Abstract
Background
Abnormally expressed in diverse cancers, circZFR has been correlated with clinical outcomes of cancer patients. Aim of this meta-analysis was to elucidate the prognostic role of circZFR in multiple human malignancies.
Methods
Literature retrieval was conducted by systematically searching on Pubmed, Web of Science, and the Cochrane Library up to December 2nd, 2021. Hazard ratios (HRs) or odds ratios (ORs) with 95% confidence intervals (CIs) were pooled to evaluate the association between circZFR expression and overall survival (OS). The reliability of the pooled results was assessed through sensitivity analysis and the publication bias was measured by Begg’s and Egger’s test.
Results
A total of seventeen studies comprising 1098 Chinese patients were enrolled in this meta-analysis. Results demonstrated that high circZFR expression was correlated with an unfavorable OS (HR = 2.14, 95% CI 1.74, 2.64). High circZFR expression predicted larger tumor size (OR = 2.79, 95% CI 1.52, 5.12), advanced clinical stage (OR = 3.38, 95% CI 1.49, 7.65), tendentiousness of lymph node metastasis (LNM) (OR = 3.08, 95% CI 2.01, 4.71), and malignant grade (OR = 3.18, 95% CI 1.09, 9.30), but not related to age, gender, and distant metastasis (DM).
Conclusions
High circZFR expression was associated with unfavorable OS and clinicopathologic parameters including tumor size, clinical stage, LNM, and histology grade, implicating a promising prognostic factor in cancers.
Similar content being viewed by others
Background
In the human genome, about 93% of DNA sequences can be transcribed into RNA, of which only less than 2% are translatable, and the rest are specified as non-coding RNA (ncRNA) [1]. Circular RNAs (circRNAs), as a member of ncRNAs, were first discovered in RNA viruses and then in eukaryotes in the 1970s [2, 3]. CircRNAs are characterized by highly conserved and the unique covalently closed loop structure without free 5′ cap and 3′ tail, which avoids degradation by exonuclease and differentiates it from other ncRNAs such as lncRNA and miRNA. The expression of circRNAs has tissue, time and disease specificity. Most circRNAs are derived from exons and located in the cytoplasm, while a few are directly cycled by introns and located in the nucleus. However, the mechanism of circRNAs formation remains unclear [4]. Currently, many studies have shown that circRNAs exert their biological functions mainly by acting as miRNA molecular sponges through competing endogenous RNA (ceRNAs), interacting with proteins and regulating gene splicing or transcription, and translating into proteins or peptides that perform epigenetic functions. CircRNAs are involved in many physiological and pathological processes, such as aging [5], diabetes [6], and various cancer [7,8,9]. It is worth noting that the circRNAs play diverse roles in tumorigenesis and development, which can regulate tumor proliferation [10], metastasis [11], and drug resistance [12, 13].
Circular RNA zinc finger RNA-binding protein (CircZFR), a transcription product of zinc finger RNA-binding protein (ZFR) gene, is mapped to chromosome 5p13.3, and has been identified as a novel oncogenic or suppressing modulator in several human cancers [14]. Recently, circZFR has been reported to be associated with the disease progression in hepatocellular carcinoma (HCC) [15], lung cancer (LC) [16], papillary thyroid cancer (PTC) [17], bladder cancer (BlC) [18], breast cancer (BrC) [19, 20], gastric cancer (GC) [21], colorectal cancer (CRC) [22], and esophageal squamous cell cancer (ESCC) [23]. Most studies reported that circZFR acted as oncogenic function to promote the cancer progression. However, the small clinical sample size limited the clinical significance of circZFR. Here, we made the meta-analysis of circZFR in various cancers to emphasize its clinical application potential.
Methods
Publication search strategy
This meta-analysis was projected, reviewed, and reported based on the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) checklist. A comprehensive research was conducted by two independent authors (ZY L and WC Z) in PubMed, Web of Science, and the Cochrane Library up to December 2nd, 2021. The Search strategy were listed as follows: “circZFR” OR “circ_ZFR” OR “circ-ZFR” OR “circRNA ZFR” OR “circular RNA ZFR” OR “circ_0072088” OR “circ_0072083” OR “Circ_103809” OR “circRNA_103809” OR “Hsa_circRNA_103809” OR “Circular RNA hsa_circRNA_103809” (the detailed search strategy for each of the databases was presented in the supplementary materials). The additional research of citation lists of included publications was manually identified for relevant articles.
Inclusion and exclusion criteria
The inclusion and exclusion criteria are presented as follows. Inclusion criteria: (1) patients definitely diagnosed with cancer by histopathology; (2) studies focusing on the clinical prognostic or clinical value of circZFR in any type of cancers; (3) circZFR were assigned to high expression group (high) or low expresssion group (low) according to its relative expression level; (4) studies providing sufficient information about the correlation between circZFR expression level and overall survival (HRs with 95% CIs) or clinical characteristics (age, gender, stage, grade, and so on). Exclusion criteria: (1) duplicate publications; (2) studies focusing on the structures or functions of circZFR and without clinical prognostic information; (3) available data are not extractable; (4) studies without original data like reviews and meta-analysis.
Data extraction and quality assessment
All the enrolled studies were independently evaluated by two investigators (ZY L and WC Z) and discrepancies were settled by consultation with a third investigator (XL R). All the included studies are non-RCT. The baseline data extracted from the included studies were as follows: (1) first author, study year, country, cancer type, clinical stage, tumor size, cut-off value, follow-up time, detection method, adjuvant therapy before surgery, survival analysis method, and outcome measure method; (2) hazard ratios (HRs) or odds ratios (ORs) with 95% confidence intervals (CIs) of circZFR for OS or clinicopathologic parameters. We used the software Engauge Digitizer (version 4.1) to calculate the HRs with 95% CIs according to Kaplan-Meier curves when they were not presented in the studies directly. The quality of the eligible literature was evaluated by NOS, whose score up to 7 indicated high quality of the study.
Data synthesis and statistical analysis
The statistical analyses were conducted by STATA software (version 12.0) and Review Manager (RevMan 5.3). HRs or ORs with corresponding 95% CIs were used to describe the relationship between circZFR expression and the prognosis or clinical characteristics. The chi-squared test and I2 statistics was used to assess the heterogeneity among the studies. A value of p < 0.05, I2 > 50% of chi-squared test demonstrated obvious heterogeneity among these studies, and a random effect model was applied in this occasion. Otherwise, a model of fixed effect was carried out to assess the pooled results with no obvious heterogeneity. Sensitivity analysis was conducted by omitting one of the included studies respectively to evaluate the certainty of the pooled HRs. Begg’s and Egger’s test was used to estimate the potential publication bias. It is considered statistically significant with p < 0.05.
Results
Selection and description of enrolled studies
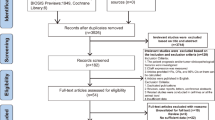
A total of 80 publications were initially searched as potential studies, and 39 duplicates of them were excluded. Then, 3 publications were excluded as review and meta-analysis by screening via the titles and abstracts. Afterwards, 38 full-text articles were thoroughly assessed, of which 18 studies without clinical analyses and 3 studies with unextractable data were removed. As a result, 17 studies comprising 1098 patients were enrolled in this meta-analysis. Of note, all the enrolled patients were from China. The selection process was concisely shown by a flow diagram in Fig. 1.
The detailed data of the enrolled publications were presented in Table 1. All of the articles were performed in China and published between 2017 and 2021. Out of the seventeen studies, seven focused on the HCC. Additionally, there were other seven cancer types including BlC, BrC, CRC, ESCC, GC, LC, and PTC. The circZFR expression level was detected by quantitative real-time polymerase chain reaction (qRT-PCR) with the sample size ranging from 30 to 170, and analyzed via univariate analyses or multivariate analyses. Among the publications, nine studies assessed the relationship between circZFR and clinical stages (TNM stage), eleven studies measured OS, and 10 of whose follow-up time were more than 60 months. Most of the studies adopted the median or mean expression of circZFR as the cut-off value. Patients in 14 studies did not accepted anti-tumor therapy (adjuvant therapy) before the specimen collected. The quality of the enrolled studies was high with all of the NOS scores ≥ 7.
Association between circZFR expression and OS
Eleven studies comprising 689 patients were included for pooled OS analysis. With the absence of the heterogeneity (I2 = 20.1%, p = 0.252), the fixed effect model was applied to analyze the OS (HR = 2.14, 95% CI 1.74, 2.64), which indicated that high expression of circZFR related to a worse OS (Fig. 2).
Additionally, four stratified analyses were further conducted on sample size (< 60 and ≥ 60), cancer type (BlC, BrC, HCC, and others), follow-up months (< 60 and ≥ 60), and cut-off value (mean, median and not available (N/A)) (Supplementary Figure 1). The detailed pooled HRs and 95% CIs were summarized in Table 2. For the sample size stratified analysis, both the more than 60 (HR = 1.90, 95% CI 1.47, 2.45) and less than 60 (HR = 2.73, 95% CI 1.90, 3.91) groups with circZFR overexpression exhibited an unfavorable OS. Of note, for studies evaluating on cancer type, the results indicated an insignificant relation of the circZFR overexpression to the OS of the BrC (HR = 1.43, 95% CI 0.54, 3.79), while high circZFR level was significantly associated with short survival of BlC (HR = 2.41, 95% CI 1.59, 3.66), HCC (HR = 2.38, 95% CI 1.66, 3.40) and other cancer (HR = 2.65, 95% CI 1.63, 4.29) patients. What is more, the high circZFR expression predicted an unfavorable OS with follow-up time more than 60 months (HR = 2.44, 95% CI 1.95, 3.05), while that less than 60 group showed no significant linkage. According to the cut-off value stratified analysis, whether the cut-off value was mean, median, or N/A, the high circZFR expression indicated worse OS.
Association between circZFR expression and clinicopathologic parameters
Seven items of clinicopathologic parameters including age, gender, tumor size, clinical stage, distant metastasis (DM), lymph node metastasis (LNM), and histology grade were further analyzed to evaluate their correlation with the circZFR expression. Notably, ten studies enrolled to explore the correlation between circZFR expression and tumor size, demonstrating that higher circZFR expression predicted larger tumor size (OR = 2.79, 95% CI 1.52, 5.12) (Fig. 3). Similarly, the upregulation of circZFR expression indicated advanced clinical stage (OR = 3.38, 95% CI 1.49, 7.65), tendency of LNM (OR = 3.08, 95% CI 2.01, 4.71) and higher histology grade (OR = 3.18, 95% CI 1.09, 9.30). As shown in Supplementary Figure 2, insignificant association found between circZFR expression and age (OR = 1.16, 95% CI 0.86, 1.57), gender (OR = 1.02, 95% CI 0.73, 1.43), and DM (OR = 1.39, 95% CI 0.49, 3.94). The detailed data were summarized in Table 3.
Sensitivity analysis and publication bias
We performed the sensitivity analysis by calculating the HRs and 95% CIs after excluding each of the enrolled study, and the result indicated the stability of the pooled HRs and corresponding 95% CIs of the association between circZFR expression and OS (Supplementary Figure 3A).
The potential publication bias was measured by Begg’s and Egger’s test. And there was no significant publication bias according to the symmetrically distributed funnel plot (Supplementary Figure 3B). Furthermore, the Begg’s and Egger’s test (p = 0.276 and p = 0.173) demonstrated the absence of the publication bias.
The regulation mechanism of circZFR in cancers
Recently, multiple studies have reported the potential mechanism of circZFR for proliferation, apoptosis, migration, invasion, and metabolism in various cancers. Currently, ceRNA mechanism has been focused on and partially illuminated in regulating cancers development. For example, the circZFR/miR-545-3p/CBLL1 axis, circZFR/miR-195-5p/KPNA4 axis, and circZFR/miR-4302/ZNF121axis promoted the growth, migration, invasion, EMT, and chem-resistance, arrested cell cycle and inhibited apoptosis in LC [16, 32, 33]. Specially, HCC tumor cell-derived exosome suppressed the metastasis of HCC mediating the degradation of miR-375 via circZFR and upregulated MMP-16 [26]. Moreover, circZFR can bind to the cellular protein directly and interfere its function. It has been reported that circZFR decreased H3K4me3 levels on the CCND1 promoter to promote the tumorigenic capacity in LC [34]. Besides, circZFR bound with SSBP1, thereby inducing the assembly of CDK2/cyclin E1 complexes, which induced p-Rb phosphorylation, thus releasing activated E2F1 leading to cell cycle progression and cell proliferation in cervical cancer [35]. We have reviewed the reported mechanism about circZFR in Table 4 and Fig. 4.
Discussion
CircRNAs have been reported as the clinical diagnostic biomarkers and treatment targets in terms of stability, specificity and conservation [44]. Isolation and detection of circRNAs from peripheral blood or tissue is feasible. A pan-cancer analysis of RNA sequencing of thousands of human cancer genomes verified that ZFR is a potential potent cancer driver gene. CircZFR is a circular RNA derived from ZFR exons via back-splicing. According to the majority of studies, circZFR was overexpressed in various cancer, including BlC, BrC, HCC, LC, and renal cell carcinoma [15,16,17, 21, 28]. But a few studies identified that circZFR was downregulated in cancer tissues like BrC and colorectal cancer compared with adjacent tissues [20, 22, 37]. It suggested the expression heterogeneity of circZFR in different tumor tissues. CircZFR has been proved the significant association with clinical characteristics and prognostic parameters in various cancer.
Here, we first estimated correlation of circZFR expression with prognosis of cancer patients. A total of 689 patients from eleven studies were included for pooled OS analysis. The pooled HR demonstrated that high circZFR expression indicated a poor OS without significant heterogeneity. In fact, the cut-off value of the included studies were inconsistent. Some cut-off values were median, some were mean, and the rest studies that did not report cut-off values were labeled ‘NA’. As the different cut-off values may affect the result, we made a stratified analysis. And according to the results, all of the three groups (mean, median, and N/A) with high circZFR expression exhibited poor OS. That means, the inconsistency of the cut-off value in this study wouldn’t affect the results of OS analysis. Then we evaluated the association between circZFR and the major clinical characteristics including age, gender, tumor size, clinical stage, DM, LNM, and histology grade. The higher circZFR expression predicted larger tumor size, advanced clinical stage, higher possibility of LNM, and higher histology grade. No significant correlation between circZFR expression and age, gender, and DM. More studies need to be included to analyze the heterogeneity of pooled HR for tumor size, clinical stage, and DM.
At present, circRNAs play their biological functions mainly through the following four patterns, including acting as miRNA molecular sponge through the mechanism of ceRNAs, interacting with RNA-binding proteins (RBPs), regulating of splicing or transcription of genes, translation into proteins or small peptides. However, the function and mechanism of circZFR in the development and progression of cancers still have not been clear deeply. We found the major mechanism of circZFR regulating development and progression in cancers was ceRNA [17, 43]. Only one study showed the function of circZFR in hepatic carcinoma cells-derived exosome suppressing migration and invasion [26]. Only a few studies reported circZFR promoted cancers via interacting with RBPs [35] or regulating the chromatin modification at the transcription start site of genes [34]. More studies are needed to put into circZFR regulation mechanism.
However, this study also has several limitations. Firstly, almost all included patients were from China, so this reduced the applicability of the results across different races and regions. Then, we only enrolled 11 studies in the prognosis meta-analysis, which limited the wide application of the meta-analysis results. Moreover, more studies needed to be included to analyze the correlation between circZFR expression and age, gender, and DM. Finally, because several studies did not report HRs with 95% CIs in the article, we had to extract and calculate them on the basis of the Kaplan-Meier curves.
Conclusions
Taken together, this study demonstrated that high circZFR expression was remarkably correlated with unfavorable OS, as well as clinicopathological parameters including larger tumor size, advanced clinical stage, higher possibility of LNM, and higher histology grade in Chinese cancer patients. Therefore, circZFR can be served as a prognostic biomarker of cancer. Nevertheless, further large-scale studies from different ethnic population are needed to verify the results.
Availability of data and materials
The dataset used and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- BlC:
-
Bladder cancer
- BrC:
-
Breast cancer
- circZFR:
-
Circular RNA zinc finger RNA-binding protein
- CI:
-
Confidence interval
- circRNAs:
-
Circular RNAs
- CeRNA:
-
Competing endogenous RNA
- CRC:
-
Colerectal cancer
- CP:
-
Clinicopathological parameters
- DFS:
-
Disease-free survival
- DM:
-
Distant metastasis
- DZF:
-
Domain associated with zinc fingers
- ESCC:
-
Esophageal squamous cell cancer
- GC:
-
Gastric cancer
- HCC:
-
Hepatocellular carcinoma
- HR:
-
Hazard ratio
- LC:
-
Lung cancer
- LNM:
-
Lymph node metastasis
- N/A:
-
Not available
- ncRNA:
-
Non-coding RNA
- NOS:
-
Newcastle-Ottawa Scale
- OR:
-
Odds ratio
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- PTC:
-
Papillary thyroid cancer
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- RBPs:
-
RNA-binding proteins
References
Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74.
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–30.
Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–60.
Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–8.
Knupp D, Miura P. CircRNA accumulation: a new hallmark of aging? Mech Ageing Dev. 2018;173:71–9.
Yang F, Chen Y, Xue Z, Lv Y, Shen L, Li K, et al. High-throughput sequencing and exploration of the lncRNA-circRNA-miRNA-mRNA network in type 2 diabetes mellitus. Biomed Res Int. 2020;2020:8162524.
Fu L, Jiang Z, Li T, Hu Y, Guo J. Circular RNAs in hepatocellular carcinoma: Functions and implications. Cancer Med. 2018;7:3101–9.
Huang W, Yang Y, Wu J, Niu Y, Yao Y, Zhang J, et al. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-beta signalling. Cell Death Differ. 2020;27:1709–27.
Shi X, Wang B, Feng X, Xu Y, Lu K, Sun M. circRNAs and Exosomes: A Mysterious Frontier for Human Cancer. Mol Ther Nucleic Acids. 2020;19:384–92.
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94.
Wei S, Zheng Y, Jiang Y, Li X, Geng J, Shen Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–93.
Kun-Peng Z, Xiao-Long M, Lei Z, Chun-Lin Z, Jian-Ping H, Tai-Cheng Z. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics. 2018;10:1327–46.
Zhao Z, Ji M, Wang Q, He N, Li Y. Circular RNA Cdr1as upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol Ther Nucleic Acids. 2019;18:24–33.
Li J, Fan R, Xiao H. Circ_ZFR contributes to the paclitaxel resistance and progression of non-small cell lung cancer by upregulating KPNA4 through sponging miR-195-5p. Cancer Cell Int. 2021;21:15.
Cedric BC, Souraka TDM, Feng YL, Kisembo P, Tu JC. CircRNA ZFR stimulates the proliferation of hepatocellular carcinoma through upregulating MAP2K1. Eur Rev Med Pharmacol Sci. 2020;24:9924–31.
Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500:846–51.
Wei H, Pan L, Tao D, Li R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochem Biophys Res Commun. 2018;503:56–61.
Zhang WY, Liu QH, Wang TJ, Zhao J, Cheng XH, Wang JS. CircZFR serves as a prognostic marker to promote bladder cancer progression by regulating miR-377/ZEB2 signaling. Biosci Rep. 2019;39:BSR20192779.
Chen Z, Wang F, Xiong Y, Wang N, Gu Y, Qiu X. CircZFR functions as a sponge of miR-578 to promote breast cancer progression by regulating HIF1A expression. Cancer Cell Int. 2020;20:400.
Liu M, Luo C, Dong J, Guo J, Luo Q, Ye C, et al. CircRNA_103809 suppresses the proliferation and metastasis of breast cancer cells by sponging microRNA-532-3p (miR-532-3p). Front Genet. 2020;11:485.
Huang SS, Guo WX, Ren MS. Circular RNA hsa_circ_103809 promotes cell migration and invasion of gastric cancer cells by binding to microRNA-101-3p. Eur Rev Med Pharmacol Sci. 2020;24:6064–71.
Zhang P, Zuo Z, Shang W, Wu A, Bi R, Wu J, et al. Identification of differentially expressed circular RNAs in human colorectal cancer. Tumour Biol. 2017;39:1010428317694546.
Fang N, Shi Y, Fan Y, Long T, Shu Y, Zhou J. Circ_0072088 promotes proliferation, migration, and invasion of esophageal squamous cell cancer by absorbing miR-377. J Oncol. 2020;2020:8967126.
Huang W, Lu Y, Wang F, Huang X, Yu Z. Circular RNA circRNA_103809 accelerates bladder cancer progression and enhances chemo-resistance by activation of miR-516a-5p/FBXL18 Axis. Cancer Manag Res. 2020;12:7561–8.
Li L, Xiao C, He K, Xiang G. Circ_0072088 promotes progression of hepatocellular carcinoma by activating JAK2/STAT3 signaling pathway via miR-375. IUBMB Life. 2021;73:1153–65.
Lin Y, Zheng ZH, Wang JX, Zhao Z, Peng TY. Tumor cell-derived exosomal circ-0072088 suppresses migration and invasion of hepatic carcinoma cells through regulating MMP-16. Front Cell Dev Biol. 2021;9:726323.
Luo L, Miao P, Ming Y, Tao J, Shen H. Circ-ZFR promotes progression of bladder cancer by upregulating WNT5A via sponging miR-545 and miR-1270. Front Oncol. 2020;10:596623.
Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/β-catenin pathway. Arch Biochem Biophys. 2019;661:196–202.
Xu R, Yin S, Zheng M, Pei X, Ji X. Circular RNA circZFR promotes hepatocellular carcinoma progression by regulating miR-375/HMGA2 axis. Dig Dis Sci. 2021;66:4361–73.
Yang X, Liu L, Zou H, Zheng Y-W, Wang K-P. circZFR promotes cell proliferation and migration by regulating miR-511/AKT1 axis in hepatocellular carcinoma. Dig Liver Dis. 2019;51:1446–55.
Zhan W, Liao X, Chen Z, Li L, Tian T, Yu L, et al. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. J Cell Physiol. 2020;235:1733–45.
Tan Z, Cao F, Jia B, Xia L. Circ_0072088 promotes the development of non-small cell lung cancer via the miR-377-5p/NOVA2 axis. Thorac Cancer. 2020;11:2224–36.
Zhang H, Wang X, Hu B, Zhang F, Wei H, Li L. Circular RNA ZFR accelerates non-small cell lung cancer progression by acting as a miR-101-3p sponge to enhance CUL4B expression. Artif Cells Nanomed Biotechnol. 2019;47:3410–6.
Ren G, Zhao Q, Yan C, Xue Q, Zhang L. Circular RNA circZFR promotes tumorigenic capacity of lung cancer via CCND1. Transl Cancer Res. 2020;9:3303–11.
Zhou M, Yang Z, Wang D, Chen P, Zhang Y. The circular RNA circZFR phosphorylates Rb promoting cervical cancer progression by regulating the SSBP1/CDK2/cyclin E1 complex. J Exp Clin Cancer Res. 2021;40:48.
Qiu X, Wang Q, Song H, Shao D, Xue J. circ_103809 promotes breast cancer progression by regulating the PI3K/AKT signaling pathway. Oncol Lett. 2020;19:3725–30.
Bian L, Zhi X, Ma L, Zhang J, Chen P, Sun S, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p / FOXO4 axis. Biochem Biophys Res Commun. 2018;505:346–52.
Liu T, Liu S, Xu Y, Shu R, Wang F, Chen C, et al. Circular RNA-ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR-130a/miR-107 and modulating PTEN. Cancer Res Treat. 2018;50:1396–417.
Cao Y, Tao Q, Kao X, Zhu X. Hsa-circRNA-103809 Promotes Hepatocellular Carcinoma Development via MicroRNA-1270/PLAG1 Like Zinc Finger 2 Axis. Dig Dis Sci. 2020;66:1524–32.
Li X, Shen M. Circular RNA hsa_circ_103809 suppresses hepatocellular carcinoma proliferation and invasion by sponging miR-620. Eur Rev Med Pharmacol Sci. 2019;23:555–66.
Zhu X, Han J, Lan H, Lin Q, Wang Y, Sun X. A novel circular RNA hsa_circRNA_103809/miR-377-3p/GOT1 pathway regulates cisplatin-resistance in non-small cell lung cancer (NSCLC). BMC Cancer. 2020;20:1190.
Li H, Liu F, Qin W. Circ_0072083 interference enhances growth-inhibiting effects of cisplatin in non-small-cell lung cancer cells via miR-545-3p/CBLL1 axis. Cancer Cell Int. 2020;20:78.
Wang M, Gao Y, Liu J. Silencing circZFR inhibits the proliferation, migration and invasion of human renal carcinoma cells by regulating miR-206. Onco Targets Ther. 2019;12:7537–50.
Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13:669–80.
Acknowledgements
None.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 81902745 and 82103228), the Natural Science Foundation of Hunan Province, China (grant number 2018JJ3716), Hunan Provincial Research and Development Program in Key Areas (2019WK2071), and China Postdoctoral Science Foundation (No. 2021M693557).
Author information
Authors and Affiliations
Contributions
ZY Liu designed the methodology of the study and performed the literature retrieval, data analysis, and interpretation. WC Zhang contributed to the study methodology, performed the literature retrieval and data analysis. WY Li, L Qi, and ZM Zhang participated in the data analysis and assisted in creating the figures. ZM Yang, L Wan, and C Tu participated in the data interpretation, critically reviewed and revised the manuscript. ZH Li and XL Ren conceptualized the study, participated in the study design, data interpretation, and manuscript revision. All authors read and approved the final manuscript as submitted.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
None.
Consent for publication
None.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Figure 1
. Forest plots of the subgroups analysis evaluating the correlation between circZFR expression and OS, including sample size (A), cancer type (B), follow-up months (C), and cut-off value (D). Supplementary Figure 2. Forest plots evaluating the correlation between circZFR expression and other clinicopathological parameters, including age (A), gender (B) and DM (C). Supplementary Figure 3. Sensitivity analysis (A) and funnel plot for publication bias (B) for circZFR on OS.
Additional file 2: Supplementary table
. Results of quality assessment using Newcastle-Ottawa Scale (NOS) score for the enrolled studies.
Additional file 3.
Search strategy.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liu, Z., Zhang, W., Tu, C. et al. Prognostic and clinicopathologic significance of circZFR in multiple human cancers. World J Surg Onc 20, 268 (2022). https://doi.org/10.1186/s12957-022-02733-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12957-022-02733-9