 and an absolute value of log2 fold changes greater than 2.0 were set as thresholds. The genes with q value smaller than 0.05 and were upregulated and downregulated by 4-fold (red dots) were considered significant differential expression genes (DEGs). With these thresholds, TPA upregulated 629 DEGs, out of which CDDO reversed 359 DEGS; in contrast, TPA downregulated 330 DEGs out of which CDDO reversed 62 DEGs (Fig. 3b). The top 20 DEGs showed the opposite expression between comparisons of TPA versus control and TPA with CDDO group versus TPA group are presented in a heatmap (Fig. 3c). These DEGs may be critical genes regulated by CDDO to neutralize TPA-induced skin carcinogenesis. The genes showing the most significant difference in gene expression in the two comparisons are LYL1 basic helix-loop-helix family member (Lyl1), ladinin 1 (Lad1), and DENN domain containing 2D (Dennd2d), and CDDO suppressed Lyl1 and Lad1 while upregulated Dennd2d. These genes may be possible therapeutic targets of CDDO to counteract TPA-induced skin JB6 cell transformation. Moreover, Gene Set Enrichment Analysis (GSEA) was performed using DEGs (p < 0.05) with log2 fold changes. The overrepresentation of pre-defined hallmark (H) and oncogenic signature (C6) gene sets from the Molecular Signatures Database (MSigDB) (42,43,44) were analyzed by GSEA software (42, 45). The gene sets which showed statistical significance (p ≤ 0.05) between TPA + CDDO versus TPA and TPA versus C comparisons are presented in Fig. S2 by the order of the NES magnitude. The magnitude represented the correlation of the gene with the phenotype (TPA + CDDO vs. TPA or TPA vs. C) with p values shown by the color gradient.
a MA plots showing differentially expressed genes (DEGs) in response to TPA or CDDO treatment. The p value less than 0.05 and absolute value of log2 fold change greater or equal to 2 (dash line) were applied as the threshold. b Venn diagrams presenting the number of DEGs significantly regulated by TPA (10 ng/mL) and CDDO (50 nM) treatment. CDDO inhibited the expression of 359 DEGs TPA upregulated and activated the expression of 62 DEGs TPA downregulated as shown in the intersections. The downregulated gene set is marked with red, and the upregulated gene set is marked with green. c Heatmap showing gene expression change of top 20 DEGs significantly upregulated or downregulated by TPA but reversed by CDDO treatment (p < 0.05, absolute value of log2 fold change ≥ 2)
CDDO Altered CpG Methylation Profile in Response to TPA Treatment
Methyl-seq was performed to understand CpG methylation at single base-pair resolution during TPA-induced transformation and CDDO treatment. It is conducted on an Illumina NextSeq 500 platform using the Agilent SureSelect Mouse Methyl-seq library. The sequences obtained from different treatment groups were aligned parallelly to a bisulfite-converted mm10 mouse genome and underwent deduplication. Then, DNA methylation profiles were established using DMRfinder based on a total of 140,640 differentially methylated regions (DMRs) and were annotated using ChIPseeker (v1.14.2). Our result notably indicated that most of the DMRs were found in the promoter regions (Fig. 4a), which are the regions that have been shown highly related to the transcriptional regulation of gene expression. The average CpG methylation level in the promoter regions was greater than in other regions (Fig. 4a). In addition, CDDO reversed the methylation status of 32 DMRs methylated by TPA and 28 DMRs demethylated by TPA (Fig. 4b). The top 21 genes with significant differences (p value < 0.05) in methylation ratio between comparisons of TPA versus control and TPA with CDDO versus TPA were identified (Fig. 4c). In counteracting TPA-induced methylation changes, several DMRs in genes including transmembrane (Tmem) 253, Beta-Carotene Oxygenase 2 (Bco2), and MAP Kinase Activating Death Domain (Madd) were significantly modified by CDDO.
a Distribution of differentially methylated regions (DMRs) annotated by gene feature. b Venn diagrams presenting the number of DMRs which TPA (10 ng/mL) and CDDO (50 nM) significantly altered. CDDO demethylate 32 DMRs methylated by TPA and methylate 28 DMRs demethylated by TPA shown by the intersection. c Top 21 genes showing significant changes in CpG methylation ratio between TPA-vs.-control and TPA with CDDO-vs.-TPA comparisons with DMRs in their promoter regions (p < 0.05)
Inverse Correlation between DNA Methylation and RNA Expression Revealed Potential Targets of CDDO Treatment
Previous literature showed that DNA methylation on cytosine bases in gene promoters was linked to gene repression (37, 38). Here, we integrated methylome with transcriptome by arranging DMRs with corresponding gene expression in TPA versus control and CDDO with TPA versus TPA comparisons. We acquired a list of DMRs with their corresponding gene expression in both comparisons and then narrowed down the results by applying the thresholds of 10% DNA methylation ratio difference and log2 twofold change for gene expression. As shown in the correlation starburst plot (Fig. 5a), the DMRs in specific genes were illustrated as dots with different colors representing annotated features/gene regions. The DMRs with an inverse relationship between the change of CpG methylation ratio and gene expression are in quadrants II and IV. A subset of DMRs in the promoter with greater than 10% DNA methylation ratio difference and log2 fold change of 2 was labeled by their gene names.
a Correlations between change in gene expression and DNA CpG methylation ratio in different gene regions. A cutoff of 10% for DNA methylation ratio difference and absolute value of log2 fold change of 2 for gene expression were applied with different colors representing annotated features/gene regions. The genes that show inverse relationship between CpG methylation status change in promoter and corresponding gene expression change are labeled with gene names on starburst plot. b Ingenuity Pathway Analysis (IPA) identified signaling pathways that were significantly regulated (p < 0.05) by TPA (10 ng/mL) or CDDO (50 nM) with TPA treatments compared with control or TPA treatment. The heatmap is based on the z-scores of IPA representing the predicted overall activation or inhibition of the pathways. By inter-comparing transcriptomes, we found that Tumor Necrosis Factor Receptor 2 signaling is a pathway that showed the most significant difference between TPA-vs.-control and TPA with CDDO-vs.-TPA comparisons
Pathway Analysis Characterizes the Effect of CDDO on Canonical Signaling Pathways during TPA-Induced Carcinogenesis
To perform pathway analysis, the DEGs were first filtered by p value less than 0.05 and an absolute value of log2 fold change greater than 0.7, which we obtained 2530 genes in the TPA versus control comparison and 2422 genes in the TPA with CDDO versus TPA comparison. Canonical pathway analysis was performed using IPA to reveal the interactions between these DEGs and the associated pathways during the treatment of TPA and CDDO. We then obtained 117 pathways that TPA regulated (compared with control) and 62 pathways that CDDO regulated in response to TPA (compared with TPA alone group) (Tables S1 and S2). The pathways significantly regulated (p value < 0.05) in both comparisons were displayed in a heatmap with the activation z-scores (Fig. 5b). In response to TPA treatment, CDDO significantly downregulates Tumor Necrosis Factor Receptor 2 (TNFR2) signaling, implying that it may be a critical pathway for CDDO to mitigate TPA-induced cell transformation. Using qPCR, we validated that CDDO with TPA treatment significantly reduced Tnfr2 mRNA expression by 38.2% after 24-h treatment compared to control and CDDO reversed the TPA-induced Tnrf2 expression by 16.4% after 2-h treatment (Fig. S1).
Pathway Network Analysis Illustrated the Interactions between CDDO-Regulated Genes with Inverse Correlation in Gene Expression and CpG Methylation
Integrating transcriptome, methylome, and significantly affected pathways, we studied if DMRs with an inverse correlation in gene expression and CpG methylation have been involved in the previously discovered 62 CDDO-regulated pathways during TPA-induced skin cell transformation. We obtained a list of five genes, and Nudix Hydrolase 14 (Nudt14), NAD(P)H quinone dehydrogenase 1 (Nqo1), and protein kinase C gamma (Prkcg) are in the promoter regions (Table I). These genes may be potential therapeutic targets of CDDO for skin cancer prevention. We further built and explored the pathway network based on the direct and indirect relationship associated with Nudt14, Nqo1, Prkcg, and a CDDO derivative, CDDO-Me, in the Ingenuity Knowledge Base (IPA, QIAGEN). The CDDO-Me molecule with a similar mechanism and biological activities to CDDO (46,47,48,49,50) was used for pathway network analysis because of the limited data availability of CDDO in Ingenuity Knowledge Base. The pathway network from signaling to transcription was characterized and arranged according to the subcellular compartments (Fig. 6). The interactions between CDDO-altered genes and CDDO-Me involved phosphorylation cascades, transcription, and protein–protein/chemical–protein/protein–DNA interactions, as consolidated in Table S3. The pathway network result shows that CDDO-Me can induce ROS, activate Ho-1, and translocate Nrf2, increasing the transcription and expression of Nqo1. In addition, CDDO-Me inhibits Tnf, downregulating the expression of Prkcg and modulating the expression of Nqo1. We further applied qPCR and validated that CDDO significantly upregulates the mRNA expression of Nqo1, Nrf2, and Ho-1 by 2.0-, 2.4-, and 20.5-fold after 2-h treatment compared to control, whereas NAC pretreatment reduced the CDDO-activated changes by 15%, 12.5%, and 43.4% (Fig. S1), implicating that CDDO potentially induced ROS acutely to activate Nrf2 pathway. In addition, CDDO significantly upregulated Nqo1 and Ho-1 by 7.5- and 2.8-fold after 24-h treatment, implying that the effect of CDDO persisted in the long term.
The CDDO-regulated pathway network from signaling to transcription presented according to the subcellular compartments. The interaction network was built and explored by IPA, characterizing the direct and indirect interactions between CDDO-altered genes (Nqo1, Prkcg, and Nudt14) and CDDO-Me including phosphorylation, activation/inhibition, and protein–protein/chemical-protein/protein-DNA interactions. The relationships between these genes and molecules were elaborated in Table S3
Discussion
Skin cancer is the most common cancer in the USA (51, 52), and the major types are basal cell carcinoma (BCC), squamous cell carcinoma (SCC), and melanoma. The development of skin cancers is highly associated with the accumulation of DNA mutations induced by UV radiation (53) and environmental carcinogens (54). To decode skin cancer treatment, profiling of the molecular events during skin carcinogenesis is a desperate need in order to improve therapeutic treatment.
In the past few decades, RNA-seq was widely applied to study skin cancer progression efficiently. Chitsazzadeh et al. performed RNA-seq on the Illumina Hi-Seq platform to profile differentially expressed genes and transcription factor motifs from normal skin, actinic keratosis, to cutaneous squamous cell carcinoma (cuSCC) in the SKH-1 mouse model and patient samples (55). Hoang et al. performed RNA-seq to determine genes consistently expressed on non-melanoma skin cancer lesions, including actinic keratosis, intraepidermal carcinoma, and SCC in clinical samples (56). Our study implemented RNA-Seq technology to determine the gene expression profile during TPA-induced JB6 P + skin cell transformation. A total of 24,421 genes from the RNA-seq library have been analyzed for TPA and CDDO treatment. The most significant DEGs dysregulated by TPA and restored by CDDO are presented (Fig. 3c), which can be potential molecular targets against skin cell transformation. Most of the genes have not been thoroughly studied yet, such as Lyl1, Lad1, and Dennd2d. Lyl1 as a member of the basic helix-loop-helix transcription factor family was known to regulate cell proliferation/differentiation (57) and lymphocytic differentiation (58, 59). Aberrant LYL1 expression triggered by super-enhancers was associated with cell growth and survival of acute myeloid leukemia (AML) in human samples (60). LAD1 (Ladinin-1) as an anchoring filament protein of basement membrane was found to strengthen the association of dermal–epidermal junction, contributing to the stability of the connection between the epithelial and the mesenchyme tissues (61, 62). The expression of LAD1 has been implicated in various cancers including lung adenocarcinoma (63), thyroid cancers (64), and prostate cancer (65) based on human and mouse models. In addition, LAD1 is a downstream phosphorylation substrate of EGFR which regulated actin polymerization controlling cell migration and proliferation as a marker of aggressive breast tumors in clinical samples (66). The elevated Lad1 expression in TPA-induced skin cell transformation is comparable to the pathology results from human skin cancer tissues in Human Protein Atlas database (67,68,69). DENND2D can function as a guanine nucleotide exchange factor (GEF) promoting the release of GDP from Rab GTPases to regulate membrane trafficking events (70,71,72). It has been reported as a candidate tumor suppressor gene in various cancers including esophagus/oral squamous cell carcinoma (73, 74), gastric cancer (75), hepatocellular carcinoma (76), and non-small cell lung cancer from analyses of in vitro samples and human specimens. The reduced expression of DENND2D in esophagus squamous cell carcinoma and gastric cancer was shown to be associated with promoter hypermethylation (73, 75).
In addition to gene mutation, the epigenetic mechanism of TPA-induced skin cancers is elusive. There is also a lack of studies on epigenomic modifications by triterpenoids in skin cancers. Our analysis is the first characterization of triterpenoid CDDO’s effect on epigenomic changes in response to TPA-induced skin cell transformation by methyl-seq. The effect of triterpenoid compounds on gene expression and epigenetic modifications in skin cancers was also discussed in our previous review (17). In this study, CDDO has been shown to reverse the methylation status of genes dysregulated by TPA (Fig. 4b, c). CDDO significantly regulated DMRs in genes of Tmem253, Bco2, and Madd, reversing TPA-induced methylation changes. The correlation between gene expression and CpG methylation was illustrated in a starburst plot (Fig. 5a). Previous literature shows that the alteration in promoter CpG methylation can modulate gene expression (77, 78). This study analyzed the DMRs in the promoter with inversed CpG methylation ratio changes and corresponding gene expression changes in TPA and CDDO treatment to understand the DMRs associated with transcription (Fig. 5a).
The signaling pathways that drive the melanoma and non-melanoma skin cancer pathogenesis include the mitogen-activated protein kinase (MAPK) pathway (p38, JNK, or ERK signaling), protein kinase B (AKT) pathway, cell cycle–related pathway, and some other pathways (79, 80). In the study, we presented the biological pathways during TPA-induced skin transformation and CDDO treatment (Fig. 5b). We showed that TNFR2 signaling induced by tumor promoter TPA was significantly downregulated by CDDO (Fig. 5b), implying inhibition of TNFR2 signaling as a potential target to block skin carcinogenesis. The result is comparable to previous studies showing that TNFR2 signaling involves in the development of melanoma and skin cancers (81, 82). TNFR2 can enhance tumor progression by promoting cell proliferation, resisting TNF-induced cell death, supporting angiogenesis, activating immunosuppressive regulatory T (Treg) cells, stabilizing myeloid-derived suppressor cells (MDSC), and enhancing metastasis in various cancers (81). A previous study showed that TNFR1 and TNFR2 regulate TNF-α signaling to promote skin carcinogenesis via multiple c-Jun containing AP-1 complexes in vivo and in vitro (83). We further studied the genes including Nudt14, Nqo1, and Prkcg with the inverse relationship between promoter methylation status and gene expression change that is associated with significantly regulated pathways (Table I). The result implies that the Nrf2-ARE pathway upregulating Nqo1 is one of the critical signaling involved in skin cancer prevention of CDDO. The results are comparable to established literature showing that phytochemicals prevent ultraviolet- and TPA-induced skin damage or carcinogenesis in vivo through activation of Nrf2 (84, 85). The Nrf2-ARE pathway is well-characterized signaling to counteract oxidative stress and is a target for cancer prevention and therapy (86,87,88,89). The translocation of the Nrf2 transcription factor from the cytoplasm to the nucleus can activate downstream targets, such as Ho-1, Nqo1, and Sod1, to exert anti-oxidative and anti-inflammatory effects (84, 85, 88). We built a pathway network illustrating that CDDO-Me can increase ROS, upregulate Ho-1, and translocate Nrf2 to enhance the expression of Nqo1, whereas N-acetylcysteine (NAC) can reverse the upregulation of Nqo1 (Fig. 6). We validated with qPCR that CDDO treatment has shown to significantly restore the TPA-induced mRNA expression changes of Nqo1 and Ho-1 after 2-h and 24-h treatment (Fig. S1). Our research has some limitations including (1) just like any model system, extrapolation to human patients required further in vivo animal studies; (2) related to #1, the use of in vitro cell line drug concentrations would need to be further supported by in vivo animal studies; (3) the utility of RNA-seq and DNA Methyl-seq would need to be better integrated with biomarkers’ studies in animal models and in human patients to allow better response prediction and target discovery.
Conclusion
Overall, our study profiled alterations of the transcriptome, CpG methylome, and signaling pathways during TPA-mediated skin cell neoplastic transformation. The treatment with CDDO, a potent triterpenoid, would modulate these alterations, potentially contributing to the overall chemopreventive effect in skin carcinogenesis.
References
Rogers HW, Weinstock MA, Feldman SR, Coldiron BM. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the U.S. population, 2012. JAMA Dermatol. 2015;151(10):1081–6.
Aggarwal P, Knabel P, Fleischer AB. United States burden of melanoma and non-melanoma skin cancer from 1990 to 2019. J Am Acad Dermatol. 2021;85(2):388–95.
Madan V, Lear JT, Szeimies RM. Non-melanoma skin cancer. Lancet. 2010;375(9715):673–85. https://doi.org/10.1016/S0140-6736(09)61196-X.
Alam M, Ratner D. Cutaneous squamous-cell carcinoma. N Engl J Med. 2001;344(13):975–83. https://doi.org/10.1056/NEJM200103293441306.
Stewart BW, Kleihues P. World cancer report. Lyons: IARC Press; 2003. p. 232–6.
Abel EL, Angel JM, Kiguchi K, DiGiovanni J. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat Protoc. 2009;4(9):1350–62.
Khan AQ, Khan R, Qamar W, Lateef A, Rehman MU, Tahir M, et al. Geraniol attenuates 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced oxidative stress and inflammation in mouse skin: possible role of p38 MAP kinase and NF-kappaB. Exp Mol Pathol. 2013;94(3):419–29. https://doi.org/10.1016/j.yexmp.2013.01.006.
Kashyap MP, Sinha R, Mukhtar MS, Athar M. Epigenetic regulation in the pathogenesis of non-melanoma skin cancer. Semin Cancer Biol. 2022;83:36–56.
Saha K, Hornyak TJ, Eckert RL. Epigenetic cancer prevention mechanisms in skin cancer. AAPS J. 2013;15(4):1064–71.
Lubecka K, Kurzava L, Flower K, Buvala H, Zhang H, Teegarden D, et al. Stilbenoids remodel the DNA methylation patterns in breast cancer cells and inhibit oncogenic NOTCH signaling through epigenetic regulation of MAML2 transcriptional activity. Carcinogenesis. 2016;37(7):656–68.
Borella R, Forti L, Gibellini L, De Gaetano A, De Biasi S, Nasi M, et al. Synthesis and anticancer activity of CDDO and CDDO-Me, two derivatives of natural triterpenoids. Molecules. 2019;24(22):4097.
Du J-R, Long F-Y, Chen C. Chapter six – research progress on natural triterpenoid saponins in the chemoprevention and chemotherapy of cancer. In: Bathaie SZ, Tamanoi F, editors. The enzymes. Academic Press; 2014. p. 95–130.
Kim H, Ramirez CN, Su Z-Y, Kong A-NT. Epigenetic modifications of triterpenoid ursolic acid in activating Nrf2 and blocking cellular transformation of mouse epidermal cells. J Nutr Biochem. 2016;33:54–62.
Yang Y, Yin R, Wu R, Ramirez CN, Sargsyan D, Li S, et al. DNA methylome and transcriptome alterations and cancer prevention by triterpenoid ursolic acid in UVB-induced skin tumor in mice. Mol Carcinog. 2019;58(10):1738–53. https://doi.org/10.1002/mc.23046.
Hudlikar RR, Sargsyan D, Wu R, Su S, Zheng M, Kong AN. Triterpenoid corosolic acid modulates global CpG methylation and transcriptome of tumor promotor TPA induced mouse epidermal JB6 P+ cells. Chem Biol Interact. 2020;321:109025.
Liby KT, Yore MM, Sporn MB. Triterpenoids and rexinoids as multifunctional agents for the prevention and treatment of cancer. Nat Rev Cancer. 2007;7:357. https://doi.org/10.1038/nrc2129.
Li S, Kuo H-CD, Yin R, Wu R, Liu X, Wang L, et al. Epigenetics/epigenomics of triterpenoids in cancer prevention and in health. Biochem Pharmacol. 2020;175:113890.
Honda T, Rounds BV, Gribble GW, Suh N, Wang Y, Sporn MB. Design and synthesis of 2-cyano-3,12-dioxoolean-1,9-dien-28-oic acid, a novel and highly active inhibitor of nitric oxide production in mouse macrophages. Bioorg Med Chem Lett. 1998;8(19):2711–4.
Suh N, Honda T, Finlay HJ, Barchowsky A, Williams C, Benoit NE, et al. Novel triterpenoids suppress inducible nitric oxide synthase (iNOS) and inducible cyclooxygenase (COX-2) in mouse macrophages. Cancer Res. 1998;58(4):717–23.
Suh N, Wang Y, Honda T, Gribble GW, Dmitrovsky E, Hickey WF, et al. A novel synthetic oleanane triterpenoid, 2-cyano-3,12-dioxoolean-1,9-dien-28-oic acid, with potent differentiating, antiproliferative, and anti-inflammatory activity. Cancer Res. 1999;59(2):336–41.
Suh N, Roberts AB, Birkey Reffey S, Miyazono K, Itoh S, ten Dijke P, et al. Synthetic triterpenoids enhance transforming growth factor beta/Smad signaling. Cancer Res. 2003;63(6):1371–6.
Place AE, Suh N, Williams CR, Risingsong R, Honda T, Honda Y, et al. The novel synthetic triterpenoid, CDDO-imidazolide, inhibits inflammatory response and tumor growth in vivo. Clin Cancer Res. 2003;9(7):2798–806.
To C, Ringelberg CS, Royce DB, Williams CR, Risingsong R, Sporn MB, et al. Dimethyl fumarate and the oleanane triterpenoids, CDDO-imidazolide and CDDO-methyl ester, both activate the Nrf2 pathway but have opposite effects in the A/J model of lung carcinogenesis. Carcinogenesis. 2015;36(7):769–81.
Liby K, Hock T, Yore MM, Suh N, Place AE, Risingsong R, et al. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 2005;65(11):4789–98. https://doi.org/10.1158/0008-5472.CAN-04-4539.
Lapillonne H, Konopleva M, Tsao T, Gold D, McQueen T, Sutherland RL, et al. Activation of peroxisome proliferator-activated receptor gamma by a novel synthetic triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid induces growth arrest and apoptosis in breast cancer cells. Cancer Res. 2003;63(18):5926–39.
Yates MS, Kwak MK, Egner PA, Groopman JD, Bodreddigari S, Sutter TR, et al. Potent protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the triterpenoid 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole. Cancer Res. 2006;66(4):2488–94. https://doi.org/10.1158/0008-5472.CAN-05-3823.
Deeb D, Brigolin C, Gao X, Liu Y, Pindolia KR, Gautam SC. Induction of apoptosis in pancreatic cancer cells by CDDO-Me involves repression of telomerase through epigenetic pathways. J Carcinog Mutagen. 2014;5:177.
Livingstone MC, Johnson NM, Roebuck BD, Kensler TW, Groopman JD. Profound changes in miRNA expression during cancer initiation by aflatoxin B1 and their abrogation by the chemopreventive triterpenoid CDDO-Im. Mol Carcinog. 2017;56(11):2382–90. https://doi.org/10.1002/mc.22635.
Kuo HD, Wu R, Li S, Yang AY, Kong AN. Anthocyanin delphinidin prevents neoplastic transformation of mouse skin JB6 P+ cells: epigenetic re-activation of Nrf2-ARE pathway. AAPS J. 2019;21(5):83.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17(1):10–2.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30.
Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–8.
Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–2.
Gaspar JM, Hart RP. DMRfinder: efficiently identifying differentially methylated regions from MethylC-seq data. BMC Bioinformatics. 2017;18(1):528.
Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31(14):2382–3.
Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916. https://doi.org/10.1126/science.1186366.
Feng S, Cokus SJ, Zhang X, Chen P-Y, Bostick M, Goll MG, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010;107(19):8689–94. https://doi.org/10.1073/pnas.1002720107.
Wei S-J, Trempus CS, Ali RC, Hansen LA, Tennant RW. 12-O-Tetradecanoylphorbol-13-acetate and UV radiation-induced nucleoside diphosphate protein kinase B mediates neoplastic transformation of epidermal cells. J Biol Chem. 2004;279(7):5993–6004.
Abel EL, Angel JM, Kiguchi K, DiGiovanni J. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat Protoc. 2009;4(9):1350.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol). 1995;57(1):289–300.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102(43):15545–50. https://doi.org/10.1073/pnas.0506580102.
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40. https://doi.org/10.1093/bioinformatics/btr260.
Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–25. https://doi.org/10.1016/j.cels.2015.12.004.
Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–73. https://doi.org/10.1038/ng1180.
Konopleva M, Tsao T, Ruvolo P, Stiouf I, Estrov Z, Leysath CE, et al. Novel triterpenoid CDDO-Me is a potent inducer of apoptosis and differentiation in acute myelogenous leukemia. Blood. 2002;99(1):326–35.
Chintharlapalli S, Papineni S, Konopleva M, Andreef M, Samudio I, Safe S. 2-Cyano-3,12-dioxoolean-1,9-dien-28-oic acid and related compounds inhibit growth of colon cancer cells through peroxisome proliferator-activated receptor gamma-dependent and -independent pathways. Mol Pharmacol. 2005;68(1):119–28.
Yates MS, Tauchi M, Katsuoka F, Flanders KC, Liby KT, Honda T, et al. Pharmacodynamic characterization of chemopreventive triterpenoids as exceptionally potent inducers of Nrf2-regulated genes. Mol Cancer Ther. 2007;6(1):154–62.
Hyer ML, Shi R, Krajewska M, Meyer C, Lebedeva IV, Fisher PB, et al. Apoptotic activity and mechanism of 2-cyano-3,12-dioxoolean-1,9-dien-28-oic-acid and related synthetic triterpenoids in prostate cancer. Cancer Res. 2008;68(8):2927–33.
Ikeda T, Sporn M, Honda T, Gribble GW, Kufe D. The novel triterpenoid CDDO and its derivatives induce apoptosis by disruption of intracellular redox balance. Cancer Res. 2003;63(17):5551–8.
Guy GP Jr, Thomas CC, Thompson T, Watson M, Massetti GM, Richardson LC. Vital signs: melanoma incidence and mortality trends and projections – United States, 1982–2030. MMWR Morb Mortal Wkly Rep. 2015;64(21):591–6.
Guy GP Jr, Machlin SR, Ekwueme DU, Yabroff KR. Prevalence and costs of skin cancer treatment in the U.S., 2002–2006 and 2007–2011. Am J Prev Med. 2015;48(2):183–7. https://doi.org/10.1016/j.amepre.2014.08.036.
de Gruijl FR, van Kranen HJ, Mullenders LHF. UV-induced DNA damage, repair, mutations and oncogenic pathways in skin cancer. J Photochem Photobiol, B. 2001;63(1):19–27. https://doi.org/10.1016/S1011-1344(01)00199-3.
Fabbrocini G, Triassi M, Mauriello MC, Torre G, Annunziata MC, De Vita V, et al. Epidemiology of skin cancer: role of some environmental factors. Cancers (Basel). 2010;2(4):1980–9.
Chitsazzadeh V, Coarfa C, Drummond JA, Nguyen T, Joseph A, Chilukuri S, et al. Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat Commun. 2016;7:12601.
Hoang VLT, Tom LN, Quek XC, Tan JM, Payne EJ, Lin LL, et al. RNA-seq reveals more consistent reference genes for gene expression studies in human non-melanoma skin cancers. PeerJ. 2017;5:e3631.
San-Marina S, Han Y, Suarez Saiz F, Trus MR, Minden MD. Lyl1 interacts with CREB1 and alters expression of CREB1 target genes. Biochim Biophys Acta. 2008;1783(3):503–17. https://doi.org/10.1016/j.bbamcr.2007.11.015.
Nagel S, Venturini L, Meyer C, Kaufmann M, Scherr M, Drexler HG, et al. Multiple mechanisms induce ectopic expression of LYL1 in subsets of T-ALL cell lines. Leuk Res. 2010;34(4):521–8. https://doi.org/10.1016/j.leukres.2009.06.020.
Zhang M-M, Liu N, Zhang Y-L, Rong B, Wang X-L, Xu C-H, et al. Destabilization of AETFC through C/EBPα-mediated repression of LYL1 contributes to t(8;21) leukemic cell differentiation. Leukemia. 2019;33(7):1822–7. https://doi.org/10.1038/s41375-019-0398-8.
Fang F, Lu J, Sang X, Tao Y-F, Wang J-W, Zhang Z-M, et al. Super-enhancer profiling identifies novel critical and targetable cancer survival gene LYL1 in pediatric acute myeloid leukemia. J Exp Clin Cancer Res. 2022;41(1):225. https://doi.org/10.1186/s13046-022-02428-9.
Teixeira JC, de Filippo C, Weihmann A, Meneu JR, Racimo F, Dannemann M, et al. Long-term balancing selection in LAD1 maintains a missense trans-species polymorphism in humans, chimpanzees, and bonobos. Mol Biol Evol. 2015;32(5):1186–96. https://doi.org/10.1093/molbev/msv007.
Motoki K, Megahed M, LaForgia S, Uitto J. Cloning and chromosomal mapping of mouse ladinin, a novel basement membrane zone component. Genomics. 1997;39(3):323–30. https://doi.org/10.1006/geno.1996.4507.
Codreanu SG, Hoeksema MD, Slebos RJC, Zimmerman LJ, Rahman SMJ, Li M, et al. Identification of proteomic features to distinguish benign pulmonary nodules from lung adenocarcinoma. J Proteome Res. 2017;16(9):3266–76. https://doi.org/10.1021/acs.jproteome.7b00245.
Rusinek D, Swierniak M, Chmielik E, Kowal M, Kowalska M, Cyplinska R, et al. BRAFV600E-associated gene expression profile: early changes in the transcriptome, based on a transgenic mouse model of papillary thyroid carcinoma. PLoS ONE. 2015;10(12):e0143688. https://doi.org/10.1371/journal.pone.0143688.
Li J, Wang Z, Tie C. High expression of ladinin-1 (LAD1) predicts adverse outcomes: a new candidate docetaxel resistance gene for prostatic cancer (PCa). Bioengineered. 2021;12(1):5749–59. https://doi.org/10.1080/21655979.2021.1968647.
Roth L, Srivastava S, Lindzen M, Sas-Chen A, Sheffer M, Lauriola M, et al. SILAC identifies LAD1 as a filamin-binding regulator of actin dynamics in response to EGF and a marker of aggressive breast tumors. Sci Signal. 2018;11(515). https://doi.org/10.1126/scisignal.aan0949.
Uhlén M, Fagerberg L, Hallström Björn M, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419.
Human Protein Atlas proteinatlas.org. https://www.proteinatlas.org/ENSG00000159166-LAD1/pathology/skin+cancer. Gene available from v21.0. proteinatlas.org.
Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):eaan2507.
Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191(2):367–81. https://doi.org/10.1083/jcb.201008051.
Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438(7068):597–604. https://doi.org/10.1038/nature04397.
Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2(2):107–17. https://doi.org/10.1038/35052055.
Hibino S, Kanda M, Oya H, Takami H, Shimizu D, Nomoto S, et al. Reduced expression of DENND2D through promoter hypermethylation is an adverse prognostic factor in squamous cell carcinoma of the esophagus. Oncol Rep. 2014;31(2):693–700. https://doi.org/10.3892/or.2013.2901.
Sakha S, Muramatsu T, Ueda K, Inazawa J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci Rep. 2016;6:38750. https://doi.org/10.1038/srep38750.
Kanda M, Shimizu D, Nomoto S, Takami H, Hibino S, Oya H, et al. Prognostic impact of expression and methylation status of DENN/MADD domain-containing protein 2D in gastric cancer. Gastric Cancer. 2015;18(2):288–96. https://doi.org/10.1007/s10120-014-0372-0.
Kanda M, Nomoto S, Oya H, Takami H, Hibino S, Hishida M, et al. Downregulation of DENND2D by promoter hypermethylation is associated with early recurrence of hepatocellular carcinoma. Int J Oncol. 2014;44(1):44–52. https://doi.org/10.3892/ijo.2013.2165.
Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–22.
Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38.
Guo W, Wang H, Li C. Signal pathways of melanoma and targeted therapy. Signal Transduct Target Ther. 2021;6(1):424.
Maner BS, Dupuis L, Su A, Jueng JJ, Harding TP, Meisenheimer J, et al. Overview of genetic signaling pathway interactions within cutaneous malignancies. J Cancer Metastasis Treat. 2020;6:37.
Sheng Y, Li F, Qin Z. TNF receptor 2 makes tumor necrosis factor a friend of tumors. Front Immunol. 2018;9:1170. https://doi.org/10.3389/fimmu.2018.01170.
Trentin L, Zambello R, Bulian P, Cerutti A, Enthammer C, Cassatella M, et al. Tumour-infiltrating lymphocytes bear the 75 kDa tumour necrosis factor receptor. Br J Cancer. 1995;71(2):240–5. https://doi.org/10.1038/bjc.1995.50.
Arnott CH, Scott KA, Moore RJ, Robinson SC, Thompson RG, Balkwill FR. Expression of both TNF-α receptor subtypes is essential for optimal skin tumour development. Oncogene. 2004;23(10):1902–10.
Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, et al. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66(16):8293–6.
Tao S, Park SL, de la Vega MR, Zhang DD, Wondrak GT. Systemic administration of the apocarotenoid bixin protects skin against solar UV-induced damage through activation of NRF2. Free Radical Biol Med. 2015;89:690–700.
Singh S, Vrishni S, Singh BK, Rahman I, Kakkar P. Nrf2-ARE stress response mechanism: a control point in oxidative stress-mediated dysfunctions and chronic inflammatory diseases. Free Radical Res. 2010;44(11):1267–88. https://doi.org/10.3109/10715762.2010.507670.
Hu R, Saw CL, Yu R, Kong AN. Regulation of NF-E2-related factor 2 signaling for cancer chemoprevention: antioxidant coupled with antiinflammatory. Antioxid Redox Signal. 2010;13(11):1679–98. https://doi.org/10.1089/ars.2010.3276.
Deshmukh P, Unni S, Krishnappa G, Padmanabhan B. The Keap1-Nrf2 pathway: promising therapeutic target to counteract ROS-mediated damage in cancers and neurodegenerative diseases. Biophys Rev. 2017;9(1):41–56.
Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–9. https://doi.org/10.1016/j.redox.2012.10.001.
Acknowledgements
This paper is dedicated to Professor Michael B. Sporn, the discoverer of CDDO series of triterpenoids, making great contribution to cancer chemoprevention research, who passed away recently. In addition, the authors are thankful to Lujing Wang in Dr. Kong’s laboratory for providing helpful insight into the study design, data analyses, and manuscript writing. The graphical abstract was created with BioRender.com.
Funding
This work is supported in part by institutional funds, R01 CA200129, from the National Cancer Institute and AT009152 from the National Center for Complementary and Integrative Health (NCCIH) to A-NTK.
Author information
Authors and Affiliations
Contributions
Participated in research design: H-CDK, A-NTK. Conducted experiments: H-CDK. Performed data analysis: H-CDK, RW, MZ, CW, DS. Wrote the manuscript: H-CDK, MSS. Reviewed the manuscript: A-NTK, NS, MSS, H-CDK.
Corresponding author
Ethics declarations
Conflict of Interest
NS is an inventor on patents dealing with chemical synthesis of triterpenoids and their application in cancer as well as inflammatory diseases. The rest of the authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kuo, HC.D., Wu, R., Sarwar, M.S. et al. DNA Methylome and Transcriptome Study of Triterpenoid CDDO in TPA-Mediated Skin Carcinogenesis Model. AAPS J 24, 115 (2022). https://doi.org/10.1208/s12248-022-00763-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-022-00763-5