Abstract
Efforts to enhance the accuracy of protein sequence classification are of utmost importance in driving forward biological analyses and facilitating significant medical advancements. This study presents a cutting-edge model called ProtICNN-BiLSTM, which combines attention-based Improved Convolutional Neural Networks (ICNN) and Bidirectional Long Short-Term Memory (BiLSTM) units seamlessly. Our main goal is to improve the accuracy of protein sequence classification by carefully optimizing performance through Bayesian Optimisation. ProtICNN-BiLSTM combines the power of CNN and BiLSTM architectures to effectively capture local and global protein sequence dependencies. In the proposed model, the ICNN component uses convolutional operations to identify local patterns. Captures long-range associations by analyzing sequence data forward and backwards. In advanced biological studies, Bayesian Optimisation optimizes model hyperparameters for efficiency and robustness. The model was extensively confirmed with PDB-14,189 and other protein data. We found that ProtICNN-BiLSTM outperforms traditional categorization models. Bayesian Optimization’s fine-tuning and seamless integration of local and global sequence information make it effective. The precision of ProtICNN-BiLSTM improves comparative protein sequence categorization. The study improves computational bioinformatics for complex biological analysis. Good results from the ProtICNN-BiLSTM model improve protein sequence categorization. This powerful tool could improve medical and biological research. The breakthrough protein sequence classification model is ProtICNN-BiLSTM. Bayesian optimization, ICNN, and BiLSTM analyze biological data accurately.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
The distinctive spherical form of proteins originates from the unusual three-dimensional structure of polypeptides. This structure imparts features of proteins. The building blocks of proteins are amino acids. Amino acid residues are joined to create peptide bonds, which are the fundamental units of polymer chains [1]. By analyzing the DNA sequence, one can learn a protein’s exact order of amino acids. Gene expression follows a predetermined sequence that is encoded in DNA. A gene is a separate DNA sequence. These locations hold the blueprints for certain genetic components, such as chromosomes, RNA molecules, or proteins [2]. Discovering and classifying protein structures and functions is tremendously challenging and complicated in bioinformatics. Conventional laboratory processes can process large amounts of RNA data. Proteins are classified into families and subfamilies meticulously, which helps researchers understand their functions in living creatures [3]. Fixing the problem will be easier if you do this.
Feature extraction approaches in traditional machine learning methodologies are responsible for protein molecule classification. However, how well manually produced features work is heavily dependent on the selection technique. Protein function prediction is achieved utilizing artificial neural networks (ANNs) parts of deep neural networks (DNNs). Discrete neural networks (DNNs) gradually enhance initial inputs as they go through the network’s levels; these networks comprise multiple hidden layers. Conventional machine-learning approaches are laborious, time-consuming, and resource-intensive [4]. The main reason for these traits, described earlier, is the exponential growth of unique protein sequences.
Many different computer methods have been created because of the need to classify proteins and guess what biological functions they fulfill. Figure 1 shows the primary, secondary, tertiary, and quaternary groups of protein structures. It also shows several different protein structures. You can better understand the complicated world of proteins and the different groups they belong to with the help of this visual aid [5].
The careful selection of features is essential for classifying proteins in protein research. The outermost layer of the protein, the arrangement of its amino acid chains, and its functionalities all contribute to its unique qualities. Techniques seek to efficiently categorize structural protein molecules to facilitate a deeper understanding of evolutionary changes and their temporal relationships. Advances in various techniques for identifying structural stability groups based on sequence details have been made possible by statistical analysis techniques [6].
DNN techniques have been widely embraced in modern scientific research, particularly in biomedical studies, to leverage recent advances in computational power. These techniques have been shown to outperform conventional bioinformatics methods in terms of effectiveness. Two areas in which they are commonly employed are the analysis of visual data and the application of machine learning to the processing of natural language. Single-task DNNs, which are responsible for making binary predictions, and multi-task DNNs, which can classify input data into multiple pre-defined classes, are the two categories used to classify DNNs [7]. Diverse classifications of DNNs are available to accomplish distinct functions in protein data modelling and analysis. The neural network architectures that have been discussed include convolutional neural networks, feed-forward neural networks, auto-encoder deep neural networks, deep belief networks, recurrent neural networks, restricted Boltzmann machines, and graph convolutional networks. Developing improved methods for accurately categorizing protein sequences in proteomics research is the main goal of this work [8].
This study tackles two key research questions: (1) overcoming difficulties in protein sequence classification approaches and (2) evaluating feature extraction methods with existing CNN, LSTM, BiLSTM, and ProtICNN-BiLSTM models [9]. Both of these questions are important in the field of research. Both of these concerns may need to be addressed in subsequent research. When advancing medical research and biological analysis, raising the bar on the precision of protein sequences is necessary. Within the scope of this investigation, an improved model known as ProtICNN-BiLSTM is presented. Attention-Based Improved Convolutional Neural Networks are paired with Bidirectional Long Short-Term Memory units in this particular instance. The model utilizes the Bayesian Optimisation technique to capture the local and global interactions within protein sequences. This is done to improve the accuracy of the protein sequence categorization process. The remarkable performance of the model in comparative investigations reveals its major influence on medical and biological research [10]. In addition to its promise for enhancing protein sequence categorization, the model also demonstrates its promise for improving classification.
A breakdown of the article’s structure is as follows: the Related Work section is where we take a detailed look at the previous research that has been done. The Materials and Methods section presents a comprehensive description of the methods and materials used for the investigation. In the Experimental Results and Discussion section, a summary of the findings and an evaluation of the significance of those findings are offered. For the time being, the study has been completed, and Conclusion and Future Directions section provides an outline of potential future research areas.
Related work
The newest protein structure analysis and classification deep learning algorithms are reviewed here. It includes hybrid models and attention mechanisms. Paper [1] used several indicators to compare deep-learning protein sequence synthesis algorithms. This research examined many cutting-edge deep-learning approaches for protein sequence generation. Many indicators were used to compare deep learning methods. We explored these strategies for synthesizing unique protein sequences from various sequences. The study’s main contribution is comparing each approach’s merits and cons. A diversified protein sequence library ensured complete research. Some approaches succeeded; however, there were few evaluation criteria and methodologies, so protein sequence design concerns may have been missed.
A deep learning-based method for predicting proteins of snake toxins using word embeddings is proposed in the article [2]. This study accomplishes the prediction of proteins containing snake venom through word embeddings within a deep learning framework. The model learned to link sequence patterns with toxin characteristics from a collection of annotated snake toxin proteins. With impressive performance indicators such as a 99.71% average 6-class classification success rate and a 99.85% binary classification accuracy for SARS-CoV-2 compared to HIV-1, Deep-STP proved to be highly accurate in predicting these proteins. Notwithstanding these encouraging findings, the model’s performance may be affected by the variety and quality of the training dataset as well as particular architectural decisions.
In [3], the authors review and investigate the many methods and instruments available for determining the locations of lysine malonylation sites in protein sequences. Machine learning and deep learning are essential to these methods and resources. We tested machine learning and deep learning algorithms to predict lysine malonylation. Researchers tested how well these algorithms predicted published protein sequence positions. This book’s most essential section evaluates the latest abilities and instruments, highlighting their benefits and cons. Every protein sequence in the collection has its lysine malonylation sites determined. Despite finding some promising solutions, the research could not evaluate their generalizability because of annotated data quality and accessibility issues.
The MaTPIP technique was initially introduced by the authors of the work described earlier [4]. Utilizing a deep learning architecture, it provides eXplainable artificial intelligence to predict sequence-driven, feature-mixed protein-protein interactions. As part of its deep learning architecture, MaTPIP makes use of explainable artificial intelligence approaches. This work aimed to attempt to predict the interactions between proteins.
The algorithm learned protein sequences and interactions. Sequence characteristics and other inputs helped it predict interactions. An explanation-based AI system is MaTPIP’s main contribution. This knowledge helps us predict protein interactions. Positive findings showed good interaction prediction accuracy during the investigation. Protein interaction complexity and annotated data may affect model efficiency. Deep learning makes protein syntheses interactive [5]. They proved that their model successfully predicted health system protein-protein interaction locations. They also acknowledged that machine learning was the primary component that led to this accomplishment. In this article, we offer a model that uses machine learning to forecast the times at which protein-protein interactions occur. Training on a set of protein sequences and their interaction sites allowed the program to predict these sites using sequence information. Including practical contact site prediction is a big improvement. This approach may benefit healthcare systems. The experiment showed exact protein-protein interaction sites. This was found throughout the investigation. However, the study found several factors may alter the model’s efficacy.
These variables include the degree of interaction complexity and the availability of data that has been annotated. Six deadly RNA viruses are being studied [6]. Their feature-engineered protein patterns classify these viruses into several types. The viruses are “Human respiratory virus type 3, influenza A, B, and C, and HIV-1”. Another respiratory virus is HIV-1. Data classification and analysis using linear complexity measures. Because of this, it provided reliable categorization, especially for large data sets. The impressive average success rate of 99.71% that the model achieved in identifying the six classes included in the data set is comparable to a published method and implemented with a high degree of precision. The SARS-CoV-2 binary classification fared better than the HIV-1 classification, with a success rate of 99.85%. This was the case across all of the success rates. Additionally, a convolutional neural network (CNN) and a gated recurrent unit (GRU) can be utilized in conjunction with a long short-term memory (LSTM) to locate proteins that bind to DNA.
This is one of several innovative ways developed in recent years. CNN-BiLG describes this combination. CNN-BiLG collects more data and analyzes protein sequence contextual relationships more thoroughly than previous approaches.
An explanation for this can be found in the enhanced capability of the CNN-BiLG technique to capture detailed information. Compared to deep learning and machine learning predictions, CNN-BiLG demonstrated superior performance, as demonstrated by the results of the studies. According to the validation findings, the reports have an incredible accuracy of 95%.
According to many investigations, the suggested model outperforms previous methods in efficiency, cost-effectiveness, and classification accuracy. The proposed model is cheaper.
NLP-based text categorization methods are widely used to classify protein sequences [8]. Deep learning and word embedding have improved text categorization. These advancements have increased protein categorization accuracy and opened new choices. Word-embedded protein sequence representations encounter many challenges in natural language processing (NLP) because amino acid sequences have different “words” than other sequences.
The longer sequences and smaller letter sizes included in the protein data bring additional difficulties for the learning models. This can be attributed to the presence of lengthier sequences in protein data. It has been established that pre-training is one way that can help boost the effectiveness of machine learning techniques. Even though it was initially proposed [9] for computer vision applications, it is currently being utilized extensively in a wide range of machine learning applications, including those connected to language.
Research shows that pre-trained models offer high generalization and convergence rates for tasks with limited training data. Pre-training methods like BERT and ELMo are important despite processing resource constraints. Data-driven neural networks like GCNs can delay hidden cell interactions. To learn and remember, GCNs act sequentially in biology. Hidden cell connections in data-driven Graph Convolution Networks (GCNs) are time-delayed. GCNs build memories and integrate knowledge by operating in biological sequences one element at a time.
Techniques such as SeqVec and ProtTrans [10] use language models and transformer frameworks to represent protein sequences as embedding vectors. This helps to contribute to the understanding of the biophysical characteristics of proteins. Pre-training can leverage comprehensive labelled datasets and transfer knowledge to smaller data problems due to shared pattern characteristics in protein sequence-based classification tasks. This is in contrast to deep learning models derived from natural language processing contexts, which require significant computing power.
Many different types of biological data [11] can be used to predict the functions of proteins. These data types include sequences, three-dimensional structures, folding information, protein-protein interactions, variations in gene expression, amino acid families, and their integration. The classification of common data has been accomplished through the development of statistical theories through the use of techniques such as decision trees, Support Vector Machines (SVM) [12], and Neural Networks (NN) [13]. A few studies that applied SVM after feature extraction from protein sequences demonstrated the potential of SVM in protein classification. Deep learning methods have shown promise in investigations focusing on relatively small groups of proteins and functional categories. Although these methods have not been extensively explored for large-scale protein function prediction pipelines [14], DNN architectures have been trained to anticipate protein operations through research investigations conducted using various protein characteristics. These investigations included both single-tasking and multi-tasking architectures. Table 1 presents a comparison of various existing research in protein sequence analysis.
Materials and methods
This section initiates by introducing the datasets employed in the model’s development. Subsequently, this article elucidates the conceptual framework and testing methodologies. Finally, a model algorithm utilized in the demonstration is presented.
Proposed hybrid model
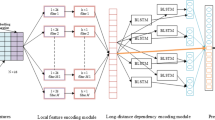
ProtICNN-BiLSTM is a proposed hybrid model that combines Bidirectional Long Short-Term Memory (BiLSTM) units with Improved Convolutional Neural Networks (ICNN) and uses Bayesian Optimization [23, 24] to improve the model parameters. Using the strengths of the ICNN and BiLSTM frameworks, the ProtICNN-BiLSTM method efficiently captures both local and global interdependencies in protein sequences. Bayesian optimization for hyperparameter modifications leads to an even greater increase in model performance. The operation of every component is explained in depth in this part, together with the pertinent equations [25]. The suggested hybrid model ProtICNN-BiLSTM is architecturally illustrated in Fig. 2. Following its receipt by the input layer, the protein sequence data is sent through a convolutional layer with 64 filters, a 3 × 3 kernel size, a stride of 1, and “same” padding. ReLU activation comes last after batch normalization. Another convolutional layer with 128 filters, a 3 × 3 kernel size, a stride of 1, and “same” padding processes the output of this layer. Batch normalization follows [26].
ICNN model
The ICNN part of the ProtICNN-BiLSTM method is supposed to extract local properties from the protein sequences. Convolutional Neural Networks can efficiently capture spatial hierarchies in data using convolutional processes [27]. ICNN applies attention methods to enhance feature extraction. Many improvements are included in the enhanced CNN architecture of the ProtICNN-BiLSTM model to increase efficiency and capture more intricate features from protein sequences. These improvements have included the remaining connections to address the disappearance of gradients and enhance gradient flow during training. Batch normalization layers are added after each convolutional layer to standardize the input of individual layers, therefore stabilizing and speeding up the training process even further [28].
ReLU activations are included following batch normalization to increase the expressiveness of the model even further and introduce non-linearity. Half-rate dropout layers train by arbitrarily removing certain input units to prevent further overfitting. Consistently handling sequences of varying lengths is easier using adaptive pooling layers, ensuring a constant output size irrespective of input dimensions. These enhancements collectively increase the overall efficacy of the ProtICNN-BiLSTM model by enhancing CNN’s comprehension of flexible and robust features from protein sequences [29].
The Improved CNN Model’s design is shown in Fig. 3. The revised CNN architecture receives protein sequences as numerical array input to do protein sequence analysis. The layered layout of the proposed paradigm is depicted in Fig. 4. 64 The first convolutional layer uses 3 × 3 filters to search for local patterns in the sequences. Next, the ReLU activation function is used to introduce non-linearity. A dropout layer with a rate of 0.5 is employed to reduce overfitting, and batch normalization is used to stabilize the activations [30].
When it comes to the second convolutional layer, there are 128 filters, each of which measures three by three measures. This particular layer is responsible for capturing more abstract and complex qualities. The ReLU activation function is utilized to achieve non-linearity. Next is a dropout layer, followed by batch normalizing. These phases promote long-term, versatile learning. Since adaptive pooling resizes feature maps to a preset size, it allows for many input periods. The 256-unit fully connected layer can combine these characteristics to build sophisticated and abstract representations using the Rectified Linear Unit (ReLU) activation function [31].
In order to compute the probability of the protein classes, the output layer of the multi-class classification process uses the application of the SoftMax activation function. This is done to achieve the desired results. The classification procedure is carried out to guarantee that it is appropriate. In addition, local and global feature extraction procedures have been incorporated into this architectural design [32] to improve the precision and continuity of the protein sequence categorization process. Along with regularization strategies, this design also includes regularization techniques.
Convolution operation process
The convolution process is used to extract local features from the sequence that is being input [33]. The mathematical form of the convolution operation is summarized in Eq. (1), which is presented below. \(\:{Z}_{i,j}^{k}\) represents the output of the convolution operation phase, (i, j) represents positions, k represents filter, \(\:{b}^{k}\:\)is a bias term, M and N filter size.
Attention mechanism
An attention technique has been implemented to concentrate on the most important features retrieved by the convolutional layers [34]. According to Eq. (2), the attention weights are calculated as a given. Here \({\sigma }_{i}\:\): attention weight, \(\:{(e}_{i})\): Energy score, L: Local feature count.
Energy score calculation
An energy score\(\:{e}_{i}\)can be calculated by Eq. (3). Here\(\:{W}_{a}\)and\(\:{b}_{a}\)are learning parameters, \(\:{h}_{i}\) : hidden state [35].
Weighted feature representation
The attention weights and the convolutional features are combined to produce the weighted feature representation \(\:{W}_{f}\) as described in Eq. (4). Here\(\:{W}_{f}\): weight feature, \(\:{h}_{i}\) : hidden state, and L: Local feature count.
Bidirectional long short-term memory (BiLSTM)
Bidirectional Long Short-Term Memory, also known as BiLSTM, is a more advanced kind of Recurrent Neural Network (RNN) that was developed to handle sequential data, for instance, sequences of protein, by taking into account dependencies in both the forward and backward directions [36]. The operation of BiLSTM is as follows.
Forward-LSTM (Fw-LSTM)
Predominantly, the Forward LSTM Layer The input sequence is processed from the beginning to the end, with the forward dependencies being captured. Each cell in this layer comprises three gates: input, forget, and output [37]. These gates control the flow of information, enabling the network to remember or forget knowledge from the past depending on the circumstances; the essential formulas are presented from Eq. (5) to (10). Table 2 presents the key symbols used in forward-BLSTM.
Backward-LSTM (Bw-LSTM)
An additional LSTM layer simultaneously processes the sequence from the end to the beginning, capturing the backward dependencies. This layer functions in a manner that is analogous to that of the forward layer, employing the same gate mechanisms to control the flow of information [38].
Cumulative output
Concatenating hidden states from forward and backward LSTM layers every time step. With information on each component and consideration for the past and future, this combination delivers a complete sequence picture every time. For the protein sequence “BHDU,” the forward LSTM would be B◊H◊ D◊U. In contrast, the reverse LSTM works as follows: U◊D◊H◊B. Combining the concealed state from both sides ensures that every point in the sequence incorporates information from the previous and next portions [39]. BiLSTM is effective for complex sequential data like protein sequences, where component interactions determine analysis and classification. Its bidirectional approach causes this.
Bayesian optimization method
Bayesian optimization allows one to modify the hyperparameters in complex processes such as deep learning models. Optimized hyperparameter setups are found by constantly updating a probabilistic model and balancing exploration and exploitation. Successfully and carefully modifying hyperparameters, Bayesian optimization improves ProtICNN-BiLSTM protein categorization [40]. Bayesian optimization operates in the model as follows.
-
Defining an Objective Function: The goal function f(x) is our attempt to maximize the performance metric for protein sequence analysis. Equation 11 is another classification metric; precision, recall, or accuracy are additional possibilities.
-
Set a Gaussian Process: To get a close approximation to the objective function, a Gaussian Process (GP) is started. A mean function µ(x) and a covariance function Cf(x, x′) are utilized by the GP model to forecast the value and uncertainty of the objective function (Eqs. 12 and 13).
-
Define an Acquisition Function: The selection of the subsequent hyperparameter point to examine is accomplished with the assistance of the acquisition function. With its ability to balance exploration and exploitation, the Expected Improvement (EI) function is a popular option (Eq. 14).Here\(\:\beta\:\left(x\right)\): Acquisition function, \(\:{x}^{+}\): Selected best hypothetical parameter at the current point.
-
Hyperparameter Update: Replace the old GP model with the updated one using the evaluation data. This makes the GP model’s goal function approximation more accurate.
Algorithm proposed hybrid model
The key steps for the proposed hybrid model are described in Algorithm 1 below.
Algorithm 1: Proposed Hybrid model for protein sequence Input: Protein dataset Output: Protein Sample categories with different classes. Step 1: Import and preprocess the data 1. Import data on protein sequences through the Protein Data Bank samples and another pertinent resource. 2. Perform preprocessing on the patterns, considering features such as amino acid structure, physicochemical characteristics, and structural details. Step 2: Divide the data 1. Partition the sample among training and testing collections with an 80:20 ratio, guaranteeing that the protein groups are evenly distributed in both sets. Step 3: Architectural Design 1. Create a Hybrid Convolution Neural Network (CNN)-Bidirectional Long Short-Term Memory (BiLSTM) model to capture short, practical- and longer-range relationships in protein patterns. 2. Convolution layers can be used to obtain spatial patterns, whereas Bi-LSTM layers, in combination, should be used to retrieve sequential data. Step 4: Hyperparameter Tuning 1. Specify a range of hyperparameter values, including learning rates, batch size, convolutional filtering size, LSTM units, and dropout rates. 2. Implement Bayesian Optimisation to systematically and effectively investigate and identify the algorithm’s most optimal hyperparameter settings. Step 5: Assessment Criterion: 1. Select the F1-score as the primary evaluation metric for precision and recall, essential in imbalanced protein sequence datasets. Step 6: Training and evaluating the model: 1. Utilise Bayesian optimization to obtain the optimal Hyperparameters and then train the model on the training set. 2. Assess the model’s performance on the test set, considering metrics such as accuracy, precision, recall, F1-score, and any metrics specific to the domain. Step 7: Analysis and depiction: 1. Observe the model’s performance and examine instances where it incorrectly classified data. 2. Utilise interpretability tools to gain insights into the specific portions of the sequences that have the most significant impact on predictions. |
Dataset
This research utilizes the standard protein online dataset PDB-14189 (Protein Data Bank-14189) [31]. The dataset comprises a heterogeneous collection of patterns of protein derived from different organisms, which includes “enzymes, antibodies, structural proteins, transport proteins, receptors, and other functional categories. The description of each protein sequence includes details on its components, operation, and biological characteristics. Such information may involve the secondary structure components, binding of legend sites, protein class categorization, organism site, the procedure utilized for organization commitment, and clarity of the empirical form. The PDB dataset is frequently utilized in bioinformatics and molecular science studies for various objectives, such as protein structural estimation, multifunctional annotation, interaction between protein and protein prediction, chemical effects target recognition, and automated tasks such as classification.
Among the 14,189 cases in the PDB-14189 dataset, there are 7,129 positive instances of DNA-binding proteins and 7,060 negative instances of proteins that do not bind DNA. When researching bioinformatics and machine learning, this dataset is frequently utilized for protein function prediction and structural analysis tasks. Table 3 presents the PDB dataset description.
Data pre-processing
Experiments and protein databases were two reliable sources from which the dataset was initially carefully assembled. Thus, the process began. We next enthusiastically launched a thorough data cleaning process. This included removing duplicate sequences, correcting errors, and meticulously handling missing data. By now, the integrity and dependability of the dataset had to be ensured for the following analysis [41].
The feature extraction was achieved using advanced techniques to convert the protein sequences into numerical representations. One-hot encoding was used to turn every amino acid in the sequence into a binary vector, achieving increased Specificity. We have recovered a large range of amino acid physicochemical properties to enhance the feature set and capture significant features of the proteins. Among these were a specific molecular mass and water repellency. Oversampling techniques were used to alleviate class imbalance. Class imbalance resulted when there were more negative than positive instances, that is, proteins that did not link to DNA than positive examples did. It needed to make fictitious data points for the minority class to achieve a balanced distribution and remove bias from the machine learning algorithms.
The dataset was next separated into training, validation, and test sets. Class distribution was carefully maintained inside each subset to ensure accuracy. The models were trained using the training set throughout the machine learning process. The validation set helped us select the best model and adjust the hyperparameters. Finally, the test set assessed the whole model’s performance. We last used feature scaling techniques like normalization to ensure that all features were equivalent. The best efficiency and ability of the machine learning algorithms to learn from the dataset were guaranteed by this stage. These preprocessing approaches ensured that the protein data was prepared and optimally suited for the training and analysis of machine learning models. Results were, therefore, more precise and trustworthy.
Performance measuring parameters
Key parameters evaluated the suggested and present models’ prediction accuracy. Precision (P), recall (R), F1-score, support, specificity (SPC), sensitivity (SNS), Matthews’ correlation coefficient (MCOC), and accuracy (ACR) are calculated using Eq. 13 to 16 [42]. Here, TP: True positive, FN: False Negative, TN: True Negative and FP: False positive.
Precision
Precision is calculated as the count of true positives divided by the overall quantity of positive cases discovered, as stated in Eq. 15.
Recall
Divide the number of successfully identified positive observations by the total positive specimens to compute recall as presented by Eq. 16.
F1-score
In binary classification (and multi-class categorization), the F1-score measures precision and recall (Eq. 17).
Specificity
As shown in Eq. 18, SPC is a binary categorization metric that measures a model’s negative case detection accuracy.
Accuracy
Eq. 18 calculates accuracy by dividing the number of successfully predicted instances by the total number of occurrences.
Experimental results and discussion
The Proposed and existing models are implemented using Python programming, and various performance-measuring parameters are calculated. Different performance metrics were computed to evaluate the efficacy of these models. The study leveraged PyTorch, an openly accessible deep-learning library. The proposed hybrid model was developed using Python Keras [43]. Evaluation of absolute parameters involved the use of ‘PDB-14189’. The dataset was divided into an 80% training sample and a 20% testing sample [44,45,46,47].
Hyperparameter specification
Table 4 represents the hyperparameter details. Table 4 defines the Hyperparameters used in experimental analysis [48,49,50]. Table 5 contains the parameters elements of CNN and LSTM in the proposed hybrid model, which affect and result from the shape of every level of a DNN in protein analysis.
Results for different parameters
The PDB-14,189 dataset was used as a key dataset in this research. Various performance measuring parameters were calculated to measure the performance of existing CNN, CNN-LSTM and the proposed hybrid model. Figure 5 presents protein sequence patterns for different classes. Figure 5 presents (a) the protein sequence frequency of attributes, 5(b) presents the protein sequence length vs. Sequences, and 5(c) presents the Protein sequence frequency count.
Experimental results
The protein classes are grouped into four categories: Hydrolase (0), Oxidoreductase (1), Ribosome (2), and Transferase (3).
Table 6 describes the experimental results of the CNN model for predicted and actual protein sequence analysis. CNN Model achieved a precision of 82.013% for Hydrolase (0), 90.631% for Oxidoreductase (1), 89.856% for Ribosome (2) and 89.163% for Transferase (3), CNN achieved a recall of 87.236% for Hydrolase (0), 88.104% for Oxidoreductase (1), 89.952% for Ribosome (2) and 87.459% for Transferase (3).F1-score results of CNN model is 86.761% for Hydrolase (0), 84.039% for Oxidoreductase (1), 83.791% for Ribosome (2) and 83.014% for Transferase (3) and Final results for accuracy is 86.286% for Hydrolase (0), 85.603% for Oxidoreductase (1), 86.786% for Ribosome (2) and 87.492% for Transferase (3).
Table 7 describes the experimental results of the CNN-LSTM model for predicted and actual protein sequence analysis. CNN Model achieved a precision of 87.949% for Hydrolase (0), 87.298% for Oxidoreductase (1), 91.791% for Ribosome (2) and 88.486% for Transferase (3), CNN-LSTM achieved a recall results of 88.659% for Hydrolase (0), 89.476% for Oxidoreductase (1), 90.856% for Ribosome (2) and 86.870% for Transferase (3). F1-score results of CNN model is 89.042% for Hydrolase (0), 86.872% for Oxidoreductase (1), 92.963% for Ribosome (2) and 87.365% for Transferase (3) and Final results for accuracy is 90.787% for Hydrolase (0), 89.321% for Oxidoreductase (1), 88.709% for Ribosome (2) and 89.326% for Transferase (3).
Table 8 describes the experimental results of the Proposed Hybrid model for predicted and actual protein sequence analysis. Proposed Hybrid model achieved a precision of 95.371% for Hydrolase (0), 96.908% for Oxidoreductase (1), 95.772% for Ribosome (2) and 93.474% for Transferase (3), Proposed model achieved a recall results of 96.375% for Hydrolase (0), 97.603% for Oxidoreductase (1), 94.667% for Ribosome (2) and 95.187% for Transferase (3). F1-score results of Proposed model is 94.874% for Hydrolase (0), 93.271% for Oxidoreductase (1), 95.337% for Ribosome (2) and 96.375% for Transferase (3) and Final results for accuracy is 96.074% for Hydrolase (0), 94.387% for Oxidoreductase (1), 96.009% for Ribosome (2) and 97.341% for Transferase (3).
Table 9 presents a comparative analysis of experimental results of existing vs. proposed models. Existing CNN achieved Specificity of 85.84% Accuracy of 89.27%, Sensitivity of 89.78%, and MCC 81.47% and Existing CNN-LSTM achieved Specificity of 87.37%, accuracy of 90.17%, Sensitivity of 88.98%, and MCC 88.35%, and Proposed Hybrid Model achieved Specificity of 94.65%, Accuracy of 96.57%, Sensitivity of 95.67% and MCC 96.85%.
Figure 6 compares the existing CNN, CNN-LSTM, and the proposed model ProtICNN-BiLSTM regarding Specificity, Accuracy, Sensitivity, and Matthews’s correlation coefficient. The proposed model exhibits outstanding performance, achieving higher results for all four parameters. This highlights the proposed model’s strength and effectiveness compared to the existing CNN and CNN-LSTM models.
Results and discussion
The fusion of Improved Convolutional Neural Networks and Bidirectional Long Short-Term Memory models, augmented with amino acid embedding techniques, presents a robust strategy for dissecting protein sequences. By harnessing the capabilities of amino acid embedding, the model can effectively exploit the feature extraction prowess inherent in BiLSTM. Subsequent processing by both ICNN and BiLSTM components enables precise prediction of protein attributes, including structural configurations and functional characteristics. However, the efficacy of this approach hinges upon factors such as dataset quality, size, and specific analytical goals.
Experimental assessments were conducted using the PDB-14189 dataset, contrasting the performance of established CNN, CNN-LSTM, and the novel ProtICNN-BiLSTM models. The training spanned 1500 epochs with a batch size 128, optimized via the ADAM optimizer. A meticulous dropout analysis encompassing metrics like Specificity, Sensitivity, Matthews’s correlation coefficient, and overall accuracy was undertaken. Results for Different Parameters section delineates binary and multi-class classification outcomes for extant and proposed methodologies. Visualization of protein sequence patterns across diverse classes, encompassing attribute frequencies, sequence length distributions, and sequence counts, is depicted in Fig. 5. The results in Table 6 outline the CNN model’s performance, while Table 7 elucidates the CNN-LSTM model’s efficacy. Notably, the proposed ProtICNN-BiLSTM model attains remarkable accuracy, recall, F1-score, and support metrics, peaking at 98.11%.
The discernible superiority of the ProtICNN-BiLSTM model over conventional CNN and LSTM variants can be ascribed to several key factors. Firstly, the fusion of ICNN with BiLSTM engenders a holistic approach to capturing local and long-range dependencies within protein sequences. Moreover, incorporating amino acid embedding techniques facilitates a nuanced representation of proteins as numerical vectors, fostering more robust feature extraction. Integrating an attention mechanism further enhances model performance by dynamically weighting the significance of various protein sequence components. Collectively, these advancements underscore the efficacy of the ProtICNN-BiLSTM model in surpassing traditional CNN and LSTM methodologies.
Conclusion and future directions
The Protein-BILSTM model demonstrates the value of collaborative deep learning and molecular biology. The proposed hybrid model surpasses existing approaches with 98.11% accuracy. Hydrolase, Oxidoreductase, Ribosome, and Transferase had similar precision and recall ratings of 87.949–91.791% and 86.870–90.856%. These results prove that the increased biological analysis method works. Though promising, protein sequence categorization needs more research to increase accuracy and application. Protein-BILSTM classifies protein sequences for the first time. CNN and BiLSTM encoding improves accuracy. Its protein structure and activity predictions are accurate enough for sophisticated biological research. This model uses NLP for feature extraction and CNN and BiLSTM for sequential associations. The model’s DNA binding prediction summarizes complex biology. The Protein-BILSTM model demonstrates the value of collaborative deep learning and molecular biology. The proposed hybrid model surpasses existing approaches with 98.11% accuracy.
Protein sequence categorization needs more research to improve accuracy and practicality. Research employing more complex datasets can improve model performance and flexibility. Model prediction benefits from protein-protein interaction and functional annotation data. Deep learning and hybrid models can improve efficiency. Protein classifications and databases improve the model. Experimental biologists can verify the model’s biological applicability by verifying predictions.
Data availability
The datasets generated and/or analysed during the current study are available in the PDBj (Protein Data Bank Japan) repository, [https://pdbj.org/emnavi/quick.php?id=EMD-14189]
References
Yu J, Mu J, Wei T, Hai-Feng Chen. Multi-indicator comparative evaluation for deep learning-based protein sequence design methods. Bioinformatics 40, no. 2 (2024): btae037.
Zulfiqar H, Guo Z, Ahmad RM, Ahmed Z, Cai P, Chen X, Zhang Y, Lin H. Deep-STP: a deep learning-based approach to predict snake toxin proteins using word embeddings. Front Med. 2024;10:1291352.
Ramazi S, Tabatabaei SAH, Khalili E, Nia AG, Motarjem K. Analysis and review of techniques and tools based on machine learning and deep learning for prediction of lysine malonylation sites in protein sequences. Database 2024 (2024): baad094.
Ghosh S, Mitra P. MaTPIP: a deep-learning architecture with eXplainable AI for sequence-driven, feature mixed protein-protein interaction prediction. Comput Methods Programs Biomed. 2024;244:107955.
He J, Wu W, Wang X. DIProT: a deep learning based interactive toolkit for efficient and effective protein design. Synth Syst Biotechnol (2024), 32–86.
Tahir M, Khan F, Hayat M, Mohammad Dahman Alshehri. An effective machine learning-based model for the prediction of protein–protein interaction sites in health systems. Neural Comput Appl. 2024;36(1):65–75.
Ali S, Sahoo B, Zelikovsky A, Chen P-Y, Patterson M. Benchmarking machine learning robustness in Covid-19 genome sequence classification. Sci Rep. 2023;13(1):4154.
Yeung W, Zhou Z, Mathew L, Gravel N, Taujale R, Boyle BO, Salcedo M, et al. Tree visualizations of protein sequence embedding space enable improved functional clustering of diverse protein superfamilies. Brief Bioinform. 2023;24(1):bbac619.
Motmaen A, Dauparas J, Baek M, Abedi MH, Baker D, Bradley P. Peptide-binding specificity prediction using fine-tuned protein structure prediction networks. Proceedings of the National Academy of Sciences 120, no. 9 (2023): e2216697120.
Yao J, Ling Y, Hou P, Wang Z, Huang L. A graph neural network model for deciphering the biological mechanisms of plant electrical signal classification. Appl Soft Comput. 2023;137:110153.
Goto K, Tamehiro N, Yoshida T, Hanada H, Sakuma T, Adachi R. Kazunari Kondo, and Ichiro Takeuchi. Novel machine learning method allerStat identifies statistically significant allergen-specific patterns in protein sequences. J Biol Chem. 2023;299:6.
Hou Z, Yang Y, Ma Z, Wong Ka-chun, Li X. Learning the protein language of proteome-wide protein-protein binding sites via explainable ensemble deep learning. Commun Biology. 2023;6(1):73.
Llinares-López F, Berthet Q, Blondel M, Teboul O. Deep embedding and alignment of protein sequences. Nat Methods. 2023;20(1):104–11.
Singh D, Roy J. A large-scale benchmark study of tools for the classification of protein-coding and non-coding RNAs. Nucleic Acids Res. 2022;50(21):12094–111.
Tripathi R, Patel S, Kumari V, Chakraborty P, Pritish Kumar V. DeepLNC, a long non-coding RNA prediction tool using deep neural network. Netw Model Anal Health Inf Bioinf. 2016;5:1–14.
Hashem, Abu MAM, Hossain AR, Marlinda MA, Mamun S, Sagadevan Z, Shahnavaz. Khanom Simarani, and Mohd Rafie Johan. Nucleic acid-based electrochemical biosensors for rapid clinical diagnosis: advances, challenges, and opportunities. Crit Rev Clin Lab Sci. 2022;59(3):156–77.
Erten M, Aydemir E, Barua PD, Baygin M, Dogan S, Tuncer T, Tan R-S. Abdul Hafeez-Baig, and U. Rajendra Acharya. Novel tiny textural motif pattern-based RNA virus protein sequence classification model. Expert Syst Appl. 2024;242:122781.
Ahmed N, Yousif WA, Alsanousi EM, Hamid MK, Elbashir KM, Al-Aidarous. Mogtaba Mohammed, and Mohamed Elhafiz M. Musa. An efficient Deep Learning Approach for DNA-Binding proteins classification from primary sequences. Int J Comput Intell Syst. 2024;17(1):1–14.
Onyema EM, Lilhore UK, Saurabh P, Dalal S, Nwaeze AS. Asogwa Tochukwu Chijindu, Lauritta Chinazaekpere Ndufeiya-Kumasi, and Sarita Simaiya. Evaluation of IoT-Enabled hybrid model for genome sequence analysis of patients in healthcare 4.0. Measurement: Sens. 2023;26:100679.
Pattnaik D, Thakur SB, Dash PM, Jena S. Sumanta Sahu, and Sulin Kumar Behera. Molecular Medical diagnosis of COVID-19 and Omicron variant. J Pharm Negat Results (2022): 6332–47.
Chhabra C. and Meghna Sharma. Machine Learning, Deep Learning and Image Processing for Healthcare: A Crux for Detection and Prediction of Disease. In Proceedings of Data Analytics and Management: ICDAM 2021, Volume 2, pp. 305–325. Springer Singapore, 2022.
Le NQ, Khanh Q-T, Ho T-T-D, Nguyen, Yu-Yen Ou. A transformer architecture based on BERT and 2D convolutional neural network to identify DNA enhancers from sequence information. Brief Bioinform. 2021;22(5):bbab005.
Tavakoli N. Seq2image: Sequence analysis using visualization and deep convolutional neural network. In 2020 IEEE 44th Annual Computers, Software, and Applications Conference (COMPSAC), pp. 1332–1337. IEEE, 2020.
Ho Q-T, Yu-Yen Ou. Classifying the molecular functions of Rab GTPases in membrane trafficking using deep convolutional neural networks. Analytical biochemistry 555 (2018): 33–41.
Sureyya Rifaioglu, Ahmet T, Doğan MJ, Martin. Rengul Cetin-Atalay, and Volkan Atalay. DEEPred: automated protein function prediction with multi-task feed-forward deep neural networks. Scientific reports 9, no. 1 (2019): 1–16.
Liu C-M, Ta V-D. Nguyen Quoc Khanh Le, Direselign Addis Tadesse, and Chongyang Shi. Deep neural network framework based on word embedding for protein Glutarylation sites prediction. Life 12, no. 8 (2022): 1213.
Yuvaraj N, Srihari K, Chandragandhi S, Raja RA, Dhiman G, Kaur A. Analysis of protein-ligand interactions of SARS-Cov-2 against selective drug using deep neural networks. Big Data Min Analytics. 2021;4(2):76–83.
Le NQ, Khanh EKY, Yapp Y-Y, Ou, Hui-Yuan Yeh. iMotor-CNN: Identifying molecular functions of cytoskeleton motor proteins using 2D convolutional neural network via Chou’s 5-step rule. Analytical biochemistry 575 (2019): 17–26.
Pu L, Govindaraj RG, Lemoine JM, Wu H-C, Brylinski M. DeepDrug3D: classification of ligand-binding pockets in proteins with a convolutional neural network. PLoS Comput Biol. 2019;15(2):e1006718.
Zhang Z, Park CY, Theesfeld CL, Olga G. Troyanskaya. An automated framework for efficiently designing deep convolutional neural networks in genomics. Nat Mach Intell. 2021;3(5):392–400.
Wang Y-B, You Z-H, Li X, Jiang T-H, Chen X. Xi Zhou, and Lei Wang. Predicting protein–protein interactions from protein sequences by a stacked sparse autoencoder deep neural network. Mol Biosyst. 2017;13(7):1336–44.
Xu M, Papageorgiou DP, Sabia Z, Abidi M, Dao. Hong Zhao, and George Em Karniadakis. A deep convolutional neural network for classification of red blood cells in sickle cell anemia. PLoS Comput Biol. 2017;13(10):e1005746.
Niu M, Lin Y, Zou Q. sgRNACNN: identifying sgRNA on-target activity in four crops using ensembles of convolutional neural networks. Plant Mol Biol. 2021;105:483–95.
Zhao T, Hu Y, Valsdottir LR. Tianyi Zang, and Jiajie Peng. Identifying drug–target interactions based on graph convolutional network and deep neural network. Brief Bioinform. 2021;22(2):2141–50.
Deng L, Liu Y, Shi Y, Zhang W, Yang C, Liu H. Deep neural networks for inferring binding sites of RNA-binding proteins by using distributed representations of RNA primary sequence and secondary structure. BMC genomics 21, no. 13 (2020): 1–10.
Wang L, Wang H-F, Liu S-R, Xin Y, Ke-Jian S. Predicting protein-protein interactions from matrix-based protein sequence using convolution neural network and feature-selective rotation forest. Sci Rep. 2019;9(1):9848.
Ben-Bassat I, Chor B, Orenstein Y. A deep neural network approach for learning intrinsic protein-RNA binding preferences. Bioinformatics. 2018;34:17.
Le NQ, Khanh, Quang-Thai H. Deep transformers and convolutional neural network in identifying DNA N6-methyladenine sites in cross-species genomes. Methods. 2022;204:199–206.
Mitra S, Saha S, Hasanuzzaman M. A multi-view deep neural network model for chemical-disease relation extraction from imbalanced datasets. IEEE J Biomedical Health Inf. 2020;24(11):3315–25.
Lv Z, Ding H, Wang L, Zou Q. A convolutional neural network using dinucleotide one-hot encoder for identifying DNA N6-methyladenine sites in the rice genome. Neurocomputing. 2021;422:214–21.
Taju S, Wellem T-T-D, Nguyen N-Q-K, Le. DeepEfflux: a 2D convolutional neural network model for identifying families of efflux proteins in transporters. Bioinformatics. 2018;34(18):3111–7. Rosdyana Mangir Irawan Kusuma, and Yu-Yen Ou.
Zhang Y, Qiao S, Ji S, Li Y. DeepSite: bidirectional LSTM and CNN models for predicting DNA–protein binding. Int J Mach Learn Cybernet. 2020;11:841–51.
Cheng J, Liu Y, Ma Y. Protein secondary structure prediction based on integration of CNN and LSTM model. J Vis Commun Image Represent. 2020;71:102844.
Pang L, Wang J, Zhao L, Wang C, Zhan H. A novel protein subcellular localization method with CNN-XGBoost model for Alzheimer’s disease. Front Genet. 2019;9:751.
Park S. GalaxyWater-CNN: prediction of water positions on the protein structure by a 3D-convolutional neural network. J Chem Inf Model. 2022;62(13):3157–68.
Zhang D, Mansur R. Kabuka. Protein family classification from scratch: a CNN based deep learning approach. IEEE/ACM transactions on computational biology and bioinformatics 18, 5 (2020): 1996–2007.
Peng Y, Rios A, Ramakanth Kavuluru, and, Lu Z. Chemical-protein relation extraction with ensembles of SVM, CNN, and RNN models. arXiv preprint arXiv:1802.01255 (2018).
Barukab O, Ali F, Alghamdi W, Bassam Y, Sher Afzal Khan. DBP-CNN: deep learning-based prediction of DNA-binding proteins by coupling discrete cosine transform with two-dimensional convolutional neural network. Expert Syst Appl. 2022;197:116729.
Jiang M, Wei Z, Zhang S, Wang S, Wang X, Li Z. Frsite: protein drug binding site prediction based on faster R–CNN. J Mol Graph Model. 2019;93:107454.
Lin Z, Lanchantin J, Qi Y. MUST-CNN: a multilayer shift-and-stitch deep convolutional architecture for sequence-based protein structure prediction. In Proceedings of the AAAI Conference on Artificial Intelligence, vol. 30, no. 1. 2016.
Acknowledgements
This Research is funded by Research Supporting Project Number (RSPD2024R553), King Saud University, Riyadh, Saudi Arabia.
Funding
This Research is funded by Research Supporting Project Number (RSPD2024R553), King Saud University, and Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
SD & UKL: Contributed to the conceptualization and design of the research project and conducted data collection and analysis. Drafted and revised the manuscript and participated in the development of the research methodology. SS & MA: Contributed to the data collection and statistical analysis and provided critical insights during manuscript drafting. NF & KA: Involved in data acquisition and preprocessing and contributed to the development and implementation of computational models, All the authors Contributed to the theoretical framework and interpretation of results and reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lilhore, U.K., Simiaya, S., Alhussein, M. et al. Optimizing protein sequence classification: integrating deep learning models with Bayesian optimization for enhanced biological analysis. BMC Med Inform Decis Mak 24, 236 (2024). https://doi.org/10.1186/s12911-024-02631-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12911-024-02631-y