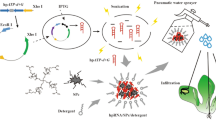
Abstract
Piercing-sucking pests are the most notorious group of pests for global agriculture. RNAi-mediated crop protection by foliar application is a promising approach in field trials. However, the effect of this approach on piercing-sucking pests is far from satisfactory due to the limited uptake and transport of double strand RNA (dsRNA) in plants. Therefore, there is an urgent need for more feasible and biocompatible dsRNA delivery approaches to better control piercing-sucking pests. Here, we report that foliar application of layered double hydroxide (LDH)-loaded dsRNA can effectively disrupt Panonychus citri at multiple developmental stages. MgAl-LDH-dsRNA targeting Chitinase (Chit) gene significantly promoted the RNAi efficiency and then increased the mortality of P. citri nymphs by enhancing dsRNA stability in gut, promoting the adhesion of dsRNA onto leaf surface, facilitating dsRNA internalization into leaf cells, and delivering dsRNA from the stem to the leaf via the vascular system of pomelo plants. Finally, this delivery pathway based on other metal elements such as iron (MgFe-LDH) was also found to significantly improve the protection against P. citri and the nymphs or larvae of Diaphorina citri and Aphis gossypii, two other important piercing-sucking hemipeteran pests, indicating the universality of nanoparticles LDH in promoting the RNAi efficiency and mortality of piercing-sucking pests. Collectively, this study provides insights into the synergistic mechanism for nano-dsRNA systemic translocation in plants, and proposes a potential eco-friendly control strategy for piercing-sucking pests.
Graphical Abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
Piercing-sucking pests, such as spider mites and the hemipteran insects aphids, psyllids, leafhoppers, whiteflies, and mealybugs, are considered as one of the most important pest groups in global agriculture [1]. These pests not only cause the wide spread of various bacterial and viral pathogens by piercing and sucking the plant sap with their slender, and sharp-pointed mouthparts [1], but such feeding characteristics also make it is difficult to be controlled. Up to date, chemical pesticides are still the primary option for the control of these pests, especially systemic pesticides, which are absorbed by plants and then transported to untreated tissues [2]. They are primarily applied through foliar spray, trunk injection, soil drench, and seed coating [2, 3]. However, there has been rapid evolution of pesticide resistance due to their massive use [4, 5]. Therefore, there is an urgent need for the development of eco-friendly management strategies for piercing-sucking pests.
RNA interference (RNAi) has been confirmed to be a powerful technology for crop protection [6, 7]. Currently, the U.S. Environmental Protection Agency (US-EPA) has registered a new foliar applied insecticide active ingredient Ledprona (Leptinotarsa decemlineata-specific recombinant double-stranded interfering Oligonucleotide GS2) that targets the Colorado potato beetle [8]. In piercing-sucking pests, double strand RNA (dsRNA) can be delivered into the body through several approaches, including body wall absorption (injection, soaking, dripping) [9,10,11], oral ingesting [12, 13], plant-induced feeding [14, 15], and foliar application [16, 17]. Among them, plant-induced feeding and foliar application are two promising delivery approaches for field trials. For plant-induced feeding, both nuclear transgenic and transplastomic RNAi can significantly reduce the survival or fecundity of the green peach aphid Myzus persicae [18] and three species of mites (Tetranychus evansi, Tetranychus truncatus, and Tetranychus cinnabarinus) [14], with the latter exhibiting better performance in plant protection. However, dsRNA is accumulated at low levels in the phloem of transplastomic plants, and piercing-sucking pests may have variable susceptibility to plastid-mediated RNAi [14, 18]. Foliar application, such as spray [17], leaf disc [19] or shoot soaking [16], has been used in laboratory and semi-field trials. However, its massive use as the nucleic acid pesticides in piercing-sucking pests is hindered by the unique barrier imposed by plant cell wall, which impedes the effective delivery of dsRNA into plant cells.
Nanoparticles can greatly improve the delivery efficacy of dsRNA, and then facilitate the development of dsRNA-based biopesticides [20, 21]. Nanoparticles are particles with diameters ranging from 1 to 100 nm with high biocompatibility and degradability but low toxicity and cost [22, 23]. Previous studies have shown that negatively charged dsRNA can be loaded onto positively charged nanoparticles with excellent properties [24,25,26]. Nanoparticle-loaded dsRNA applied through foliar spraying has been shown to have superior effects in controlling piercing-sucking pests on herbaceous plants, such as soybean [26], cotton [27, 28] and rice [29]. For instance, foliar spraying of layered double hydroxide (LDH)-dsRNA could disrupt Bemisia tabaci at multiple developmental stages by enhancing the delivery of dsRNA to cotton leaves and into whitefly [27]. Foliar application (soaking) of chitosan (CS)-dsRNA enhanced the RNAi efficiency of T. cinnabarinus by improving the stability of dsRNA in the pest body and environment [30]. Notably, some studies have demonstrated that trunk injection (similar to shoot soaking approach in herbaceous plants) is also an ideal approach to deliver dsRNA into woody plants such as citrus tree, the host of several notorious piercing-sucking pests, but the RNAi efficiency remains unknown [31, 32]. Additionally, exogenous RNA molecules have been found to be strictly restricted to the xylem and apoplast in woody plants such as fruit trees and herbaceous plants [33, 34]. To date, it remains unclear yet how nanomaterials systematically enhance the RNAi efficiency of dsRNA from the plants to the pests.
Panonychus citri, Diaphorina citri and Aphis gossypii are important piercing-sucking pests for many plants, especially citrus trees [1]. In this study, four nanoparticles were selected as delivery carriers of dsRNA, including chitosan-csodium tripolyphosphate pentabasic (CS-STPP) [35], carbon quantum dot (CQD) [36], LDH [37] and star polycation (SPC) [26, 38], which have been used to enhance the RNAi efficiency in pests. Chitinase (Chit) and Chitin synthase (Chs) gene, which are crucial for arthropod growth and development by participating in chitin biosynthesis and metabolic pathway [39], were selected as the target genes. The results showed that MgAl-LDH nanoparticles enhanced the stability of dsRNA in P. citri gut, promoted the absorption of dsRNA by leaf cells, and facilitated the transmission and diffusion of dsRNA in plant tissues, thereby enhancing the RNAi efficiency and mortality of pests. Similar results were obtained for D. citri and A. gossypii, two other important piercing-sucking pests. Overall, this study systematically elaborated the mechanism for nanoparticles to improve the RNAi efficiency and insecticidal effects, and provided a promising approach for controlling piercing-sucking pests in the field.
Results
Characterization of nano-dsRNA
To obtain nanoparticles with small sizes and high monodispersity and positive charge for loading dsRNA, we firstly synthesized CS-STPP, CQD and MgAl-LDH nanoparticles in the laboratory. The TEM and DLS results showed that the obtained nanoparticles were positively charged with an approximately spherical or hexagonal morphology, and the mean particle diameter was smaller than 80 nm with a polydispersity index lower than 0.4 (Fig. S1 A-C).
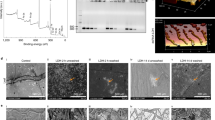
To determine the optimal mass ratio for the combination of dsRNA and nanoparticles, we evaluated the loading efficiency of nanoparticles for dsRNA at different mass ratios by a retardation assay through agarose gel electrophoresis. The results showed that the optimal mass ratio for the loading of dsRNA onto CS-STPP, CQD-STPP, MgAl-LDH, and SPC was 3.0:1, 1/100:1, 5.0:1, and 1/3:1, respectively (Fig. 1A). The morphological examination of nano-dsRNA complex by TEM showed that CS-STPP-dsRNA displayed an approximately spherical shape, CQD-STPP-dsRNA and SPC-dsRNA were spherical, and MgAl-LDH-dsRNA showed a shape of hexagonal nanosheet (Fig. 1B).
Generation and characterization of nano-dsRNA. (A) Gel images of nano-dsRNA at the different mass ratios of dsRNA and CS-STPP, CQD-STPP, MgAl-LDH and SPC nanoparticles; (B) TEM images of nano-dsRNA at the optimal mass ratio; (C) Particle size and polydispersity index of nano-dsRNA and (D) Zeta potential of nano-dsRNA by DLS analysis; (E) Loading efficiency (%) of dsRNA onto CS-STPP, CQD-STPP, MgAl-LDH and SPC nanoparticles by UV-visible spectrophotometer analysis. The data were expressed as a mean ± SEM. Different uppercase and lowercase letters indicate significant differences among biological replicates using of One-way ANOVA and Tukey’s multiple comparison tests (P < 0.05)
Size distribution and zeta potential of nano-dsRNA have significant effects on the transport efficiency of dsRNA in plants and insects [40]. Therefore, we examined the average particle size, polydispersity index, and zeta potential of nano-dsRNA at the optimal mass ratio by DLS. The results showed that complexation between dsRNA and nanoparticles significantly reduced the particle size of dsRNA from 366.55 nm to below 200 nm and the polydispersity index from 0.41 to below 0.3 (Fig. 1C). Four positively charged nanoparticles were bound with negatively charged dsRNA (-25.03 mV) to form positively or negatively charge nano-dsRNA complexes (Fig. 1D).
To determine the dsRNA loading efficacy of nanoparticles, we measured the loading efficiency of different nanoparticles for dsRNA at the optimal mass ratio using UV-visible spectrophotometry. The results showed that the loading efficiency of CS-STPP, CQD-STPP, MgAl-LDH and SPC nanoparticles for dsRNA was 84.68%, 68.29%, 86.40% and 63.12%, respectively (Fig. 1E), among which CS-STPP and MgAl-LDH nanoparticles exhibited relatively higher dsRNA loading efficiencies.
Nanoparticles enhance dsRNA stability in P. Citri nymph gut
To investigate the effects of nanoparticles on RNAi efficiency in nymphs, we directly fed nymphs with an artificial diet containing naked dsPcChit or nano-dsRNA complexes, including CS-STPP-dsPcChit, CQD-STPP-dsPcChit, MgAl-LDH-dsPcChit and SPC-dsPcChit. Firstly, we identified that the optimal concentration of dsPcChit to induce effective RNAi in nymphs was 320 ng/µl (100 µl) (Fig. S2 A-B) by artificial diet feeding bioassay (Fig. 2A). Then, we analyzed the RNAi efficiency induced by naked dsPcChit or nano-dsPcChit. The results showed that compared with that of naked dsPcChit, the knockdown efficiency of the Chit gene induced by CS-STPP-dsPcChit and MgAl-LDH-dsPcChit was significantly increased by 23.35% (P < 0.01) and 10.85% (P < 0.05), respectively (Fig. 2B), and correspondingly the mortality was significantly increased by 24.88% (P < 0.01) and 12.49% (P < 0.01), respectively (Fig. 2C). CQD-STPP-dsPcChit and SPC-dsPcChit showed no significant enhancement of RNAi efficiency compared with naked dsPcChit (P > 0.05). However, CQD-STPP nanoparticles dramatically increased the mortality of nymphs (Fig. S3).
Nanoparticles enhance dsRNA stability in P. citri nymph gut to improve RNAi efficiency. (A) Schematic diagram of artificial diet feeding bioassay; (B) Relative expression level of Chitnase and (C) Corrected mortality (%) after nymphs were fed on artificial diet containing 320 ng/µl naked dsPcChit or nano-dsPcChit at 48 h; (D) Confocal microscopy images of nymphs feeding on artificial diet containing naked dsPcChit-Cy3 or nano-dsPcChit-Cy3 at 24 and 48 h. BF, bright field. Scale bar: 50 μm. (E) Quantitative analysis of fluorescence intensities of nano-dsChit in nymph gut by Image J software. GAPDH was used as the reference gene for P. citri. Nymphs feeding on artificial diet without dsRNA or nano-dsRNA were used as a blank control. Data represent mean ± SEM for four biological replicates, *P < 0.05, **P < 0.01, ***P < 0.001 with a Student’s t test; ns, not significant (P > 0.05). Different lowercase letters indicate the significant difference using of One-way ANOVA and Tukey’s multiple comparison tests (P < 0.05)
To elucidate how nano-dsRNA enhances RNAi efficiency in nymph, we first detected the dsRNA stability in nymph gut. The images revealed that CS-STPP-dsRNA-Cy3 and MgAl-LDH-dsRNA-Cy3 resulted in stronger red fluorescence signals in nymph guts than naked dsRNA-Cy3 at 24 and 48 h (Fig. 2D-E). We further found that MgAl-LDH nanoparticles protect dsRNA from degradation in the gut environment for more than 120 h (Fig. S4 A). The fluorescence intensities were higher in MgAl-LDH-dsRNA-Cy3 group than naked dsRNA-Cy3 and control groups, and there was no significant difference between the latter two (Fig. S4 B). The naked dsRNA-Cy3 group showed gradual decreases in Cy3 fluorescence intensity with the extension of treatment time (Fig. 2D-E, Fig. S4 A-B). However, the fluorescence intensities of CQD-STPP-dsRNA-Cy3 group did not show significant difference with that of the naked dsRNA group. Additionally, the SPC-dsRNA-Cy3 group showed very weak red fluorescence signals compared with other treatment groups at 24 and 48 h. In short, CS-STPP and MgAl-LDH nanoparticles enhanced the stability of dsRNA in nymph gut, and significantly enhanced the acaricidal effect of dsPcChit based on artificial diet bioassay.
Nanoparticles facilitate dsRNA entry into pomelo leaf cells
To clarify whether nanoparticles promote the entry of dsRNA into leaves, the leaf disc soaking bioassay (Fig. 3A) was employed for the RNAi experiments in P. citri nymphs. We clarified that the optimal concentration of dsPcChit to induce effective RNAi was 480 ng/µl (150 µl) by this method (Fig. S2 A-B). Next, we analyzed the RNAi efficiency induced by naked dsPcChit, CS-STPP-dsPcChit or MgAl-LDH-dsPcChit. The results showed that compared with naked dsPcChit, MgAl-LDH-dsPcChit significantly increased the gene knockdown efficiency and nymph mortality by 31% (P < 0.001) and 16.72% (P < 0.01), respectively. However, CS-STPP-dsPcChit did not effectively trigger RNAi (P > 0.05) (Fig. 3B-C).
Nanoparticles facilitate dsRNA entry into pomelo leaf cells to improve RNAi efficiency in P. citri nymphs. (A) Schematic diagram of leaf disc soaking bioassay; (B) Relative expression level of Chitnase and (C) Corrected mortality (%) after nymphs fed on leaves separately soaked with 480 ng/µl naked dsPcChit or nano-dsPcChit at 48 h; (D) Confocal microscopy images of leaves separately soaked with naked dsPcChit-Cy3 and nano-dsPcChit-Cy3 at 24 h, magnifications of 200 × were used. BF, bright field. Scale bar: 20 μm. (E) Quantitative analysis of the fluorescence intensities of nano-dsPcChit in leaf surface and leaf cell by Image J software. The blank control leaf disc was soaked with RNase-free water. Data represent mean ± SEM for four biological replicates, *P < 0.05, **P < 0.01, ***P < 0.001 with a Student’s t test; ns, not significant (P > 0.05). Different lowercase letters indicate the significant difference using of One-way ANOVA and Tukey’s multiple comparison tests (P < 0.05)
To elucidate how nano-dsRNA enhances RNAi efficiency in nymphs with leaf disc bioassay, the delivery efficiency of nanoparticles-loaded dsRNA into leaf cells was determined by using confocal microscopy assay. The images showed that nanoparticles enhanced the adhesion of dsRNA onto the leaf surface (Fig. 3D-E). CS-STPP-dsRNA-Cy3 and MgAl-LDH-dsRNA-Cy3 were not easily washed off from the leaf, and displayed very strong red fluorescence signals on the leaf surface. In contrast, most of the naked dsRNA-Cy3 was easily washed off. Significantly, MgAl-LDH nanoparticles facilitated the entry of dsRNA-Cy3 into leaf cells, with the strongest red fluorescence signals being observed in the leaf cells (Fig. 3D-E). Compared with the MgAl-LDH-dsRNA-Cy3 group, the CS-STPP-dsRNA-Cy3 and naked dsRNA-Cy3 groups showed much weaker fluorescence signals. Furthermore, the naked dsRNA-Cy3 group showed accumulation of most fluorescence signals around the cells. In short, CS-STPP and MgAl-LDH nanoparticles significantly enhanced the adhesion of dsRNA onto the leaf surface, but only MgAl-LDH could result in rapid entry of dsRNA into leaf cells, thereby significantly enhance the RNAi efficiency and the acaricidal effect of dsPcChit in the leaf disc bioassay. Therefore, MgAl-LDH was used for further study.
MgAl-LDH nanoparticles promote dsRNA transport in pomelo plants
To determine whether MgAl-LDH promote dsRNA transport in citrus plants, we first investigated the RNAi effects in P. citri by shoot soaking bioassay (Fig. 4A). The optimal concentration of dsPcChit for inducing effective RNAi was 640 ng/µl (300 µl) by this method (Fig. S2 A-B). Then, the RNAi efficiency of nymphs induced by naked dsPcChit or MgAl-LDH-dsPcChit was analyzed. The results showed that compared with naked dsPcChit, MgAl-LDH-dsPcChit increased the knockdown efficiency of Chit gene (by 25.00%) (P < 0.01) and nymph mortality (by 9.90%) (P < 0.05) (Fig. 4B-C).
MgAl-LDH promote dsRNA transport in pomelo plants to enhance RNAi efficiency in P. citri nymph. (A) Schematic diagram of shoot soaking bioassay; (B) Relative expression level of Chitnase and (C) Corrected mortality (%) after nymphs feeding on shoots separately soaked with 640 ng/µl naked dsPcChit or MgAl-LDH-dsPcChit at 48 h; (D) Confocal microscopy images of shoots separately soaked with naked dsPcChit-Cy3 or MgAl-LDH-dsPcChit-Cy3 at 24 h. BF, bright field. Scale bar: 200 μm. (E) Quantitative analysis of the fluorescence intensities of nano-dsPcChit in stem, vein and leaf by Image J software. Shoot soaked with RNase-free water was used as control. The copy number of PcChit dsRNA molecules in 1 µg total RNA from the stem top (F) and leaf (G) using ddPCR analysis after shoots separately soaked with naked dsRNA or MgAl-LDH-dsRNA at 24 h. Data represent mean ± SEM for biological replicates, *P < 0.05, **P < 0.01, ***P < 0.001 with a Student’s t test. Different lowercase letters indicate the significant difference using of One-way ANOVA and Tukey’s multiple comparison tests (P < 0.05)
To clarify how MgAl-LDH promotes dsRNA transport in citrus plants, we analyzed the transport efficiency of nano-dsRNA from the stem to the leaf tissues by confocal microscopy assay. Confocal microscopy images showed that MgAl-LDH nanoparticles greatly promoted the transport of dsRNA from the stem to the leaf (Fig. 4D-E). In the base and middle parts of the stem, comparable and strong fluorescence signals were observed in the vascular bundle of both naked dsRNA-Cy3 and MgAl-LDH-dsRNA-Cy3 groups. Notably, in the top part of the stem, main vein and leaf, the MgAl-LDH-dsRNA-Cy3 group showed stronger fluorescence intensities in the vascular bundle and cells than the naked dsRNA-Cy3 group (Fig. 4D-E).
To further determine the transport efficiency of MgAl-LDH-dsRNA in plant tissues, we detected the number of dsRNA molecules in the stem and leaf tissue by ddPCR. In the MgAl-LDH-dsRNA group, the copy number of PcChit dsRNA molecules in 1 µg of total RNA from the stem top and leaf was 1.95 times (Fig. 4F) and 1.23 times (Fig. 4G) that of the naked dsRNA group, respectively. Taken together, MgAl-LDH nanoparticles performed as a vector for dsRNA, and improved its long-distance transport from the stem to the leaf in citrus plants.
MgAl-LDH-dsRNA enhances control of P. Citri at other developmental stages
To investigate the controlling effect of MgAl-LDH-dsPcChit on other stages of P. citri, the RNAi efficiency of MgAl-LDH-dsPcChit on larvae and adults was evaluated by the above-mentioned three bioassays (Text S1). For knockdown experiments in larvae, compared with naked dsPcChit, MgAl-LDH-dsPcChit significantly increased the knockdown efficiency of the Chit gene by 28.00% (P < 0.01), 28.00% (P < 0.05), 32.00% (P < 0.001) (Fig. 5A), and significantly enhanced the corrected mortality by 25.48% (P < 0.01), 29.68% (P < 0.001), 25.66% (P < 0.05) (Fig. 5B) in artificial diet feeding, leaf disc soaking, and shoot soaking bioassay, respectively.
RNAi efficiency and mortality of P. citri larvae and adults induced by MgAl-LDH-dsRNA. (A) Relative expression level of Chitnase and (B) Corrected mortality (%) after larvae feeding on naked dsPcChit or MgAl-LDH-dsPcChit using artificial diet, leaf disc soaking and shoot soaking bioassay at 48 h; (C) Relative expression level of Chitnase and (D) Corrected mortality (%) after adults feeding on naked dsPcChit or MgAl-LDH-dsPcChit using artificial diet, leaf disc soaking and shoot soaking bioassay at 48 h. The final concentrations of naked dsPcChit or MgAl-LDH-dsPcChit were 320, 480 and 640 ng/µl in artificial diet, leaf disc soaking and shoot soaking bioassay, respectively. Data represent mean ± SEM for four biological replicates. Different lowercase letters indicate significant differences within the same bioassay groups using of One-way ANOVA and Tukey’s multiple comparison tests (P < 0.05)
For knockdown experiments in adults, compared with naked dsPcChit, MgAl-LDH-dsPcChit significantly increased the knockdown efficiency of the Chit gene by 36.00% (P < 0.01), 24.88% (P < 0.05), and 22.00% (P < 0.01) (Fig. 5C), as well as significantly enhanced the corrected mortality by 26.84% (P < 0.01), 19.58% (P < 0.01), and 16.40% (P < 0.05) (Fig. 5D) in artificial diet feeding, leaf disc soaking, and shoot soaking bioassay, respectively. Taken together, MgAl-LDH-dsPcChit could effectively control P. citri at different developmental stages.
In addition, we also investigate the dsRNA release efficiency from MgAl-LDH-dsRNA complex in PBS buffer to simulate the dsRNA release in P. citri. Our results showed that the dsRNA release efficiency decreased with the increase of pH (Fig. S5 A-B). The results showed that dsRNA was quickly released from MgAl-LDH-dsRNA in pH 3.0 PBS buffer, followed by in pH 5.0 PBS buffer, and then in pH 7.0 PBS buffer (Fig. S5B). In pH 3.0 PBS buffer, MgAl-LDH begin to dissolve within the first several minutes rapidly, leading to a 56.29% cumulative dsRNA release during the first 48 h, while it was released slowly from 48 to 96 h, with a 72.22% cumulative release of dsRNA at 96 h. However, in pH 5.0 and pH 7.0 PBS buffer, dsRNA was released uniformly, with 60.01% and 50.24% cumulative release of dsRNA at 96 h, respectively (Fig. S5B).
Universality of nanoparticles LDH in promoting the RNAi efficiency and mortality of piercing-sucking pests
Some metal elements such as iron are essential nutrients for plant growth [40]. To investigate whether those beneficial metal elements have similar function to enhance transport of dsRNA in the plants. We investigated the RNAi efficiency and insecticidal effects of MgFe-LDH-loaded dsRNA against P. citri nymphs by shoot soaking bioassay (Text S2). Firstly, we synthesized MgFe-LDH nanoparticles (Fig. S6 A), and obtained negatively charge MgFe-LDH-dsRNA complexes with the smaller particle sizes (< 80 nm) (Fig. S6 B-C) and high loading efficiencies (> 90%) (Fig. S6 D). The RNAi results revealed that compared with naked dsRNA (PcChs, PcChit, PcChs + PcChit), MgFe-LDH-dsRNA markedly increased the knockdown efficiency of the target genes by 28.76% (P < 0.05), 18.62% (P < 0.05) and 24.66% (P < 0.05) (Fig. S6 E-F), and enhanced the corrected mortality of nymphs by 16.01% (P < 0.05), 6.51% (P < 0.05), 19.19% (P < 0.05), respectively (Fig. S6 G).
To determine the universality that nanoparticles LDH promotes the RNAi efficiency and the mortality of piercing-sucking pests, this delivery pathway was also validated in more piercing-sucking pests, including D. citri and A. gossypii, two hemipeteran pests (Text S2). Firstly, we synthesized MgAl-LDH-dsRNA (Chs, Chit) and MgFe-LDH-dsRNA (Chs, Chit) complex with excellent properties (Fig. S7 A-B, Table S1). Then, the optimal concentration of dsRNA (Chs, Chit) that could induce effective RNAi in 5th instar D. citri nymphs (Fig. S8 A-B) and 3rd instar A. gossypii larvae (Fig. S8 C-D) was determined as 160 ng/µl in the shoot soaking bioassay.
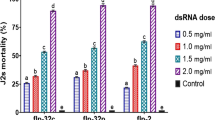
In the shoot soaking bioassay of D. citri (Fig. 6A), compared with naked dsRNA (DcChs, DcChit, DcChs + DcChit), MgAl-LDH-dsRNA and MgFe-LDH-dsRNA significantly increased (P < 0.05) the knockdown efficiency of the target genes by 16.05%, 30.63%, 24.46%, 30.44%, 22.67%, and 29.90% (Fig. 6B-C), and significantly increased (P < 0.05) the corrected mortality of nymphs by 15.32%, 12.15%, 11.67%, 15.32%, 23.15%, and 19.89%, respectively (Fig. 6D).
RNAi efficiency of D. citri and A. gosspii induced by MgAl-LDH-dsRNA and MgFe-LDH-dsRNA. (A) Schematic of 5th instar D. citri nymph and M. paniculata shoot soaked with naked dsRNA or nano-dsRNA; Relative expression level of (B)Chitin synthase, (C)Chitnase and (D) Corrected mortality (%) after D. citri fed on shoots separately soaked with 160 ng/µl naked dsRNA or nano-dsRNA at 48 h; (E) Schematic of 3rd instar A. gossypii larvae and cotton shoot soaked with naked dsRNA or nano-dsRNA; Relative expression level of (F)Chitin synthase, (G)Chitnase and (H) Corrected mortality (%) after A. gossypii fed on shoots separately soaked with 160 ng/µl naked dsRNA or nano-dsRNA at 48 h. Data represents mean ± SEM for four biological replicates. Different lowercase letters indicate significant differences within the same concentration groups using of One-way ANOVA and Tukey’s multiple comparison tests (P < 0.05)
In the shoot soaking bioassay of A. gossypii (Fig. 6E), compared with naked dsRNA (AgChs, AgChit, AgChs + AgChit), MgAl-LDH-dsRNA and MgFe-LDH-dsRNA markedly increased (P < 0.05) the knockdown efficiency of the target genes by 16.42%, 27.02%, 34.75%, 18.34%, 17.53% and 65.58% (Fig. 6F-G), and significantly increased (P < 0.05) the corrected mortality of larvae by 6.50%, 16.77%, 26.04%, 23.59%, 26.25% and 38.13%, respectively (Fig. 6H). Taken together, MgAl-LDH-dsRNA and MgFe-LDH-dsRNA could enhance the RNAi efficiency of the piercing-sucking pests D. citri and A. gossypii.
Discussion
In this study, we identified LDH nanoparticles as an ideal carrier for systemic delivery of dsRNA and facilitating dsRNA transport in plants tissues. LDH-dsRNA could provide effective control of the piercing-sucking pests P. citri, D. citri, and A. gosspii. Our study revealed that LDH nanoparticles can systematically transport dsRNA from the stem to the leaf and then to the gut of piercing-sucking pests.
Herein, we found that CS-STPP and MgAl-LDH nanoparticles enhanced the stability of dsRNA in nymph gut compared with naked dsRNA in artificial diet feeding bioassay. Previous studies have demonstrated that oral uptake of SPC-dsRNA, CS-dsRNA or CS-STPP-dsRNA could increase the RNAi efficiency in pests with an alkaline gut pH, such as Agrotis ypsilon [41], Chilo suppressalis [42, 43], Bactrocera dorsalis [44] and Anopheles aegypti [35]. Mites generally have a weakly alkaline (from 6.6 to 7.6) gut pH [45]. The dsRNA was more quickly released from CS-dsRNA in pH 7.4 PBS buffer than in pH 5.4 PBS buffer [30], indicating that the dsRNA release rate from the nano-dsRNA is pH-dependent [25, 30]. MgAl-LDH nanoparticles can be dissolved in solutions with pH below 7.52, and the dissolution rate increases with decreasing of pH [25]. Our results showed that dsPcChit is slowly and uniformly released from MgAl-LDH-dsPcChit complex in pH 5.0 and pH 7.0 PBS buffer, which then enhances the RNAi efficiency of P. citri. The dsRNA and nanoparticles complex can protect the dsRNA from unfavorable external environment, such as nucleases, rainwater, high temperature and other factors [27, 46,47,48]. The stability of dsRNA in other environments expect for pest gut deserves our further investigation. Additionally, CQD nanoparticles showed the lethal effect on P. citri nymphs, which is similar with the study in Spodoptera frugiperda larvae [49], but the reason for this lethal effect needs further study.
In this study, MgAl-LDH nanoparticles enhanced the adhesion of dsRNA onto the leaf surface, and facilitated the entry of dsRNA into leaf cells of pomelo plants. In the foliar application of nanoparticles, positively charged nanoparticles are more readily adsorbed on leaf surface, whereas negatively charged nanoparticles can more efficiently enter cells [40, 50], and nanoparticles with smaller particle sizes can better enter the plant tissues through stomatal, wounds and cuticular pathways [40, 51]. These findings further supported our confocal results that MgAl-LDH-dsRNA complex successfully entered into the leaf cells of pomelo. Negatively charged MgAl-LDH-dsRNA-Cy3 with the smallest particle size showed the strongest fluorescence signals in leaf cells, whereas positively charged CS-STPP-dsPcChit with larger particle sizes were mainly adsorbed on the leaf surface. The efficient adhesion of MgAl-LDH-dsRNA onto leaves was also reported in cowpea plant against virus [25]. Piercing-sucking P. citri could not feed on the CS-STPP-dsPcChit on the leaf surface, and therefore RNAi was not effectively triggered in the leaf disc soaking bioassay.
MgAl-LDH-dsRNA complex is usually applied in the form of foliar spray to control plant diseases caused by pepper mild mottle virus and cucumber mosaic virus [25], pathogenic fungi Fusarium oxysporum [52], and Botrytis cinerea [53]. In this study, we found a mechanism that MgAl-LDH nanoparticles enhance the RNAi efficiency to target pests by facilitating long-distance transport of dsRNA from the stem to the leaf via the vascular system of pomelo plants, thereby effectively controlling the piercing-sucking pests P. citri. In the foliar or root application of nano dsRNA-based biopesticides, nanoparticles improve the stability and adhesion of dsRNA, promote the absorption, translocation, and processing of dsRNA by plant tissues, and increase the absorption and uptake of pest cells, and improve its insecticidal efficiency [27, 38, 52, 54]. Systemic pesticides, which are absorbed by plants and then transported to various untreated tissues [2], are most suitable for controlling the piercing-sucking pests [55], due to their feeding characteristics of piercing and sucking plant sap [1]. Therefore, delivery of MgAl-LDH-dsRNA by facilitating transport of dsRNA through the vascular system may be a promising strategy to controlling piercing-sucking pests on plants, particularly woody plants such as citrus in the field.
We found that MgAl-LDH-dsRNA complex could enhance control over the life cycle of P. citri, and the RNAi efficiency in the larvae and nymphs was greater than that in adult mites. Those results are similar to previously reports that RNAi efficiency is associated with the developmental stage of the pests, and the immature pests were more responsive to environmental RNAi than their counterpart teneral or mature adults, including Drosophila suzukii [56], Bactericera cockerelli [57], Leptinotarsa decemlineata [58], which was potentially attributed to physiology of younger instars (larvae or nymphs) that experience rapid growing and molting [59].
In this study, MgFe-LDH nanoparticles-loaded dsChit or dsChs also enhanced the RNAi efficiency of P. citri nymphs. This is similar to the previous study that MgFe-LDH nanoparticles-loaded dsRNA effectively disrupted Bemisia tabaci at multiple developmental stages in cotton plant [27]. For management of tomato fungal pathogen Botrytis cinerea, MgFe-LDH nanoparticles-loaded dsRNA achieved a similar RNAi efficiency to MgAl-LDH nanoparticles-loaded dsRNA [53]. Metal elements such as iron and magnesium are essential nutrients for plant growth [40], thus revealing good prospects for field applications. Furthermore, we also found that the universality of the RNAi efficiency enhanced by MgAl-LDH and MgFe-LDH nanoparticles in other piercing-sucking hemipteran pests like D. citri and A. gossypii, providing further evidences for the effective application of metal elements-based nanoparticles against piercing-sucking pests.
Conclusion
In conclusion, LDH nanoparticles can facilitate long-distance transport of dsRNA in plant tissues, improve the adhesion of dsRNA onto the leaf surface and entry into leaf cells, and also enhance the stability of dsRNA in pest gut. LDH-dsRNA provides effective control of more piercing-sucking pests P. citri, D. citri and A. gossypii. This study provides insights into the synergistic mechanism underlying the systemic transport of nano-dsRNA in plants, and proposes a potential eco-friendly control strategy against piercing-sucking pests in the field.
Materials and methods
Pest rearing
P. citri was collected from citrus orchards in the Huazhong Agricultural University, Wuhan, China. They were reared on pomelo plants in a glass greenhouse at 25 ± 2 ℃ with 70 ± 5% relative humidity.
D. citri was continuously reared in a greenhouse at 26 ± 2 ℃ with 65 ± 5% RH and 14:10 h (light: dark) photoperiod in the Gannan Normal University, Ganzhou, China. The D. citri colonies were reared on Murraya paniculata plants and caged using insect rearing cages (60 × 60 × 90 cm3).
A. gossypii was continuously reared in a greenhouse at 22 ± 2 ℃ with 70 ± 5% RH and 14:10 h (light: dark) photoperiod in the Huazhong Agricultural University, Wuhan, China. The A. gossypii colonies were reared on cotton plants and caged using insect rearing cages (60 × 60 × 90 cm3).
Synthesis of dsRNA and dsRNA-Cy3 in vitro
Total RNA was extracted from P. citri, D. citri, and A. gossypii with the Trizol Reagent (TaKaRa) following the manufacturer’s instructions. Briefly, approximately 100 surviving P. citri nymphs, D. citri nymphs and A. gossypii larvae were quickly transferred to 0.25 ml trizol reagent, and the samples were ground for 30 s on ice with electric tissue lapping instrument (TGrinder). After grinding, 0.75 ml of trizol reagent was added immediately, and the sample tissues were lysed at room temperature for 10 min. The cDNA was synthesized using a First Strand cDNA synthesis kit (Simgen) following manufacturer’s instructions with 1 µg total RNA used as template.
Fragments of PcChs, PcChit, DcChs, DcChit, AgChs and AgChit genes were amplified from the cDNA by RT-PCR using primers containing a T7 polymerase promoter region (Table S2). The purified PCR products were cloned into pTOPO-T Simple vector (Aidlab). The accuracy of sequences was verified by Quintara sequencing. The plasmids were extracted using a plasmid extraction kit (Omega). The dsRNA was synthesized in vitro using 1 µg PCR product as the specific-template on the T7 Ribomax Express RNAi System (Promega). DsRNA-Cy3 was synthesized using the HyperScribeTM T7 High Yield Cy3 RNA Labeling kit (APExBIO). The quality and integrity of dsRNA and dsRNA-Cy3 were determined on 1.2% (w/v) agarose gel, and their concentrations were measured with a NanoDrop 2000 (Thermo).
Preparation of nano-dsRNA
CS-STPP-dsRNA
CS nanoparticles were prepared by the ionic gelation method with slight modifications [24]. Specifically, 0.02 g chitosan (Sigma-Aldrich, C3646) was dissolved in 100 ml sodium acetate buffer (pH 4.5), and the reaction mixture was stirred (700 rpm) overnight at room temperature (25 ± 2 ℃, 60 ± 5% RH).
About 8 µg dsRNA was suspended in 25 µl sodium tripolyphosphate pentabasic (STPP, Sinopharm, 72061; 200 µg/ml) solutions, which was then added to 25 µl chitosan solutions. The mixture was heated in a water bath at 55℃ for 1 min, immediately mixed by vortexing for 30 s, and allowed to stand at room temperature for 10 min. To obtain optimal CS-STPP-dsRNA complex with complete dsRNA loading, the mass ratio of CS-STPP to in vitro transcribed dsRNA was set as 1:1, 3:1, 5:1, 7:1, and 9:1.
CQD-STPP-dsRNA
CQD nanoparticles were prepared by microwave-mediated caramelization of aqueous PEG-200 solution [36]. First, 3 ml polyethylene glycol-200 (PEG-200, Coolaber, CP8186; 9 ml) aqueous solution was added to 2 ml aqueous solution of polyethylenimine (PEI, Sigma-Aldrich, 408727; 100 mg), and the mixture was heated in a microwave for nearly 3 min at 800 W under reflux condensing conditions.
About 8 µg of dsRNA was suspended in 25 µl STPP solutions, and then added to 25 µl cooled PEG-PEI solution, followed by incubation at 4 ℃ overnight [60]. Subsequently, CQD-STPP was conjugated with the fixed amount of dsRNA at the mass ratio of 1/50:1, 1/100:1, 1/150:1, 1/200:1, 1/300:1, 1/500:1, 1/700:1, 1/900:1.
LDH-dsRNA
MgAl-LDH nanoparticles were prepared by the non-aqueous precipitation method with slight modifications [61]. Specifically, 6 mmol Mg(NO3)2·6H2O (Sinopharm, C217560010) and 2 mmol Al(NO3)3·9H2O (Sinopharm, 80003661) dissolved in 10 ml methanol (Sinopharm, 10014118) were added into 16 mmol NaOH (Sinopharm, 10019718; 40 ml) solution and stirred for 30 min with N2 bubbling. After reaction, the precipitate was collected by centrifugation, and washed three times with 40 ml methanol. Then the precipitates were transferred to a Teflon-lined autoclave and treated at 100℃ for 16 h. The precipitates were collected by centrifugation, washed three times with 40 ml deionized water, and re-suspended in 40 ml deionized water.
About 8 µg dsRNA was suspended in 25 µl RNase-free water. Subsequently, the dsRNA suspension was added to an aliquot of MgAl-LDH nanoparticles, and incubated at 37℃ with shaking at 220 rpm for 30 min. MgAl-LDH was loaded with in vitro transcribed dsRNA at the mass ratio of 1:1, 3:1, 5:1, 7:1, and 9:1, respectively, to obtain optimal MgAl-LDH-dsRNA complex with complete dsRNA loading.
Following the procedure mentioned above, MgFe-LDH nanoparticles were prepared under optimized reaction conditions (unpublished data). MgFe-LDH nanoparticles with the smallest particle size were used as dsRNA carriers in the shoot soaking of P. citri, D. citri, and A. gossypii. MgFe-LDH nanoparticles was loaded with in vitro transcribed dsRNA (Chs, Chit) of P. citri, D. citri and A. gossypii at the mass ratio of 1:1, 3:1, 5:1, 7:1, 9:1 and 11:1, respectively, to obtain optimal MgFe-LDH-dsRNA complex with complete dsRNA loading.
SPC-dsRNA
Star polycation (SPC, size: 100.60 ± 6.47 nm, Pdi: 0.35 ± 0.02; zeta potential: + 22.27 ± 0.30 mV) nanoparticles were provided by Prof. Jie Shen, China Agricultural University. First, 8 µg dsRNA was suspended in 25 µl RNase-free water. Then, dsRNA suspension was added to an aliquot of SPC nanoparticles. The mixture was vortexed for 3 s, and then allowed to stand at room temperature for 30 min [41]. The SPC nanoparticles were conjugated with in vitro transcribed dsRNA at the mass ratio of 1:1, 1/3:1, 1/5:1, 1/7:1, 1/9:1, respectively.
For all the above nano-dsRNA complex treatments, complete nano-dsRNA loading was assessed by their retention in the wells of a 1.2% (w/v) agarose gel.
Characterization of nano-dsRNA
Transmission electron microscopy
The morphologies of nanoparticles and nano-dsRNA with complete loading of dsRNA were observed by transmission electron microscopy (TEM, JEOL 2100 F, Japan). Nanoparticles and nano-dsRNA were drop-cast onto plasma-treated carbon-coated copper mesh grids and imaged at an acceleration voltage of 100 kV (H-7650).
Dynamic light scattering (DLS) measurement
The particle size, polydispersity index, and zeta potential of nanoparticles and nano-dsRNA were determined by Zetasizer Nano ZS instrument (Malvern, UK). Nanoparticles and nano-dsRNA were dispersed in distilled water, respectively, diluted to an optical density of 1, and placed in cuvettes (Malvern) for DLS measurement. All measurements were conducted in triplicates at 25℃.
Calculation of loading efficiency of nano-dsRNA
The dsRNA loading efficiency of nanoparticles was evaluated by a previously described method [35] with minor modifications. To determine the dsRNA loading efficiency onto CS-STPP, CQD-STPP, MgAl-LDH, MgFe-LDH and SPC nanoparticles at the optimal mass ratio, the nano-dsRNA was separated by centrifugation at 15, 000 rpm for 30 min at 4℃. The absorbance at 260 nm of free dsRNA in the supernatants was determined with a NanoDrop 2000. Supernatant recovered from naked nanoparticles (without dsRNA) was used as the blank control. The dsRNA loading efficiency was calculated according to the following equation:
Nano-dsRNA-mediated knockdown of Chitinase in P. Citri nymphs with three methods
To determine the minimum effective dsRNA concentration of RNAi in artificial diet feeding, leaf disc soaking, and shoot soaking bioassay, five diluted concentrations of dsRNA (160, 320, 480, 640, and 800 ng/µl) were prepared using RNase-free water, and the same concentrations of dsGFP were used as controls.
Artificial diet feeding bioassay
The artificial diet feeding system consisted of a glass tube (3 cm in height and 2 cm in diameter) with an artificial diet (7.5% sucrose solutions) contained in a double layer of Parafilm. Approximately 200 nymphs were starved for 9 h, and then fed on 100 µl artificial diet containing naked dsPcChit, CS-STPP-dsPcChit, CQD-STPP-dsPcChit, MgAl-LDH-dsPcChit or SPC-dsPcChit, respectively. Nymphs fed on 100 µl artificial diet separately containing naked dsGFP, CS-STPP-dsGFP, CQD-STPP-dsGFP, MgAl-LDH-dsGFP or SPC-dsGFP were used as controls. The final concentration of naked dsRNA or nano-dsRNA was 320 ng/µl. Nymphs fed on 100 µl artificial diet was used as a blank control.
To evaluate the effect of nanoparticles in nymphs, starved nymphs were fed on 100 µl artificial diet containing CS-STPP, CQD-STPP, MgAl-LDH or SPC nanoparticles, respectively. The mortality of nymphs was recorded at 48 h post feeding.
Pomelo leaf disc soaking bioassay
The leaf disc soaking was performed by a previously described method with minor modifications [62]. Young leaves were collected from HB pomelo trees, rinsed with RNase-free water for 5 min to reduce bacterial contamination. A leaf was made into leaf disc (2.1 cm diameter) using a leaf puncher, and dried for 2 h at room temperature. The abaxial side of leaf disc was soaked with 150 µl of naked dsPcChit, CS-STPP-dsPcChit or MgAl-LDH-dsPcChit, respectively. The control leaf disc was soaked with 150 µl of naked dsGFP, CS-STPP-dsGFP, MgAl-LDH-dsGFP and RNase-free water (blank control), respectively. The final concentration of naked dsPcChit or nano-dsPcChit was 480 ng/µl. After the test solutions were completely absorbed, each leaf disc was put in a Petri dish (5 cm diameter). The adaxial side of leaf disc was attached to the medium, and the edges of the leaf disc were covered with dampened cotton to avoid the escape of nymphs. A total of 150 nymphs (starved for 16 h) on each leaf disc were fed with treated leaves.
Pomelo shoot soaking bioassay
The shoot soaking was performed as previously described [16] with minor modifications. Shoots of HB pomelo plants about 6–8 cm long with three leaves were collected, and rinsed with RNase-free water for 5 min to reduce bacterial contamination. The base of the shoot was cut at a 45° angle, and dried for 2 h at room temperature. After that, the shoot was immersed in a 0.6 ml RNase-free tube separately filled with 300 µl (640 ng/µl) of naked dsPcChit, MgAl-LDH-dsPcChit or MgFe-LDH-dsPcChit as treated groups, and that soaked with 300 µl (640 ng/µl) of naked dsGFP and MgAl-LDH-dsGFP, MgFe-LDH-dsPcChit, and 300 µl RNase-free water (blank control) were used as control groups. The treated shoot was placed at room temperature for 12 h to stimulate absorption of the tested solutions. Shoot treated with test solutions was transferred to a box (6.5 cm in diameter, 10 cm in high) with sterile agar medium to fix the tube containing the shoot. A total of 200 nymphs (starved for 24 h) were released into the box to feed on the shoot.
In all treated and control groups, nymphs were reared under controlled growth conditions of 25 ± 1 °C, 70 ± 5% RH, with a photoperiod of 14 h light :10 h dark. All treatments were conducted in four replications for 48 h. The mortality was recorded and surviving individuals were collected for RNA isolation. qRT-PCR was performed to detect the mRNA levels of PcChit gene. The percentage of nymph mortality was corrected using Abbott′s correction [63] as corrected mortality (%) = (% mortality in treatment - % mortality in control) / (100 - % mortality in control).
Transfer of nano-dsRNA into nymphs, leaf cells, and shoots
We performed nano-dsRNA transfer experiments with the prepared nanoparticles to deliver Cy3-labeled dsRNA, and analyzed the transfer efficiency of nano-dsRNA into nymph guts, leaf cells, and shoots based on Cy3 fluorescence signal intensities. Fluorescence intensities were quantified by the Image J software. All treatments and controls were performed in triplicate, and maintained in a dark incubator at 25 ± 1 ℃ and 70 ± 5% RH.
Transfer of nano-dsRNA into P. Citri nymphs
We tested the protective effect of nanoparticles on dsRNA in the mite guts. Briefly, approximately 100 starved nymphs were separately fed with 100 µl artificial diet containing 320 ng/µl of CS-STPP-dsRNA-Cy3, CQD-STPP-dsRNA-Cy3, MgAl-LDH-dsRNA-Cy3 or SPC-dsRNA-Cy3, respectively. Nymphs fed on 100 µl (320 ng/µl) naked dsPcChit-Cy3 was used as the control, and those fed on 100 µl artificial diet was used as a blank control. After 24 and 48 h, fluorescent images of nymphs were taken with a Leica M205FA microscope equipped with a RFP filter using the DFC7000GT system. Bright-field images were collected using the same system with an exposure time of 440 ms without filters. We further tested the protective effect of MgAl-LDH nanoparticles on dsRNA in the mite guts at 72, 96 and 120 h under the same conditions. Additionally, the dsRNA release efficiency from MgAl-LDH-dsRNA complex was detected in different PBS buffer in vitro as previously described [30] with minor modifications (Text S3).
Transfer of nano-dsRNA into pomelo leaf cells
The abaxial side of leaf disc (7.5 mm diameter) was soaked with 50 µl (80 ng/µl) of CS-STPP-dsRNA-Cy3, MgAl-LDH-dsRNA-Cy3 and naked dsRNA-Cy3 (control), respectively. The blank control leaf disc was soaked with 50 µl RNase-free water. After the tested solutions were completely absorbed, leaf discs were rinsed with 3 ml RNase-free water to remove fluorescence on the surface. After 24 h, leaf discs were observed under a confocal laser scanning microscopy (Leica TCS SP8 DLS). The images of the abaxial side and cells of leaves were taken with a Cy3 filter (excitation 552 nm, emission 560–620 nm), and bright-field images were taken using the same system without filters.
Transfer of nano-dsRNA into pomelo shoots
The shoot (6–8 cm in length) was separately soaked with 300 µl (160 ng/µl) of MgAl-LDH-dsRNA-Cy3 complex, dsRNA-Cy3 (control), and RNase-free water (blank control), respectively. After 24 h, transverse slices (30–50 μm thickness) from the stem, main vein, and leaf were cut with a vibroslicer (VT1200S, Bio-rad). All samples of transverse slices were observed under a confocal laser scanning microscopy (Leica SP8). The images of transverse sections of the stem and leaves were collected with a Cy3 filter (excitation 552 nm, emission 560–620 nm), and bright-field images were taken using the same system without filters. Gene expression analysis by qPCR.
Further, we investigated the expression levels of PcChs, PcChit, DcChs, DcChit, AgChs and AgChit genes by qPCR according to the manufacturers’ instructions. Fluorescence qPCR was performed with SYBR Green Master Mix (Bio-Rad). The reaction system was 20 µl in volume, with each consisting of 10 µl SYBR Green Master Mix (Hieff UNICON®), 0.8 µl forward and reverse primers, 6.4 µl sterile water, and 2 µl template. The reaction conditions for fluorescence quantification were as follows: 95 °C for 30 s; 39 cycles of 95 °C for 10 s and 60 °C for 30 s. Primer sequences are shown in Table S2, and the qPCR experiment was conducted with at least three independent biological replicates and three technical replicates for each sample. Relative gene expression was calculated using the 2−ΔΔCT method. For normalization of gene expression levels, PcGAPDH [19], DcGAPDH [64] and AgGAPDH [65] were employed to be used as internal reference gene for P. citri, D. citri and A. gossypii, respectively, as described in previously related literatures. The data were expressed as mean ± standard error.
Droplet digital PCR (ddPCR) to quantify dsRNA in stem leaf tissues
To compare dsRNA expression in the shoot away from the application site, the total RNA of stem and leaf (about 150 mg of each sample) was extracted using the plant polysaccharide polyphenol RNA kit (Simgen). cDNA was synthesized using 1 µg RNA per sample as described in the previous Sect. 2.3. The region used for primers and probes designing of ddPCR are shown in Fig. S9. The reaction system was 22 µl in volume, with each consisting of 11 µl 2x dPCR Probe Master mix Plus (cy5.5) (Sniper), 1 µl forward and reverse primers, 0.5 µl probe, 7.5 µl sterile water, and 1 µl template. Droplets were generated with a DQ24 (Sniper) at 60 °C for 5 min, and PCR was performed using a DQ24 with the following temperature profile of initial denaturation 95 °C for 5 min, 45 cycles of 95 °C for 20 s, 60 °C for 30 s. After thermal cycling, the plates were transferred to a Droplet reader (Sniper), and the digital PCR data were analyzed with the Sight Pro software (V0.3, Sniper).
Statistical analysis
Statistical analysis was performed by using SPSS 21.0 version and GraphPad Prism version 8.0 software. The t-test and one-way ANOVA (with Tukey multiple tests) were conducted to determine the differences in mean particle diameter and polydispersity of nanoparticles and nano-dsRNA complex, dsRNA loading efficiency of nanoparticles, expression levels of PcChs, PcChit, DcChs, DcChit, AgChs and AgChit genes, and corrected mortality of P. citri (larvae, nymphs and adults), D. citri 5th instar nymphs and A. gossypii 3rd instar larvae among groups. P-value smaller than 0.05 were considered as significantly different statistically.
Data availability
No datasets were generated or analysed during the current study.
References
Omkar. Sucking pests of crops. Springer Singap. 2020;1–515.
Akashe MM, Pawade UV, Nikam AV. Classification of pesticides: a review. Int J Res Ayurveda Pharm. 2018;9:144–50.
Delso NS, Rogers VA, Belzunces LP, Bonmatin JM, Chagnon M, Downs C, Furlan L, Gibbons DW, Giorio C, Girolami V, Goulson D, Kreutzweiser DP, Krupke CH, Liess M, Long E, McField M, Mineau P, Mitchell EAD, Morrissey CA, Noome DA, Pisa L, Settele J, Stark JD, Tapparo A, Van Dyck H, Van Praagh J, Van der Sluijs JP, Whitehorn PR, Wiemers M. Systemic insecticides (neonicotinoids and fipronil): trends, uses, mode of action and metabolites. Environ Sci Pollut Res. 2014;22:5–34.
De Rouck S, İnak E, Dermauw W, Van Leeuwen T. A review of the molecular mechanisms of acaricide resistance in mites and ticks. Insect Biochem Molec. 2023;159:103981.
Carletto J, Martin T, Vanlerberghe-Masutti F, Brévault T. Insecticide resistance traits differ among and within host races in Aphis gossypii. Pest Manag Sci. 2009;66:301–7.
Price DRG, Gatehouse JA. RNAi-mediated crop protection against insects. Trends Biotechnol. 2008;26:393–400.
Liu SS, Jaouannet M, Dempsey DA, Imani J, Coustau C, Kogel KH. RNA-based technologies for insect control in plant production. Biotechnol Adv. 2020;39:107463.
Messina E. Registration decision for the new active ingredient Ledprona (Leptinotarsa decemlineata-specific recombinant double-stranded interfering Oligonucleotide GS2) (CAS Number: 2433753-68-3). U.S. Environmental Protection Agency; 2023. pp. 1–21.
Jaubert-Possamai S, Le Trionnaire G, Bonhomme J, Christophides GK, Rispe C, Tagu D. Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC Biotechnol. 2007;7:63.
Rosa C, Kamita SG, Falk BW. RNA interference is induced in the glassy winged sharpshooter Homalodisca vitripennis by actin dsRNA. Pest Manag Sci. 2012;68:995–1002.
Wu ZZ, Zhang WY, Lin YZ, Li DQ, Shu BS, Lin JT. Genome-wide identification, characterization and functional analysis of the chitianse and chitinase-like gene family in Diaphorina citri. Pest Manag Sci. 2022;78:1740–8.
Li FQ, Di ZJ, Tian JH, Dewer Y, Qu C, Yang SY, Luo C. Silencing the gustatory receptor BtGR11 affects the sensing of sucrose in the whitefly Bemisia tabaci. Front Bioeng Biotechnol. 2022;10:1054943.
Li TC, Chen JS, Fan XB, Chen WW, Zhang WQ. MicroRNA and dsRNA targeting chitin synthase a reveal a great potential for pest management of the hemipteran insect Nilaparvata lugens. Pest Manag Sci. 2017;73:1529–37.
Wu MT, Zhang Q, Dong Y, Wang ZC, Zhan WQ, Ke ZB, Li SC, He L, Ruf S, Bock R, Zhang J. Transplastomic tomatoes expressing double-stranded RNA against a conserved gene are efficiently protected from multiple spider mites. New Phytol. 2023;237:1363–73.
Gong C, Yang ZZ, Hu Y, Wu QJ, Wang SL, Guo ZJ, Zhang YJ. Silencing of the BtTPS genes by transgenic plant-mediated RNAi to control Bemisia tabaci MED. Pest Manag Sci. 2021;78:1128–37.
de Andrade EC, Hunter WB. Integrated pest management: current concepts and ecological perspective, in 19 ‘: RNA interference – natural gene-based technology for highly specific pest control (HiSPeC), ed. by Smart LE, Aradottir GL and Bruce TJA, pp.93–109 (2016).
Chakraborty P, Ghosh A. Topical spray of dsRNA induces mortality and inhibits Chilli leaf curl virus transmission by Bemisia tabaci Asia II 1. Cells. 2022;11:833–52.
Dong Y, Wu MT, Zhang Q, Fu JQ, Loiacono FV, Yang Y, Wang ZC, Li SC, Chang L, Bock R, Zhang J. Control of a sap-sucking insect pest by plastid-mediated RNA interference. Mol Plant. 2022;15:1176–91.
Xia WK, Shen XM, Ding TB, Niu JZ, Zhong R, Liao CY, Feng YC, Dou W, Wang JJ. Functional analysis of a chitinase gene during the larval-nymph transition in Panonychus citri by RNA interference. Exp Appl Acarol. 2016;70:1–15.
Yan S, Ren BY, Shen J. Nanoparticle-mediated double-stranded RNA delivery system: a promising approach for sustainable pest management. Insect Sci. 2020;28:21–34.
Nitnavare RB, Bhattacharya J, Singh S, Kour A, Hawkesford MJ, Arora N. Next generation dsRNA-based insect control: success so far and challenges. Front Plant Sci. 2021;12:673576.
Sadeghi R, Rodriguez RJ, Yao Y, Kokini JL. Advances in nanotechnology as they pertain to food and agriculture: benefits and risks. Annu Rev Food Sci Technol. 2017;8:467–92.
Li P, Huang Y, Fu C, Jiang SX, Peng W, Jia Y, Peng H, Zhang P, Manzie N, Mitter N, Xu ZP. Eco-friendly biomolecule‐nanomaterial hybrids as next‐generation agrochemicals for topical delivery. EcoMat. 2021;3:e12132.
Zhang X, Zhang J, Zhu KY. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol Biol. 2010;19:683–93.
Mitter N, Worrall EA, Robinson KE, Li P, Jain RG, Taochy C, Fletcher SJ, Carroll BJ, Lu GQ, Xu ZP. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat Plants. 2017;9:16207.
Yan S, Qian J, Cai C, Ma ZZ, Li JH, Yin MZ, Ren BY, Shen J. Spray method application of transdermal dsRNA delivery system for efficient gene silencing and pest control on soybean aphid Aphis glycines. J Pest Sci. 2019;93:449–59.
Jain RG, Fletcher SJ, Manzie N, Robinson KE, Li P, Lu E, Brosnan CA, Xu ZP, Mitter N. Foliar application of clay-delivered RNA interference for whitefly control. Nat Plants. 2022;8:535–48.
Lv HX, Li XC, Li JQ, Yu C, Zeng QH, Ning GG, Wan H, Li JH, Ma KS, He S. Overcoming resistance in insect pest with a nanoparticle-mediated dsRNA and insecticide co-delivery system. Chem Eng J. 2023;475:146239.
Yu C, Li JQ, Zhang ZY, Zong M, Qin CW, Mo ZY, Sun D, Yang DS, Zeng QH, Wang JY, Ma KS, Li JH, Wan H, He S. Metal–organic framework-based insecticide and dsRNA codelivery system for insecticide resistance management. ACS Appl Mater Interfaces. 2023;15:48495–505.
Zhou H, Wan FL, Jian YF, Guo FY, Zhang M, Shi SY, Yang L, Li SL, Liu Y, Ding W. Chitosan/dsRNA polyplex nanoparticles advance environmental RNA interference efficiency through activating clathrin-dependent endocytosis. Int J Biol Macromol. 2023;253(Pt 4):127021.
Wise JC, Wise AG, Rakotondravelo M, Vandervoort C, Seeve C, Fabbri B. Trunk injection delivery of dsRNA for RNAi-based pest control in apple trees. Pest Manag Sci. 2022;78:3528–33.
Hunter WB, Glick E, Paldi N, Bextine BR. Advances in RNA interference: dsRNA treatment in trees and grapevines for insect pest suppression. Southwest Entomol. 2012;37:85–7.
Liu S, Ladera-Carmona MJ, Poranen MM, van Bel AJE, Kogel K-H, Imani J. Evaluation of dsRNA delivery methods for targeting macrophage migration inhibitory factor MIF in RNAi-based aphid control. J Plant Dis Protect. 2021;128:1201–12.
Dalakouras A, Jarausch W, Buchholz G, Bassler A, Braun M, Manthey T, Krczal G, Wassenegger M. Delivery of hairpin RNAs and small RNAs into woody and herbaceous plants by trunk injection and petiole absorption. Front Plant Sci. 2018;9:01253.
Dhandapani RK, Gurusamy D, Howell JL, Palli SR. Development of CS-TPP-dsRNA nanoparticles to enhance RNAi efficiency in the yellow fever mosquito, Aedes aegypti. Sci Rep-UK. 2019;9:8775.
Das S, Debnath N, Cui YJR, Unrine J, Palli SR. Chitosan, carbon quantum dot, and silica nanoparticle mediated dsRNA delivery for gene silencing in Aedes aegypti: a comparative analysis. ACS Appl Mater Interfaces. 2015;7:19530–5.
Yong JX, Wu MM, Zhang R, Bi SN, Mann CWG, Mitter N, Carroll BJ, Xu ZP. Clay nanoparticles efficiently deliver small interfering RNA to intact plant leaf cells. Plant Physiol. 2022;190:2187–202.
Ma ZZ, Zhang YH, Li MS, Chao ZJ, Du XG, Yan S, Shen J. A first greenhouse application of bacteria-expressed and nanocarrier-delivered RNA pesticide for Myzus persicae control. J Pest Sci. 2022;96:181–93.
Zhu KY, Merzendorfer H, Zhang W, Zhang J, Muthukrishnan S. Biosynthesis, turnover, and functions of chitin in insects. Annu Rev Entomol. 2016;61:177–96.
Wu HH, Li ZH. Nano-enabled agriculture: how do nanoparticles cross barriers in plants? Plant Commun. 2022;3:100346.
Li JH, Qian J, Xu YY, Yan S, Shen J, Yin MZ. A facile-synthesized star polycation constructed as a highly efficient gene vector in pest management. ACS Sustain Chem Eng. 2019;7:6316–22.
Dong CL, Zhu F, Lu MX, Du YZ. Characterization and functional analysis of Cshsp19.0 encoding a small heat shock protein in Chilo Suppressalis (Walker). Int J Biol Macromol. 2021;188:924–31.
Wang KX, Peng YC, Chen JS, Peng Y, Wang XS, Shen ZH, Han ZJ. Comparison of efficacy of RNAi mediated by various nanoparticles in the rice striped stem borer (Chilo Suppressalis). Pestic Biochem Physiol. 2020;165:104467.
Guo SK, Guo XY, Zheng LY, Zhao ZH, Liu LJ, Shen J, Li ZH. A potential genetic control by suppression of the wing developmental gene wingless in a global invasive pest Bactrocera dorsalis. J Pest Sci. 2020;94:517–29.
Culo Z. The role of V-ATPase in regulating pH in the digestive tract of Tetranychus urticae Koch. Electronic thesis and dissertation repository, Western University. 2020.
Mitter N, Worrall EA, Robinson KE, Li P, Jain RG, Taochy C, Fletcher SJ, Carroll BJ, Lu GQ, Xu ZP. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat Plants. 2017;3:16207.
Kolge H, Kadam K, Galande S, Lanjekar V, Ghormade V. New frontiers in pest control: chitosan nanoparticles-shielded dsRNA as an effective topical RNAi spray for Gram Podborer biocontrol. ACS Appl Bio Mater. 2021;4:5145–57.
Pal G, Ingole KD, Yavvari PS, Verma P, Kumari A, Chauhan C, Chaudhary D, Srivastava A, Bajaj A, Vemanna RS. Exogenous application of nanocarrier-mediated double‐stranded RNA manipulates physiological traits and defence response against bacterial diseases. Mol Plant Pathol. 2024;25:e13417.
Yao Y, Lin DJ, Cai XY, Wang R, Hou YM, Hu CH, Gao SJ, Wang JD. Multiple dsRNases involved in exogenous dsRNA degradation of fall armyworm Spodoptera frugiperda. Front Physiol. 2022;13:850022.
Su YM, Ashworth V, Kim C, Adeleye AS, Rolshausen P, Roper C, White J, Jassby D. Delivery, uptake, fate, and transport of engineered nanoparticles in plants: a critical review and data analysis. Environ Sci-Nano. 2019;6:2311–31.
Schwab F, Zhai GS, Kern M, Turner A, Schnoor JL, Wiesner MR. Barriers, pathways and processes for uptake, translocation and accumulation of nanomaterials in plants-critical review. Nanotoxicology. 2016;10:257–78.
Mosa MA, Youssef K. Topical delivery of host induced RNAi silencing by layered double hydroxide nanosheets: an efficient tool to decipher pathogenicity gene function of Fusarium crown and root rot in tomato. Physiol Mol Plant P. 2021;115:101684.
Niño-Sánchez J, Sambasivam PT, Sawyer A, Hamby R, Chen A, Czislowski E, Li P, Manzie N, Gardiner DM, Ford R, Xu ZP, Mitter N, Jin H. BioClay™ prolongs RNA interference‐mediated crop protection against Botrytis Cinerea. J Integr Plant Biol. 2022;64:2187–98.
Xu X, Jiao Y, Shen L, Li Y, Mei Y, Yang W, Li C, Cao Y, Chen F, Li B, Yang J. Nanoparticle-dsRNA treatment of pollen and root systems of diseased plants effectively reduces the rate of tobacco mosaic virus in contemporary seeds. ACS Appl Mater Interfaces. 2023;15:29052–63.
Warneke B, Pscheidt JW, Nackley L. Pesticide redistribution and its implications on pesticide efficacy. Oregon State University. 2023;PNW 722.
Taning CNT, Christiaens O, Berkvens N, Casteels H, Maes M, Smagghe G. Oral RNAi to control Drosophila suzukii: laboratory testing against larval and adult stages. J Pest Sci. 2016;89:803–14.
Mondal M, Carver M, Brown JK. Characteristics of environmental RNAi in potato psyllid, Bactericera cockerelli (Sulc) (Hemiptera: Psylloidea: Triozidae). Front Physiol. 2022;13:931951.
Shen CH, Jin L, Fu KY, Guo WC, Li GQ. RNA interference targeting Ras GTPase gene Ran causes larval and adult lethality in Leptinotarsa decemlineata. Pest Manag Sci. 2022;78:3849–58.
George J, Ammar E-D, Hall DG, Lapointe SL. Sclerenchymatous ring as a barrier to phloem feeding by Asian citrus psyllid: evidence from electrical penetration graph and visualization of stylet pathways. PLoS ONE. 2017;12:e0173520.
Kaur R, Gupta M, Singh S, Joshi N, Sharma A. Enhancing RNAi efficiency to decipher the functional response of potential genes in Bemisia tabaci AsiaII-1 (Gennadius) through dsRNA feeding assays. Front Physiol. 2020;11:00123.
Chen M, Cooper HM, Zhou JZ, Bartlett PF, Xu ZP. Reduction in the size of layered double hydroxide nanoparticles enhances the efficiency of siRNA delivery. J Colloid Interf Sci. 2013;390:275–81.
Suzuki T, Nunes MA, Espana MU, Namin HH, Jin P, Bensoussan N, Zhurov V, Rahman T, De Clercq R, Hilson P, Grbic V, Grbic M. RNAi-based reverse genetics in the chelicerate model Tetranychus urticae: a comparative analysis of five methods for gene silencing. PLoS ONE. 2017;12:e0180654.
Abbot WS. A method of computing the effectiveness of an insecticide. J Econ Entomol. 1925;18:265–7.
Yu HZ, Huang YL, Lu ZJ, Zhang Q, Su HN, Du YM, Yi L, Zhong BL, Chen CX. Inhibition of trehalase affects the trehalose and chitin metabolism pathways in Diaphorina citri (Hemiptera: Psyllidae). Insect Sci. 2020;28:718–34.
Ullah F, Gul H, Yousaf HK, Qian D, Gao XW, Tariq K, Han P, Desneux N, Song DL. Impact of low lethal concentrations of buprofezin on biological traits and expression profile of chitin synthase 1 gene (CHS1) in melon aphid, Aphis gossypii. Sci Rep-UK. 2019;9:12291.
Acknowledgements
We thank Prof. Zhiping Xu (Shenzhen Bay Laboratory, China), Shun He (Huazhong Agricultural University, China) for insightful advice and comments that improved the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2021YFD1400800), the China Agriculture Research System of MOF and MARA (No. CARS-26) and Hubei Hongshan Laboratory.
Author information
Authors and Affiliations
Contributions
H.Z. and X.C. conceived and designed the project. X.C. performed experiments and analyzed data. Q.Z. reared the mites and aphids. J.X. synthesized MgFe-LDH nanoparticles. X.Q. and Y.Z. involved in characterizing nanoparticles. X.L. involved in revising it critically for important intellectual content. H.Z., W.Z., and X.C. involved in writing-reviewing and editing. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors agree to be published.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cheng, X., Zhou, Q., Xiao, J. et al. Nanoparticle LDH enhances RNAi efficiency of dsRNA in piercing-sucking pests by promoting dsRNA stability and transport in plants. J Nanobiotechnol 22, 544 (2024). https://doi.org/10.1186/s12951-024-02819-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12951-024-02819-4