Abstract
Background
Gonadotropin-releasing hormone receptor (GnRHR) transmits its signal via two major Gα-proteins, primarily Gαq and Gαi. However, the precise mechanism underlying the functions of Gαs signal in prostate cancer cells is still unclear. We have previously identified that GV1001, a fragment of the human telomerase reverse transcriptase, functions as a biased GnRHR ligand to selectively stimulate the Gαs/cAMP pathway. Here, we tried to reveal the potential mechanisms of which GV1001-stimulated Gαs-cAMP signaling pathway reduces the migration and metastasis of prostate cancer (PCa) cells.
Methods
The expression of epithelial-mesenchymal transition (EMT)-related genes was measured by western-blotting and spheroid formation on ultra-low attachment plate was detected after GV1001 treatment. In vivo Spleen-liver metastasis mouse model was used to explore the inhibitory effect of GV1001 on metastatic ability of PCa and the transwell migration assay was performed to identify whether GV1001 had a suppressive effect on cell migration in vitro. In order to demonstrate the interaction between androgen receptor (AR) and YAP1, co-immunoprecipitation (co-IP), immunofluorescence (IF) staining, chromatin immunoprecipitation (ChIP) were performed in LNCaP cells with and without GV1001 treatment.
Results
GV1001 inhibited expression of EMT-related genes and spheroid formation. GV1001 also suppressed in vivo spleen-liver metastasis of LNCaP cells as well as cell migration in vitro. GV1001 enhanced the phosphorylation of AR and transcription activity of androgen response element reporter gene through cAMP/protein kinase A pathway. Moreover, GV1001 increased Ser-127 phosphorylation of YAP1 and its ubiquitination, and subsequently decreased the levels of AR-YAP1 binding in the promoter region of the CTGF gene. In contrast, both protein and mRNA levels of NKX3.1 known for tumor suppressor gene and AR-coregulator were upregulated by GV1001 in LNCaP cells. YAP1 knockout using CRISPR/Cas9 significantly suppressed the migration ability of LNCaP cells, and GV1001 did not affect the cell migration of YAP1-deficient LNCaP cells. On the contrary, cell migration was more potentiated in LNCaP cells overexpressing YAP5SA, a constitutively active form of YAP1, which was not changed by GV1001 treatment.
Conclusions
Overall, this study reveals an essential role of AR-YAP1 in the regulation of PCa cell migration, and provides evidence that GV1001 could be a novel GnRHR ligand to inhibit metastasis of PCa via the Gαs/cAMP pathway.
Similar content being viewed by others
Background
Gonadotropin-releasing hormone (GnRH), a decapeptide hormone released from the hypothalamus, is the central activator of the reproductive hormonal network [1, 2]. GnRH receptor (GnRHR) is a G-protein-coupled cell surface receptor (GPCR) with seven-transmembrane-spanning domains connected by extracellular and intracellular loops [3, 4]. As one of the GPCR families, GnRHR transduces the intracellular GPCR signals via multiple heterotrimeric G proteins [5, 6]. In pituitary gonadotropes, GnRHR primarily interacts with Gαq and exerts reproductive effects such as synthesis and secretion of pituitary gonadotropin hormones, luteinizing hormone (LH), and follicle-stimulating hormone (FSH) [7,8,9]. Conversely, it has been demonstrated that coupling of the GnRHR to Gαi abolishes neuronal function and hormonal secretion of GnRH [10]. In many types of human tissues and immortalized cell lines, it has been proposed that GnRHR interacts with different G-proteins to mediate the diverse physiological effects. Protein or mRNA expression of GnRH and GnRHR was detected not only in pituitary cells but also in extra-pituitary cells; ovarian, breast, prostate tissues, and various cancer cell lines including breast (MCF-7 and MDA-MB-468) and prostate cancer (PCa) cells (PC-3 and LNCaP) [11,12,13,14]. Several reports suggest that GnRHR expressed in cancer cells directly contributes to cancer progression, and GnRH analogs exert direct anti-cancer actions by modulating Gα subunits. GnRH analogs induced apoptosis of several prostate cancer cells presumably via Gαi coupling [15,16,17,18,19]. Also, GnRH analogs counteracted the tumor growth of androgen receptor (AR)-negative DU145 xenografts, further supporting their direct and reproductive system-independent anti-cancer effects [20]. Based on the evidence, cancer cell-expressing GnRHR is considered as a novel receptor-targeting anti-cancer strategy.
GV1001, a 16 amino acid peptide derived from the human telomerase reverse transcriptase (hTERT), has been proposed as a cancer vaccine to boost immune responses of CD8 and CD4 T cells [21]. We have recently demonstrated that GV1001 functions as a GnRHR ligand and selectively stimulates the Gαs-cAMP pathway [22]. Although it has been proposed that GnRH analogs regulate not only well-established Gαq but also Gαs pathway in several extra-pituitary cells [23,24,25], pharmacological effects of Gαs-coupled GnRHR activation has not been studied in cancer cells. In an attempt to elucidate the underlying molecular mechanisms of anti-migration and metastatic effects of GV1001 in PCa, we investigated the functions of Gαs-cAMP signaling under GV1001-stimulated GnRHR activation, focusing on AR and its coregulator, yes-associated protein1 (YAP1).
Materials and methods
Materials
Antibodies recognizing YAP1, phospho-YAP1 (Ser-127 p-YAP1)(Cell Signaling Technology, Beverly, MA, USA), matrix metalloproteinase2 (MMP2), slug, twist, vimentin, AR, NK3 Homeobox 1 (NKX3.1), Myc, connective tissue growth factor (CTGF, Santa Cruz Biotechnology, Santa Cruz, CA, USA), E-cadherin, N-cadherin (BD Biosciences, San Jose, CA, USA), phospho-AR (ser-81 p-AR)(Milipore, Billerica, MA, USA), and ß-actin, GAPDH (Sigma-Aldrich, St. Louis, MO, USA) were used for immunoblottings. Horseradish peroxidase (HRP)-conjugated donkey anti-rabbit and anti-mouse IgGs were obtained from Cell Signaling Technology. GV1001 peptide was supplied by GemVax & KAEL (Seongnam, Kyeonggi-do, South Korea). Leuprolide acetate (LA) and cetrorelix acetate (CA) were synthesized by AnyGen (Gwangju, South Korea). KT5720 (Cat# K3761), forskolin (Cat# F6886), dihydrotestosterone (Cat# A8380), flutamide (Cat# F9397), pertussis toxin (Cat# P2980) and SQ22536 (Cat# S153) were purchased from Sigma-Aldrich (St. Louis, MO, USA). KH7 (Cat# 13243, Cayman Chemical, Ann Arbor, MI, USA) and 1,2-Bis(2-aminophenoxy) ethane-N,N,N’,N’-tetraacetic acid tetrakis(acetoxymethyl ester, BAPTA/AM)(Biovision, Mountain View, CA, USA) were used as signaling inhibitors for Gα-proteins.
Cell culture and establishment of stably GnRHR-overexpressing HEK293 cells
LNCaP (androgen receptor positive human PCa cell line) cells were cultured in RPMI-1640 medium with 10% fetal bovine serum (Hyclone, Logan, UT, USA) and 1% penicillin/streptomycin (100 U/ml, Hyclone). YAP1 knockout LNCaP cells were established by CRISPR/Cas9 gene editing system. U6-YAP1/Cas9-2A-RFP plasmid or CRISPR Universal Negative Control plasmid were transfected with LNCaP cells and the red fluorescence protein (RFP)-positive cells were further sorted by FACS Aria II flow cytometer (BD Biosciences, San Jose, CA, USA). HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum and 1% penicillin/streptomycin. All cultures were maintained in a humidified 5% CO2 environment at 37 °C. To obtain GnRHR-overexpressing HEK293 (HEK293-GnRHR) cells, transfection of pcDNA3.1(+)-GnRHR vector was performed by using Lipofectamine 2000 as specified by the manufacturer's instruction (Invitrogen, Carlsbad, CA, USA). Geneticin-resistant colonies were selected by G418 (800 μg/ml, Thermo Fisher Scientific, Waltham, MA, USA) treatment. HEK293-pcDNA3.1(+) vector was also used as mock-transfection group.
Establishment of AR and YAP1 knockout cells
AR- or YAP1-knockout LNCaP cells were established by using U6-AR/Cas9-2A-RFP plasmid (HS0000000952), U6-YAP1/Cas9-2A-RFP plasmid (HS0000121498) or CRISPR Universal Negative Control plasmid (Sigma-Aldrich). Lipofectamine 2000 (Thermo Fisher Scientific) was used as the manufacturer’s protocol. 48 h after transfection, AR or YAP1 knockout cells were selected. For the selection of RFP-positive-knockout cells, the transfected LNCaP cells were sorted using BD FACS Aria II flow cytometer (BD Biosciences, San Jose, CA, USA).
Western blot analysis
After washing with PBS, cells were lysed with lysis buffer containing 20 mM Tris–Cl (pH 7.5), 1% Triton X-100, 137 mM sodium chloride, 10% glycerol, 2 mM EDTA, 1 mM sodium orthovanadate, 25 mM ß-glycerophosphate, 2 mM sodium pyrophosphate, 1 mM phenylmethylsulfonylfluoride and 1 μg/ml leupeptin. The cell lysates were centrifuged at 13,000×g for 15 min to remove insoluble material, the supernatants were fractionated using 10, 12 or 15% separating polyacrylamide gel, and electrophoretically transferred to nitrocellulose membranes, which were incubated overnight at 4 °C with one of the primary antibodies. HRP-conjugated anti-IgG antibodies were used as the secondary antibodies. The nitrocellulose papers were developed using an ECL chemiluminescence system (Milipore, Billerica, MA, USA). For ECL chemiluminescence detection, LAS-3000 mini system (Fujifilm, Tokyo, Japan) was used.
Reporter gene assay
LNCaP, HEK293-pcDNA3.1( +) and HEK293-GnRHR cells (1 × 105 cells/well) were cultured in 48-well plates, then transfected with luciferase reporter plasmid containing cAMP response element (CRE) or androgen response element (ARE). In order to evaluate the transcriptional activity of YAP1, YAP1 knockout- and wildtype-LNCaP cells were cultured in 48-well plates and transfected with YAP/TAZ-responsive luciferase reporter plasmid (TEAD reporter). CRE-luciferase plasmid was purchased from Stratagene (La Jolla, CA, USA) and ARE-luciferase plasmid) [61] was kindly donated from Dr. Young Joo Lee (Department of Bioscience and Biotechnology, Sejong University, Republic of Korea. Lipofectamine 2000 (Thermo Fisher Scientific) was used per the manufacturer’s protocol. The transfected cells were exposed to compounds for 24 h, and the promoter activity was measured using a dual-luciferase reporter assay system (Promega, Madison, WI, USA). The firefly and hRenilla luciferase activities in the cell lysates were measured using a luminometer (LB960, Berthold Technologies, Bad Wildbad, Germany). The relative luciferase activities were calculated by normalizing the promoter-driven firefly luciferase activity to phRL-SV (hRenilla) luciferase (Promega, Madison, WI, USA).
Spheroid formation assay
Cells were detached into a single-cell suspension in RPMI medium containing 10% FBS at a density of 5 × 103 cells/mL. The suspended cells (1 × 103 cells/well) were seeded and allowed to form spheroids for 4 days onto ultra-low-attachment (ULA) plates (Costar, Corning Inc., Corning, NY, USA). The number of spheroids (average diameter > 100 μm, Scale bar = 300 μm) was determined.
Transwell migration assay
LNCaP cells were seeded in the upper chamber of the transwell plate (Essen BioScience, Ann Arbor, MI, USA) and the lower chamber was filled with 200 μl serum-containing media. The cells were incubated at 37ºC for 24 h and the migrated cell numbers were counted using IncuCyte ZOOM live-cell analysis system (Essen BioScience, Ann Arbor, MI, USA).
Immunocytochemistry
LNCaP cells were cultured overnight on coverslips. After treatment with 10 μM GV1001 or vehicle, the cells were fixed with 4% paraformaldehyde and blocked with 1% bovine serum albumin at room temperature for 1 h. The cells were incubated with rabbit monoclonal YAP/TAZ or phospho-YAP antibody (1:200 dilution) in 0.1% Tween20-containing PBS at 4 °C overnight. After twice washing with PBS, the coverslips were incubated with goat anti-rabbit Alexa Fluor-488 or -568 IgG (1:2000 dilution, Thermo Fisher Scientific, Waltham, MA, USA) at room temperature with ProLong Gold Antifade reagent with 4’,6-diamidino-2-phenylindole (DAPI, Invitrogen, Carlsbad, CA, USA). Images were obtained using SP8-STED Confocal Microscope System (Leica Microsystems, Heidelberg, Germany).
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was isolated from LNCaP cells using Trizol® reagent (Invitrogen, Carlsbad, CA, USA), and cDNA was synthesized by reverse transcriptase using an oligo(dT) primer. PCR was performed using the selective primers for human MMP2 (sense: 5′-CTTCCAAGTCTGGAGCGATGTG-3′, antisense: 5′-ATGAGCCAGGAGTCCGTCCTTA-3′), human AR (sense: 5′-AGGATGCTCTACTTCGCCCC-3′, antisense: 5′-ACTGGCTGTACATCCGGGAC-3′), human prostate specific antigen (PSA, sense: 5′-ACCAGAGGAGTTCTTGACCCCAAA-3′, antisense: 5′-CCCCAGAATCACCCGAGCAG-3′), human CTGF (sense: 5′-CAAGGGCCTCTTCTGTGACT-3′, antisense: 5′-ACGTGCACTGGTACTTGCAG-3′), human NKX3.1 (sense: 5′-GCCGCACGAGCAGCCAGAGACA-3′, antisense: 5′-TTCAGGGCCGGCAAAGAGGAGTG-3′), human GAPDH (sense: 5′-AAGGCTGAGAACGGGAAG-3′, antisense: 5′-GCCCCACTTGATTTTGGA-3′). The SYBR Green real-time PCR amplifications were conducted with MiniOpticon real time PCR detection system (Bio-Rad laboratories Inc., Hercules, CA, USA).
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed according to the EZ-ChIP kit (Milipore, Bedford, MA, USA). An anti-AR antibody (Cat# sc-819, Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used to precipitate DNA–protein complexes. ChIP-derived DNA was quantified using qRT-PCR with SYBR Green real-time PCR Mastermix (Bio-Rad laboratories Inc., Hercules, CA, USA). The primers specific for the promoters were as follows: human PSA (sense: 5′-GCTAGCACTTGCTGTTCTGC-3′, antisense: 5′-GGGATCAGGGAGTCTCACAA-3′), human CTGF (sense: 5′-TGTGCCAGCTTTTTCAGACG-3′, antisense: 5′-TGAGCTGAATGGAGTCCTACACA-3′) and human NKX3.1 (sense: 5′-CCGAGCCAGAAAGGCACTTG-3′, antisense: 5′-CTTAGGGGTTTGGGGAAGCC-3′).
Spleen-liver metastasis
5-Week-old male BALB/c-nu mice (Jung Ang Lab Animal Inc., Seoul, South Korea) were anaesthetized and an abdominal incision was made in the left flank and the spleen was isolated. 1 × 105 LNCaP cells in 100 μl PBS were injected into the spleen with a 26-gauge needle, and the incision was closed with stitches. After 3 days, mice were divided into three groups: vehicle, GV1001 (10 mg/kg, daily, sc) and LA (0.1 mg/kg, daily, sc) treatment groups. After 4 weeks, the mice were sacrificed and the liver and spleen samples were analyzed by hematoxylin & eosin (H&E) staining. For pathological assessment, specimens were cut into 2 mm thick sections after formalin fixation. All abnormal regions (nodules or suspicious regions) were observed under a microscope, and the absolute metastatic tumor area was analyzed. Basically all abnormal regions were analyzed (3–4 slides per liver tissue) when nodules are found.
Protein–protein interaction (PPI) network
The STRING database (http://string-db.org) [26] was used to analyze the protein–protein interactions. The STRING database was applied to predict the PPIs by DEGs, and the parameter of combined score > 0.4 was set as the threshold value for choosing significant interactions. The nodes in the PPI network were ranked by their connectivity degrees, which correspond to the number of interactions by other proteins.
Statistics
Statistical analysis was performed using one-way ANOVA and Tukey’s post hoc multiple comparisons to determine the differences. The statistical significance was accepted at *P < 0.05 and **P < 0.01.
Results
Anti-migratory and anti-metastatic effects of GV1001 on human malignant PCa
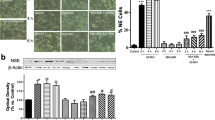
We recently reported that Gαs-cAMP signaling is selectively activated by GV1001 binding to GnRHR, but not by LA, a representative GnRHR agonist [22]. We compared cAMP-dependent transcriptional activities of CA (a representative GnRHR antagonist), LA and GV1001 in GnRHR-overexpressing HEK293 cells. Among the three ligands, only GV1001 increased the reporter activity of the cAMP-responsive element, which confirms that GV1001 is a unique GnRHR ligand to selectively activate Gαs coupling (Fig. 1A). GV1001 also increased the CRE reporter activity in LNCaP cells (Fig. 1B) [22]. We then raised a question that which phenotype is under the control of cAMP increase by GnRHR/Gαs coupling. In our previous study, we revealed that the proliferation of PCa cells was slightly inhibited by both GV1001 and LA, but cell migration was only suppressed by GV1001 [22]. As we found the reduced mRNA levels of matrix metalloproteinase (MMP) 2 and MMP9 in tumor tissues from LNCaP xenografts injected with GV1001 [22], we further assessed both mRNA and protein expressions of MMP2 and MMP9 in LNCaP cells exposed to vehicle or GV1001. The MMP2 mRNA expression was significantly decreased by 10 μM GV1001, but MMP9 mRNA was not detected (data not shown) in LNCaP cells (Fig. 1C). Moreover, cleavage of MMP2 indicating the activation of MMP2 was also reduced by 10 μM GV1001 (Fig. 1D). From the above results and previous findings, we hypothesized that the anti-migratory effect of GV1001 is related to Gαs activation via GnRHR binding. To clarify whether the reduced cell motility by GV1001 is associated with the epithelial mesenchymal transition (EMT) process, we examined the levels of mesenchymal markers in LNCaP cells. We found that the protein levels of N-cadherin, vimentin, and the major transcription factors of EMT, slug and twist in LNCaP cells were significantly reduced by GV1001 (0.1–10 μM) (Fig. 1E). Since spheroid formation on a 3D-culture or ultra-low attachment (ULA) plate is a whole marker for EMT progress of cancer cells [27, 28], we then performed the spheroid formation assay. Spheroid formation on the ULA plate was significantly inhibited by 10 μM GV1001, but not by 100 nM LA (Fig. 1F). The data indicate that GV1001 inhibits migration of LNCaP cells presumably via targeting the EMT process. To determine the in vivo efficacy of GV1001, anti-metastasis activity was assessed in the spleen-liver metastasis model. GV1001 (10 mg/kg, daily sc)-injected group showed the lower incidence rate of liver metastasis (5/10, P value = 0.044) compared to vehicle (sterile water, daily sc) (7/7)- or LA (0.1 mg/kg, daily sc)-injected group, with no liver weight loss (Fig. 1G, H). Moreover, tumor area in liver tissues was also potently diminished in GV1001 injected mice compared to vehicle-injected mice (Fig. 1I).
Effects of GV1001 on LNCaP-cell migration and tumor metastasis. A Effects of CA, LA, and GV1001 on CRE-reporter activity in HEK293-pcDNA3.1(+) and HEK293-GnRHR cells. The cells were exposed to CA (100 nM), LA (100 nM), or GV1001 (10 μM) for 24 h. Data represent means ± SD (n = 6, ***P < 0.005 vs vehicle-treated control). B Effect of 10 μM GV1001 on CRE-reporter activity in LNCaP cells. Data represent means ± SD (n = 6, ***P < 0.005 vs vehicle-treated control). C mRNA levels of MMP2. RT-qPCR was performed in LNCaP cells treated with LA (100 nM) or GV1001 (10 μM) for 24 h. D Determination of pro-MMP2 and cleaved-MMP2 proteins in LNCaP cells. LNCaP cells were incubated with or without GV1001 (0.1–10 μM) or LA (1–100 nM) for 24 h, and the cell lysates were subjected to immunoblotting using specific antibodies. E Protein expressions of EMT-related genes in LNCaP cells. LNCaP cells were incubated with GV1001 (0.1–10 μM) for 24 h. F Effects of GV1001 or LA on the spheroid formation of LNCaP cells. LNCaP cells were seeded in a 96-well ULA plate and treated with LA (100 nM) or GV1001 (10 μM) for 96 h. The number of spheroids (average diameter > 100 μm) was determined. Scale bar = 300 μm. Data represent means ± SD (n = 4, *P < 0.05 vs vehicle-treated control). G–I Spleen-liver metastasis in BALB/c-nude mice implanted with LNCaP cells into their spleens. After implantation, the mice were divided into 3 groups (n = 7–10) and then injected with vehicle (sterile water 100 μl, daily, sc), GV1001 (10 mg/kg, daily, sc) or LA (0.1 mg/kg, daily, sc) for 4 weeks. G Representative pictures of the metastatic liver tumors from each group are shown. Scale bar = 10 mm. H Statistical analysis of the incidence of liver metastatic colonization (the percentage of mice with metastatic tumor burden in the liver) and weights of tumor-bearing livers. Data represent means ± SD (n = 7–10, P = 0.044 vs vehicle-treated control, The P-value was calculated by two-sided Fisher exact test.). I Histological analysis of the metastasized-liver section. Data represent means ± SD (n = 7–10, **P < 0.01 vs vehicle-treated control)
Inhibition of PCa cell migration by AR activation
EMT phenotype is closely correlated with AR signaling in castration-resistant PCa cells [29,30,31]. When we analyzed microarray data (GSE66852) of four different PCa cell lines, mRNA expressions of AR in all PCa cell lines were significantly decreased by developmental stem-transition reprogramming (Fig. 2A). We then examined the effects of AR activation on the migration of LNCaP cells. The number of migrated cells was significantly increased by 1 μM flutamide, AR antagonist compared to vehicle-treated cells, and the enhanced cell migration by flutamide was reversed by dihydrotestosterone, an active form of androgen (Fig. 2B). To confirm these results, we established AR knockout LNCaP cells using the CRISPR/Cas9 gene-editing system (Fig. 2C). The number of migrated AR knockout LNCaP cells was 2.2-fold higher than that of control cells (Fig. 2D). The data support the notion that AR activation negatively regulates the migratory capacity of AR-positive PCa cells.
Role of AR activity in LNCaP cell migration. A Gene expression of AR in therapy-induced developmental reprogrammed-PCa cells (n = 3) as compared to parental PCa cell lines (GSE66852). Data were shown as box and whisker plots. Box, interquartile range (IQR); whiskers, 5–95 percentiles; and horizontal line within the box, median. B AR-mediated inhibition of LNCaP cell migration. LNCaP cells were treated with flutamide (AR antagonist, 1 μM) in the presence or absence of dihydrotestosterone (DHT, AR agonist, 2 nM) for 24 h. Migratory cell numbers were determined by the IncuCyte ZOOM live-cell analysis system. Data represent means ± SD (n = 4, ***P < 0.005 vs vehicle-treated control, #P < 0.05 vs flutamide-treated group). C Development of AR knockout LNCaP cells by CRISPR/Cas9 system. Expression of AR was determined by immunoblot analysis. D Migration of AR knockout LNCaP cells. Migratory cell numbers were determined by IncuCyte ZOOM live-cell analysis system for 24 h. Data represent means ± SD (n = 4, *P < 0.05, **P < 0.01 vs vehicle-treated control)
AR activation by GV1001 in PCa cells
We determined Ser-81 phosphorylation AR in LNCaP cells to examine the involvement of AR activation in GV1001-mediated inhibition of cell migration. Notably, we observed that 10 μM GV1001increased the phosphorylation of AR 12 h after exposure, and the pattern was similar to dihydrotestosterone treatment (Fig. 3A). We also found that AR protein expression, as well as AR phosphorylation, concentration-dependently increased by GV1001 (0.1–10 μM) (Fig. 3B). To assess the effect of GV1001 on AR-mediated transcriptional activity, androgen response element (ARE)-driven reporter activity was determined. Although the induction intensity was lower than dihydrotestosterone, GV1001 (0.1–10 μM) increased the ARE-reporter activity (Fig. 3C). Moreover, the additive increase in ARE-reporter activity was seen in LNCaP cells cotreated with GV1001 (10 μM) and 100 nM dihydrotestosterone (Fig. 3D), which imply that GV1001 acts as a ligand-independent AR activator. Because we previously revealed that GV1001 is a novel ligand of GnRHR, AR phosphorylation effects were assessed by using classical GnRHR ligands. Both GnRH and LA, representative endogenous and synthetic GnRHR ligands mainly activating the Gαq pathway, did not cause any significant phosphorylation of AR (Fig. 3E). We next assessed the effect of GV1001 on flutamide-induced cell migration in LNCaP cells. The enhanced migration was significantly attenuated by 10 μM GV1001 (Fig. 3F). These results indicate that AR activation may contribute to the inhibitory effects of GV1001 on cell migration.
GV1001-induced AR activation in LNCaP cells. A Time-dependent AR phosphorylation (Ser-81) by dihydrotestosterone (DHT, 100 nM) or GV1001 (10 μM) in LNCaP cells. All of the results were confirmed by multiple experiments. B Total AR expression and AR phosphorylation in LNCaP cells treated with GV1001 (0.1–10 μM) for 24 h. C Effect of GV1001 on ARE-reporter activity. LNCaP cells were transiently transfected with ARE-luciferase plasmid (1 μg/ml) and the cells were exposed to DHT (100 nM) or GV1001 (0.1–10 μM) for 24 h. Luciferase activity was determined to assess ARE responsiveness. Data represent means ± SD (n = 6, **P < 0.01 vs vehicle-treated control). D An additive increase in ARE-reporter activity by GV1001 and DHT. GV1001 (0.1–10 μM) was simultaneously treated with dihydrotestosterone (100 nM) in LNCaP cells for 24 h. Data represent means ± SD (n = 8, **P < 0.01 vs vehicle-treated control, #P < 0.05 vs DHT-treated group). E Expression and phosphorylation of AR by GnRH analogs. LNCaP cells were treated with GnRH (0.1–10 μM) or LA (1–100 nM) for 24 h. F Inhibitory effect of GV1001 on flutamide-stimulated LNCaP cell migration. LNCaP cells were treated with flutamide (1 μM) and GV1001 (10 μM) for 24 h. Migratory cell numbers were determined by the IncuCyte ZOOM live-cell analysis system. Data represent means ± SD (n = 4, **P < 0.01 vs vehicle-treated control, #P < 0.05 vs flutamide-treated group)
Gαs/cAMP-dependent AR phosphorylation by GV1001
Although most GnRHR ligands preferentially activate Gαq-calcium signaling, our previous study demonstrated that GV1001 binding to GnRHR selectively activates Gαs-cAMP signaling [22]. To identify the upstream signaling for Ser-81 phosphorylation of AR, we used diverse inhibitors targeting G protein signaling. LNCaP cells were treated with 10 μM BAPTA/AM (a cell-permeable calcium chelator, Gαq inhibitor), 5 μM KH7 (a specific soluble adenylate cyclase inhibitor, Gαs inhibitor), 100 ng/ml pertussis toxin (PTX, an ADP-ribosylating toxin, an inhibitor of Gαi) in the presence or absence of 10 μM GV1001. KH7 blocked GV1001-stimulated AR phosphorylation (Fig. 4A). Moreover, KH7 almost completely suppressed ARE-driven reporter activity induced by 10 μM GV1001 (Fig. 4B). Vice versa, forskolin (0.1–10 μM), an adenylate cyclase activator, increased AR phosphorylation in LNCaP cells (Fig. 4C), and the enhanced AR phosphorylation and ARE reporter activity by 10 μM forskolin was inhibited by KH7 (Fig. 4D, E). As shown in Fig. 4A, cotreatment with BAPTA/AM and GV1001 potently increased AR phosphorylation. Thus, in order to determine whether Gαq-calcium signaling is linked to AR activity, we further tested the effect of LA, a representative Gαq-stimulating ligand of GnRHR on AR phosphorylation. Interestingly, 100 nM LA abrogated dihydrotestosterone-stimulated AR phosphorylation (Fig. 4F) and ARE-reporter activity (Fig. 4G). The data suggest that AR phosphorylation and its transcriptional activity are reciprocally controlled by GnRHR-coupled Gαs and Gαq (Fig. 4H).
Involvement of Gαs-cAMP signaling pathway in GV1001-stimulated AR activation. A Effects of Gα signaling inhibitors on GV1001-stimulated AR phosphorylation in LNCaP cells. LNCaP cells were exposed to various Gαs inhibitors, a cell-permeable cytosolic calcium chelator BAPTA/AM (10 μM), a specific adenylyl cyclase inhibitor KH7 (5 μM) and an ADP-ribosyltransferase of Gαi, pertussis toxin (100 ng/ml) with 10 μM GV1001. All of the results were confirmed by multiple experiments. B Inhibitory effect of KH7 on GV1001-induced ARE-reporter activity. LNCaP cells were transiently transfected with ARE-luciferase plasmid (1 μg/ml) and the cells were exposed to KH7 (5 μM) and GV1001 (10 μM) for 24 h. Luciferase activity was determined to assess ARE responsiveness. Data represent means ± SD (n = 5, **P < 0.01 vs vehicle-treated control, ###P < 0.005 vs GV1001-treated group). C AR activation by forskolin. LNCaP cells were treated with forskolin, a cAMP activator (0.1–10 μM) for 24 h, and protein expression of AR and phosphorylated AR was assessed by immunoblotting. D Inhibitory effect of KH7 on forskolin-stimulated AR expression and phosphorylation. LNCaP cells were exposed to 5 μM KH7 and 10 μM forskolin for 24 h. E Inhibitory effect of KH7 on forskolin-induced ARE-reporter activity in LNCaP cells. LNCaP cells were transiently transfected with ARE-luciferase plasmid (1 μg/ml) and the cells were exposed to KH7 (5 μM) and forskolin (10 μM) for 24 h. Luciferase activity was determined to assess ARE responsiveness. Data represent means ± SD (n = 6, **P < 0.01 vs vehicle-treated control, ##P < 0.01 vs forskolin-treated group). F, G Inhibitory effect of LA on DHT-induced AR activation. F Inhibitory effect of LA on DHT-induced AR phosphorylation. LNCaP cells were pretreated with LA (100 nM) for 1 h and treated with DHT (100 nM) for 24 h. G Inhibitory effect of LA on DHT-induced ARE-reporter activity. LNCaP cells were transiently transfected with ARE-luciferase plasmid (1 μg/ml) and the cells were exposed to DHT (100 nM) and LA (100 nM) for 24 h. Luciferase activity was determined to assess ARE responsiveness. Data represent means ± SD (n = 6, **P < 0.01 vs vehicle-treated control, ###P < 0.005 vs DHT-treated group). H Hypothetical diagram for the differential effects of GV1001 and LA on AR
AR coregulator switching by GV1001
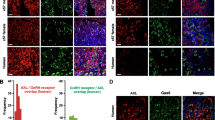
The transcriptional activity of AR is modulated by numerous AR coregulators [32]. To explore the molecular mechanism by which AR activation suppresses PCa cell migration, we performed a network analysis of AR using STRING, a well-known database of both known and predicted protein–protein interactions (Fig. 5A). Because we have focused on the tumor-suppressive function of AR, NKX3.1, a transcriptional repressor (Combined Score: 0.981) was identified (Fig. 5A). We then analyzed the public ChIP-seq database (ChIP-Atlas) and identified colocalization of AR and NKX3.1 at the promoter region of NKX3.1 (Fig. 5B). These results imply that the AR regulates transcription of the target gene in combination with NKX3.1, a coregulator of AR, as previously reported [33, 34]. It has been reported that YAP1, a transcription factor regulated by the Hippo pathway, interacts with AR and contributes to the growth in PCa [35]. When we analyzed NCBI GEO data set (GSE6919) of normal prostate tissues, prostate tumor tissues, and metastatic prostate tumor tissues in PCa patients, the mRNA expression of YAP1 was significantly increased in metastatic prostate tumor tissues compared to primary ones (Fig. 5C). In contrast, expression of NKX3.1 was reduced in metastatic tissues compared to primary ones (Fig. 5C). Immunoprecipitation analyses were performed to evaluate if YAP1 or NKX3.1 was involved in the anti-migratory action of GV1001. The protein level of YAP1 binding to AR was reduced by GV1001 treatment in LNCaP cells (Fig. 5D). On the contrary, the binding of NKX3.1 with AR was sharply increased by GV1001 (Fig. 5D), and western blot analyses further indicated that the total protein expression of NKX3.1 was increased by GV1001 in LNCaP cells (Fig. 5E). Subcellular fractionation also showed the cytoplasmic retention and the subsequent nuclear loss of YAP1 in GV1001-exposed cells (Fig. 5F). Immunocytochemistry further confirmed the localization of total and Ser-127 phosphorylated YAP1 in the cytoplasm (Fig. 5G). Hippo pathway-induced YAP1 phosphorylation eventually leads to YAP/TAZ ubiquitination and degradation [36]. We also found that GV1001 accelerated the ubiquitination of YAP1 in LNCaP cells (Fig. 5H). To test whether GV1001-stimulated Gαs-cAMP signaling affects the phosphorylation of YAP1, we used adenylate cyclase inhibitors, KT5720 and KH7. GV1001-induced YAP1 phosphorylation was attenuated by treatment with KT5720 (10 μM) or KH7 (5 μM) (Fig. 5I). We then determined mRNA expressions of AR target genes, AR, PSA, and NKX3.1, and a representative YAP target gene, CTGF using quantitative PCR analyses. We revealed that the mRNA expression of AR target genes, AR, PSA, and NKX3.1, was highly upregulated by 10 μM GV1001 (Fig. 5J). In contrast, GV1001 potently reduced the mRNA level of CTGF (Fig. 5J). These data indicate that GV1001-stimulated Gαs signaling evokes both inhibition of YAP1 transcriptional activity and upregulation of AR transcriptional activity via common cAMP/PKA pathway. We further assessed promoter binding activities of the AR complex using ChIP analyses (Fig. 5K). AR complex enrichments at the promoter region of AR target genes, PSA and NKX3.1, were significantly enhanced by GV1001 treatment (Fig. 5K). However, AR complex enrichment at the CTGF gene promoter, a representative YAP1 target gene, was decreased by GV1001 (Fig. 5K). Collectively, these results suggest that GV1001 inactivates YAP1 via Gαs-cAMP signaling and subsequently suppresses the AR interaction with YAP1, and consequently switches AR-coregulator to NKX3.1.
AR coregulator switching by GV1001. A Protein–protein interaction network analysis of predicted and reported functional partners of AR. Gene interactions were constructed using the STRING online database (http://string-db.org/). Network nodes represent proteins and the edges represent protein–protein associations. B Binding probability of AR and NKX3.1 at the transcription initiation site of the human NKX3.1 and AR genes. Experimental data from the chromatin immunoprecipitation (ChIP) atlas (http://chip-atlas.org/) are shown using Integrative Genomics Viewer (IGV) software. C Expressions in normal prostate tissues, primary prostate tumor tissues, and metastatic prostate tumor tissues from prostate cancer patients (n = 164) (GSE6919). DATA were shown as box and whisker plots. Box, interquartile range (IQR); whiskers, 5–95 percentiles; and horizontal line within the box, median. D Protein interaction of YAP1 or NKX3.1 with AR. LNCaP cells were treated with GV1001 (10 μM) for 24 h, and reciprocal immunoprecipitation and immunoblotting were performed. All of the results were confirmed by multiple experiments. E GV1001 or LA-induced NKX3.1 expression. LNCaP cells were treated with GV1001 (0.1–10 μM) or LA (1–100 nM) for 24 h. F Cytoplasmic retention of YAP1 by GV1001. LNCaP cells were treated with GV1001 (10 μM) for 24 h, and subcellular fractionation and subsequent immunoblot analyses were performed. G Immunocytochemistry of YAP1 and phospho-YAP1 proteins in LNCaP cells. LNCaP cells were treated with 10 μM GV1001 for 4 h. Alexa Fluor 568 stained YAP1 (red), Alexa Fluor 488 stained phospho-YAP1 (green) and DAPI stained nuclei (blue). Magnification: 60X. H Ubiquitination induction by GV1001. LNCaP cell lysates were immunoprecipitated with YAP1 antibody and analyzed by immunoblotting with anti-ubiquitin antibody to detect polyubiquitinated YAP1. I Inhibitory effects of KT5720 (upper) and KH7 (lower) on GV1001-induced YAP1 phosphorylation. KT5720 (10 μM) or KH7 (5 μM) was added 1 h before 10 μM GV1001 treatment in LNCaP cells, and 24 h after total cell lysates were subjected to immunoblotting for phospho-YAP1 and YAP1. J mRNA levels of target genes for AR/coregulator complex. RT-qPCR was performed in LNCaP cells treated with DHT (100 nM) or GV1001 (10 μM) for 24 h. K ChIP-qPCR analysis of AR recruitment to the promoter region of PSA, CTGF or NKX3.1 gene. LNCaP cells were treated with DHT (100 nM), GV1001 (10 μM) for 24 h. Input and immunoprecipitated samples were analyzed by qPCR for PSA, CTGF, or NKX3.1 promoter region. ChIP-qPCR data are presented as a percentage of input. Data represent means ± SD (n = 6, **P < 0.01 vs vehicle-treated control)
Involvement of YAP1 in the migration and proliferation of PCa cells
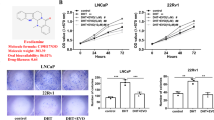
To further define whether GV1001-mediated YAP1 inactivation is related to the reduced PCa cell migration, YAP1 knockout LNCaP cells were established by CRISPR/Cas9 gene editing. Although we did not identify any significant changes in YAP1 protein levels in all clones (Additional file 1: Fig. S1A), T7E1 analysis showed that the YAP1 gene was edited in YAP1 knockout #7 clone (Additional file 1: Fig. S1B). Moreover, the cell migration intensity of the YAP1 knockout #7 clone (LNCaP-YAP1 sgR/Cas9 cells) was diminished compared to negative control-transfected LNCaP cells (LNCaP-control sgR/Cas9 cells) (Fig. 6A). YAP/TAZ-responsive reporter gene assay using TEAD promoter confirmed that the YAP1-dependent gene transcription was reduced in LNCaP-YAP1 sgR/Cas9 cells both in the presence or absence of serum (Fig. 6B). The migratory capacity of LNCaP-YAP1 sgR/Cas9 cells was also decreased both in the presence or absence of serum (Fig. 6C) We then tested the effects of GV1001 on the cell migration of YAP1 knockout cells. Exposure of LNCaP-control sgR/Cas9 cells to 10 μM GV1001 suppressed the cell migration, but the LNCaP-YAP1 sgR/Cas9 cells did not show any significant response in the presence of GV1001 (Fig. 6D). We then established LNCaP cells expressing mutant YAP1 in which the serine residue targeted by LATS1/2 and necessary for cytoplasmic retention are substituted with alanine (YAP5SA) (Fig. 6E)[37]. Notably, the transwell migration assay demonstrated that the basal cellular migration capacity of LNCaP-YAP5SA cells was increased (Fig. 6F). However, GV1001 treatment (10 μM) did not inhibit cell migration of LNCaP-YAP5SA cells expressing constitutively active YAP1 (Fig. 6F). Thus, the anti-migratory effect of GV1001 seems to be related to the suppression of Hippo pathway-dependent YAP1 activation. To check if cAMP/PKA activation by GV1001 is essential for the YAP1 phosphorylation and subsequent inactivation, we further used an adenylate cyclase activator, forskolin. Forskolin (0.3–10 μM) increased Ser-127 phosphorylation of the YAP1 (Fig. 6G). Moreover, forskolin also suppressed the LNCaP cell migration (Fig. 6H). We further confirmed that the Gαs-cAMP signaling inhibitors, KT5720 (1 μM) and KH7 (1 μM), restored the GV1001-induced inhibition of cell migration (Fig. 6I). Taken together, these findings support the concept that GV1001 phosphorylates YAP1 via Gαs-cAMP-mediated hippo pathway activation and inhibits the migration and metastasis of PCa cells.
Involvement of YAP1 in LNCaP cell migration. A Migration of YAP1 knockout LNCaP cells. Migratory cell number was determined by IncuCyte ZOOM live-cell analysis system. Data represent means ± SD (n = 4, *P < 0.05 vs control cell). B YAP/TAZ-reporter activity of YAP knockout LNCaP cells. Control and YAP knockout LNCaP cells were transiently transfected with YAP/TAZ-reporter plasmid (TEAD reporter, 1 μg/ml) and the cells were incubated with or without 10% FBS for 24 h. Luciferase activity was determined to assess YAP/TAZ-mediated gene transcription. Data represent means ± SD (n = 6, **P < 0.01 vs control LNCaP cells). C Diminished cell migration by YAP knockout in serum-containing and serum-deprived conditions. Data represent means ± SD (n = 4, *P < 0.05, **P < 0.01 vs vehicle-treated control). D Effect of GV1001 on YAP1 knockout LNCaP cell migration. Control- and YAP1-knockout LNCaP cells were treated with 10 μM GV1001 for 24 h, and cell migration was quantified by IncuCyte ZOOM live-cell analysis system. Data represent means ± SD (n = 4, *P < 0.05 vs vehicle-treated control). E Establishment of constitutive active YAP5SA-LNCaP cells. Protein expression of YAP1, Myc, and CTGF were determined by immunoblot analyses. F Effect of GV1001 on the migration of YAP5SA-overexpressing LNCaP cells. Control and YAP-5SA cells were exposed to 10 μM GV1001 for 24 h. Data represent means ± SD (n = 4, **P < 0.01 vs vehicle-treated control, ##P < 0.01 vs control-LNCaP cell group). G Forskolin-induced YAP1 phosphorylation. LNCaP cells were treated with 0.1–10 μM Forskolin for 1 h, and protein levels of YAP1 and p-YAP1 were determined by immunoblot analysis. H Effect of forskolin on LNCaP cell migration. LNCaP cells were treated with forskolin (10 μM) for 24 h. Data represent means ± SD (n = 4, **P < 0.01 vs vehicle-treated control). I Reversal of GV1001-induced anti-migratory activity by KT5720 and KH7. LNCaP cells were incubated with 10 μM GV1001 in the presence or absence of KT5720 (1 μM) or KH7 (1 μM) for 24 h. Data represent means ± SD (n = 4, *P < 0.05, **P < 0.01 vs vehicle-treated control, ###P < 0.005 vs GV1001-treated group)
Discussion
Ligand-stimulated GnRHR triggers multiple signaling pathways through diverse Gα proteins. Among the G protein pathways, the activated Gαq mediates reproductive function in pituitary cells and is considered as the primary pathway for GnRHR [7,8,9]. It has been also reported that GnRH analogs possess inhibitory effects in several cancer cell types via Gαi protein. GnRHR ligands-stimulated Gαi activation inhibited cell proliferation of endometrial (HEC-1A and Ishikawa) and ovarian cancer cells (EFO-21 and EFO-27) through the suppression of mitogenic signal transduction [38,39,40,41,42]. However, the Gαs-mediated pharmacological function of the GnRHR ligand is still unclear. In our previous study, we identified GV1001 as a novel GnRHR ligand to selectively activate Gαs/cAMP pathway and demonstrated that the peptide showed an inhibiting effect on tumor growth in LNCaP cells-implanted xenograft model [22]. It has been reported that the anti-proliferation effect of GnRH-II in PCa cells is associated with GnRHR and Gαs/cAMP pathway [43]. However, the precise role of GnRHR-mediated Gαs-cAMP signaling in cancer progression needs to be further clarified. In the present study, we showed for the first time that GV1001 suppressed EMT of PCa cells and cancer cell migration in vitro. In addition, GV1001 potently inhibited liver metastasis of PCa cells implanted in the spleen in vivo.
Because AR plays a pivotal role in the development of PCa, AR targeting by inhibiting either biosynthesis of androgens or ligand-receptor interaction has been the mainstream of current therapeutic intervention for PCa malignancy [44, 45]. However, AR has been suggested as a key negative factor for the EMT process and metastasis of prostate cancer. A series of recent reports have demonstrated that inhibition of AR activity contributes to the cell migration and EMT process of PCa [29, 31, 46].
Here, we also revealed that both AR inactivation and AR deletion promote the migratory capacity of LNCaP cells. Unexpectedly, we found that GV1001 treatment stimulates AR transcriptional activation with an increase in AR phosphorylation in LNCaP cells. GV1001 as a selective ligand for Gαs/cAMP pathway phosphorylates Ser-81 of AR, which is required for the inhibition of EMT and cell migration of LNCaP cells. Conversely, we found that LA, a GnRHR-Gαq activator, inhibits AR phosphorylation via the Gαq-calcium signaling pathway. There is still much that remains unknown about the relationship between AR phosphorylation and Gαs-cAMP signaling. One possible explanation for this is that PKA could phosphorylate not only AR but also HSP90, which is a binding partner of AR. It has been reported that AR phosphorylation in the hinge region, which is critical for nuclear import and transcriptional activity, could be induced by PKA [47]. However, whether PKA directly phosphorylates AR or not is still unclear since this process involves multiple other molecules. Nevertheless, PKA could be potential candidate for mediating AR phosphorylation and transactivation in PCa cells.
Coregulator recruitment is an important step for the control of AR-dependent gene transcription. Depending on the type of coregulator, the transcription of the AR target genes and the cancer progression could be affected [48]. For example, transcription intermediary factor2 (TIF2), a coregulator of AR, is upregulated in response to interleukin-6 and promotes resistance to bicalutamide, an AR antagonist [49]. NKX3.1, a prostate epithelial factor is a homeodomain protein that plays an important role in the advance of PCa [50]. NKX3.1 is known to be one of the target genes of AR activation and inhibit the proliferation and tumorigenesis of PCa [51]. NKX3.1 deficient LNCaP cells showed enhanced cell proliferation, metastasis to the lymphatic system and tumor growth in vivo [52]. Hippo pathway functions as a tumor suppressor signal in normal cells and regulates cell proliferation, apoptosis, and progenitor cell development [53, 54]. The Hippo signaling consists of the mammalian ste2-like kinases (MST1/2) and large tumor suppressor kinase (LATS1/2) in mammals [55]. Activation of the Hippo pathway leads to the degradation of YAP1 after LATS1/2-mediated Ser-127 phosphorylation of YAP1. In an environment where the Hippo pathway is suppressed, the accumulated YAP1 enters the nucleus and promotes the transcription of various oncogenic genes such as cyr61 and ctgf [56]. Overexpression of the YAP1 target genes, including CTGF and Cyr61, is actively involved in cell proliferation, reprogramming, stemness, EMT, anti-apoptosis, and chemoresistance acquisition in cancer cells [54, 57, 58]. Recent studies have identified that YAP1 is a coregulator of AR in PCa cells [35, 59]. Here, we identified that GV1001 treatment inactivated YAP1 through Gαs/cAMP-dependent phosphorylation, vice versa induced protein expression of NKX3.1. Immunoprecipitation and ChIP analyses confirmed that coregulator binding of AR was switched from YAP1 to NKX3.1 in PCa cells exposed to GV1001. The switch of AR coregulators consequentially leads to changes in transcriptional regulation of AR target genes, which seems to be related to inhibition of EMT and cell migration in PCa cells. We have previously shown that GV1001 treatment suppresses proliferation and migration of AR-negative PC-3 cells [22]. Hence, it could be possible that YAP1 inactivation by GV1001 inhibits cell migration in an AR-independent manner.
A recent study reported that the activity of the Hippo-YAP pathway could be controlled by GPCRs coupled with the individual G protein. Selective agonists on GPCRs activating G12/13, Gαq, or Gαi protein stimulate the transcriptional activity of YAP1, but Gαs coupled GPCR signaling inhibits YAP/YAZ [60].
Conclusion
Collectively, these observations suggest a new paradigm that GnRHR-driven inactivation of the YAP1-AR axis could be a therapeutic target for prostate cancer. We have previously shown that GV1001 as a GnRHR biased ligand, suppresses testosterone production as well as tumor growth of LNCaP xenograft in vivo [22]. Because GV1001, as a new GnRHR ligand and a YAP1 inhibitor, represents a potential therapeutic for prostate cancer without causing cancer cell migration and metastasis by AR depletion (Fig. 7).
Schematic diagram for anti-metastatic effect of GV1001. GV1001-stimulated GnRHR/Gαs/cAMP pathway simultaneously causes both the activation of AR and the inactivation of YAP via phosphorylation of Ser-81 AR and Ser-127 YAP1, respectively. NKX3.1 upregulation by AR activation and YAP1 degradation in GV1001-treated PCa cells switches AR-YAP1 binding to AR-NKX3.1 binding
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Abbreviations
- ADT:
-
Androgen deprivation therapy
- AR:
-
Androgen receptor
- ARE:
-
Androgen-response element
- CA:
-
Cetrorelix acetate
- CRE:
-
CAMP response element
- CRPC:
-
Castration-resistant prostate cancer
- DAPI:
-
4′6-Diamidino-2-phenylindole
- DMF:
-
Dimethylformamide
- EDC:
-
1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride
- EDTA:
-
Ethylenediamine tetraacetic acid
- EGTA:
-
Ethyleneglycol tetraacetic acid
- FITC:
-
Fluorescein isothiocyanate
- FSH:
-
Follicle-stimulating hormone
- GnRH:
-
Gonadotropin-releasing hormone
- GnRHR:
-
Gonadotropin-releasing hormone receptor
- HOBt:
-
Hydroxybenzotriazole
- HRP:
-
Horseradish peroxidase
- hTERT:
-
Human telomerase reverse transcriptase catalytic subunit
- LA:
-
Leuprolide acetate
- LH:
-
Luteinizing hormone
- MMP2:
-
Matrix metalloproteinase-2
- MMP9:
-
Matrix metalloproteinase-9
- NSCLC:
-
Non-small cell lung cancer
- PBS:
-
Phosphate buffered saline
- PCa:
-
Prostate cancer
- PSA:
-
Prostate-specific antigen
- SDS:
-
Sodium dodecyl sulfate
References
Millar RP, Lu ZL, Pawson AJ, Flanagan CA, Morgan K, Maudsley SR. Gonadotropin-releasing hormone receptors. Endocr Rev. 2004;25(2):235–75.
Naor Z, Huhtaniemi I. Interactions of the GnRH receptor with heterotrimeric G proteins. Front Neuroendocrinol. 2013;34(2):88–94.
Reinhart J, Mertz LM, Catt KJ. Molecular cloning and expression of cDNA encoding the murine gonadotropin-releasing hormone receptor. J Biol Chem. 1992;267(30):21281–4.
Tsutsumi M, Zhou W, Millar RP, Mellon PL, Roberts JL, Flanagan CA, Dong K, Gillo B, Sealfon SC. Cloning and functional expression of a mouse gonadotropin-releasing hormone receptor. Mol Endocrinol (Baltimore, Md). 1992;6(7):1163–9.
Hawes BE, Barnes S, Conn PM. Cholera toxin and pertussis toxin provoke differential effects on luteinizing hormone release, inositol phosphate production, and gonadotropin-releasing hormone (GnRH) receptor binding in the gonadotrope: evidence for multiple guanyl nucleotide binding proteins in GnRH action. Endocrinology. 1993;132(5):2124–30.
Stanislaus D, Ponder S, Ji TH, Conn PM. Gonadotropin-releasing hormone receptor couples to multiple G proteins in rat gonadotrophs and in GGH3 cells: evidence from palmitoylation and overexpression of G proteins. Biol Reprod. 1998;59(3):579–86.
Hille B, Tse A, Tse FW, Almers W. Calcium oscillations and exocytosis in pituitary gonadotropes. Ann N Y Acad Sci. 1994;710:261–70.
Voliotis M, Garner KL, Alobaid H, Tsaneva-Atanasova K, McArdle CA. Gonadotropin-releasing hormone signaling: an information theoretic approach. Mol Cell Endocrinol. 2018;463:106–15.
Grosse R, Schmid A, Schoneberg T, Herrlich A, Muhn P, Schultz G, Gudermann T. Gonadotropin-releasing hormone receptor initiates multiple signaling pathways by exclusively coupling to G(q/11) proteins. J Biol Chem. 2000;275(13):9193–200.
Krsmanovic LZ, Mores N, Navarro CE, Arora KK, Catt KJ. An agonist-induced switch in G protein coupling of the gonadotropin-releasing hormone receptor regulates pulsatile neuropeptide secretion. Proc Natl Acad Sci USA. 2003;100(5):2969–74.
Kang SK, Choi KC, Yang HS, Leung PC. Potential role of gonadotrophin-releasing hormone (GnRH)-I and GnRH-II in the ovary and ovarian cancer. Endocr Relat Cancer. 2003;10(2):169–77.
Kakar SS, Grizzle WE, Neill JD. The nucleotide sequences of human GnRH receptors in breast and ovarian tumors are identical with that found in pituitary. Mol Cell Endocrinol. 1994;106(1–2):145–9.
Bono AV, Salvadore M, Celato N. Gonadotropin-releasing hormone receptors in prostate tissue. Anal Quant Cytol Histol. 2002;24(4):221–7.
Borroni R, Di Blasio AM, Gaffuri B, Santorsola R, Busacca M, Vigano P, Vignali M. Expression of GnRH receptor gene in human ectopic endometrial cells and inhibition of their proliferation by leuprolide acetate. Mol Cell Endocrinol. 2000;159(1–2):37–43.
Maudsley S, Davidson L, Pawson AJ, Chan R, Lopez de Maturana R, Millar RP. Gonadotropin-releasing hormone (GnRH) antagonists promote proapoptotic signaling in peripheral reproductive tumor cells by activating a Galphai-coupling state of the type I GnRH receptor. Cancer Res. 2004;64(20):7533–44.
Kraus S, Levy G, Hanoch T, Naor Z, Seger R. Gonadotropin-releasing hormone induces apoptosis of prostate cancer cells: role of c-Jun NH2-terminal kinase, protein kinase B, and extracellular signal-regulated kinase pathways. Cancer Res. 2004;64(16):5736–44.
Angelucci C, Iacopino F, Lama G, Capucci S, Zelano G, Boca M, Pistilli A, Sica G. Apoptosis-related gene expression affected by a GnRH analogue without induction of programmed cell death in LNCaP cells. Anticancer Res. 2004;24(5a):2729–38.
Kraus S, Naor Z, Seger R. Gonadotropin-releasing hormone in apoptosis of prostate cancer cells. Cancer Lett. 2006;234(2):109–23.
Limonta P, Moretti RM, Marelli MM, Dondi D, Parenti M, Motta M. The luteinizing hormone-releasing hormone receptor in human prostate cancer cells: messenger ribonucleic acid expression, molecular size, and signal transduction pathway. Endocrinology. 1999;140(11):5250–6.
Dondi D, Moretti RM, Montagnani Marelli M, Pratesi G, Polizzi D, Milani M, Motta M, Limonta P. Growth-inhibitory effects of luteinizing hormone-releasing hormone (LHRH) agonists on xenografts of the DU 145 human androgen-independent prostate cancer cell line in nude mice. Int J Cancer. 1998;76(4):506–11.
Kyte JA. Cancer vaccination with telomerase peptide GV1001. Expert Opin Investig Drugs. 2009;18(5):687–94.
Kim JW, Yadav DK, Kim SJ, Lee MY, Park JM, Kim BS, Kim MH, Park HG, Kang KW. Anti-cancer effect of GV1001 for prostate cancer; function as a ligand of GnRHR. Endocr Relat Cancer. 2018. https://doi.org/10.1530/ERC-18-0454.
Collado B, Carmena MJ, Cortes J, Schally AV, Prieto JC. Luteinizing hormone-releasing hormone antagonist Cetrorelix regulates the expression of Galphas and Galphai protein subunits and adenylate cyclase activity in rat ovary, breast and pituitary. Int J Oncol. 2004;24(3):725–30.
Salvador A, Garcia-Paramio MP, Sanchez-Chapado M, Carmena MJ, Prieto JC. Effects of the luteinising hormone-releasing hormone (LH-RH) agonist leuprolide on adenylyl cyclase regulation through G-protein coupled receptors in rat ventral prostate. Eur J Cancer (Oxford, England: 1990). 2001;37(5):641–8.
Garcia-Fernandez MO, Solano RM, Sanchez-Chapado M, Ruiz-Villaespesa A, Prieto JC, Carmena MJ. Low expression of Galpha protein subunits in human prostate cancer. J Urol. 2001;166(6):2512–7.
von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33(Database issue):D433-437.
Sutherland RM, McCredie JA, Inch WR. Growth of multicell spheroids in tissue culture as a model of nodular carcinomas. J Natl Cancer Inst. 1971;46(1):113–20.
Jeon S, Lee HS, Lee GY, Park G, Kim TM, Shin J, Lee C, Oh IH. Shift of EMT gradient in 3D spheroid MSCs for activation of mesenchymal niche function. Sci Rep. 2017;7(1):6859.
Sun Y, Wang BE, Leong KG, Yue P, Li L, Jhunjhunwala S, Chen D, Seo K, Modrusan Z, Gao WQ, Settleman J, Johnson L. Androgen deprivation causes epithelial-mesenchymal transition in the prostate: implications for androgen-deprivation therapy. Cancer Res. 2012;72(2):527–36.
Chen WY, Tsai YC, Siu MK, Yeh HL, Chen CL, Yin JJ, Huang J, Liu YN. Inhibition of the androgen receptor induces a novel tumor promoter, ZBTB46, for prostate cancer metastasis. Oncogene. 2017;36(45):6213–24.
Izumi K, Fang LY, Mizokami A, Namiki M, Li L, Lin WJ, Chang C. Targeting the androgen receptor with siRNA promotes prostate cancer metastasis through enhanced macrophage recruitment via CCL2/CCR2-induced STAT3 activation. EMBO Mol Med. 2013;5(9):1383–401.
Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev. 2007;28(7):778–808.
He WW, Sciavolino PJ, Wing J, Augustus M, Hudson P, Meissner PS, Curtis RT, Shell BK, Bostwick DG, Tindall DJ, Gelmann EP, Abate-Shen C, Carter KC. A novel human prostate-specific, androgen-regulated homeobox gene (NKX31) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics. 1997;43(1):69–77.
Tan PY, Chang CW, Chng KR, Wansa KD, Sung WK, Cheung E. Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival. Mol Cell Biol. 2012;32(2):399–414.
Kuser-Abali G, Alptekin A, Lewis M, Garraway IP, Cinar B. YAP1 and AR interactions contribute to the switch from androgen-dependent to castration-resistant growth in prostate cancer. Nat Commun. 2015;6:8126.
Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30(1):1–17.
Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21(21):2747–61.
Imai A, Horibe S, Takagi A, Tamaya T. Gi protein activation of gonadotropin-releasing hormone-mediated protein dephosphorylation in human endometrial carcinoma. Am J Obstet Gynecol. 1997;176(2):371–6.
Grundker C, Volker P, Emons G. Antiproliferative signaling of luteinizing hormone-releasing hormone in human endometrial and ovarian cancer cells through G protein alpha(I)-mediated activation of phosphotyrosine phosphatase. Endocrinology. 2001;142(6):2369–80.
White CD, Stewart AJ, Lu ZL, Millar RP, Morgan K. Antiproliferative effects of GnRH agonists: prospects and problems for cancer therapy. Neuroendocrinology. 2008;88(2):67–79.
Imai A, Takagi H, Horibe S, Fuseya T, Tamaya T. Coupling of gonadotropin-releasing hormone receptor to Gi protein in human reproductive tract tumors. J Clin Endocrinol Metab. 1996;81(9):3249–53.
Emons G, Muller V, Ortmann O, Schulz KD. Effects of LHRH-analogues on mitogenic signal transduction in cancer cells. J Steroid Biochem Mol Biol. 1998;65(1–6):199–206.
Montagnani Marelli M, Moretti RM, Mai S, Januszkiewicz-Caulier J, Motta M, Limonta P. Type I gonadotropin-releasing hormone receptor mediates the antiproliferative effects of GnRH-II on prostate cancer cells. J Clin Endocrinol Metab. 2009;94(5):1761–7.
Shafi AA, Yen AE, Weigel NL. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther. 2013;140(3):223–38.
Denmeade SR, Isaacs JT. A history of prostate cancer treatment. Nat Rev Cancer. 2002;2(5):389–96.
El-Sayed IY, Daher A, Destouches D, Firlej V, Kostallari E, Maille P, Huet E, Haidar-Ahmad N, Jenster G, de la Taille A, Abou Merhi R, Terry S, Vacherot F. Extracellular vesicles released by mesenchymal-like prostate carcinoma cells modulate EMT state of recipient epithelial-like carcinoma cells through regulation of AR signaling. Cancer Lett. 2017;410:100–11.
Zhou ZX, Kemppainen JA, Wilson EM. Identification of three proline-directed phosphorylation sites in the human androgen receptor. Mol Endocrinol. 1995;9(5):605–15.
Lonergan PE, Tindall DJ. Androgen receptor signaling in prostate cancer development and progression. J Carcinog. 2011;10:20.
Feng S, Tang Q, Sun M, Chun JY, Evans CP, Gao AC. Interleukin-6 increases prostate cancer cells resistance to bicalutamide via TIF2. Mol Cancer Ther. 2009;8(3):665–71.
Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461(7263):495–500.
Lei Q, Jiao J, Xin L, Chang CJ, Wang S, Gao J, Gleave ME, Witte ON, Liu X, Wu H. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell. 2006;9(5):367–78.
Zhang H, Muders MH, Li J, Rinaldo F, Tindall DJ, Datta K. Loss of NKX3.1 favors vascular endothelial growth factor-C expression in prostate cancer. Cancer Res. 2008;68(21):8770–8.
Mo JS, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15(6):642–56.
Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19(4):491–505.
Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114(4):445–56.
Janse van Rensburg HJ, Yang X. The roles of the Hippo pathway in cancer metastasis. Cell Signal. 2016;28(11):1761–72.
Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13(1):63–79.
Hsu PC, You B, Yang YL, Zhang WQ, Wang YC, Xu Z, Dai Y, Liu S, Yang CT, Li H, Hu B, Jablons DM, You L. YAP promotes erlotinib resistance in human non-small cell lung cancer cells. Oncotarget. 2016;7(32):51922–33.
Thoma C. Prostate cancer: targetable YAP1-AR interaction key to disease progression. Nat Rev Urol. 2015;12(11):596.
Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, Fu XD, Mills GB, Guan KL. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150(4):780–91.
Park J, Kim DH, Ahn HN, Song YS, Lee YJ, Ryu JH. Activation of estrogen receptor by Bavachin from Psoralea corylifolia. Biolmol Ther (Seoul). 2012;20(2):183–8.
Acknowledgements
We appreciate KAEL GemVex for the supply of GV1001.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (Kim HS, 2018M3A9C8021792, Kang KW, 2021R1A2C2093196).
Author information
Authors and Affiliations
Contributions
KWK and JWK conceived and designed the experiments; JWK, MP, SK, SCL and HSK performed the experiments; SCL and HSK analyzed the data; KWK and JWK wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All experiments and methods were performed in accordance with relevant guidelines and regulations. All animal procedures were approved by the Institutional Animal Care and Use Committee of Seoul National University. We took adequate steps to ensure that animals did not suffer unnecessarily at any stage of an experiment, whether acute or chronic.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Establishment of YAP1 knockout LNCaP cells. (A) Validation of YAP1 knockout LNCaP cells by CRISPR/Cas9 system. YAP1 expression was determined by immunoblot analysis. All of the results were confirmed by multiple experiments. (B) Validation for CRISPR/Cas9 editing. Representative gel images of T7E1-treated PCR products amplified from the target sites of YAP1. Cleaved-products are designated by arrowheads.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kim, J.W., Park, M., Kim, S. et al. Anti-metastatic effect of GV1001 on prostate cancer cells; roles of GnRHR-mediated Gαs-cAMP pathway and AR-YAP1 axis. Cell Biosci 11, 191 (2021). https://doi.org/10.1186/s13578-021-00704-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13578-021-00704-3