Abstract
Background
Continuous monitoring of antimicrobial resistance (AMR) in Uganda involves testing bacterial isolates from clinical samples at national and regional hospitals. Although the National Microbiology Reference Laboratory (NMRL) analyzes these isolates for official AMR surveillance data, there's limited integration into public health planning. To enhance the utilization of NMRL data to better inform drug selection and public health strategies in combating antibiotic resistance, we evaluated the trends and spatial distribution of AMR to common antibiotics used in Uganda.
Methods
We analyzed data from pathogenic bacterial isolates from blood, cerebrospinal, peritoneal, and pleural fluid from AMR surveillance data for 2018–2021. We calculated the proportions of isolates that were resistant to common antimicrobial classes. We used the chi-square test for trends to evaluate changes in AMR resistance over the study period.
Results
Out of 537 isolates with 15 pathogenic bacteria, 478 (89%) were from blood, 34 (6.3%) were from pleural fluid, 21 (4%) were from cerebrospinal fluid, and 4 (0.7%) were from peritoneal fluid. The most common pathogen was Staphylococcus aureus (20.1%), followed by Salmonella species (18.8%). The overall change in resistance over the four years was 63–84% for sulfonamides, fluoroquinolones macrolides (46–76%), phenicols (48–71%), penicillins (42–97%), β-lactamase inhibitors (20–92%), aminoglycosides (17–53%), cephalosporins (8.3–90%), carbapenems (5.3–26%), and glycopeptides (0–20%). There was a fluctuation in resistance of Staphylococcus aureus to methicillin (60%-45%) (using cefoxitin resistance as a surrogate for oxacillin resistance) Among gram-negative organisms, there were increases in resistance to tetracycline (29–78% p < 0.001), ciprofloxacin (17–43%, p = 0.004), ceftriaxone (8–72%, p = 0.003), imipenem (6–26%, p = 0.004), and meropenem (7–18%, p = 0.03).
Conclusion
The study highlights a concerning increase in antibiotic resistance rates over four years, with significant increase in resistance observed across different classes of antibiotics for both gram-positive and gram-negative organisms. This increased antibiotic resistance, particularly to commonly used antibiotics like ceftriaxone and ciprofloxacin, makes adhering to the WHO's Access, Watch, and Reserve (AWaRe) category even more critical. It also emphasizes how important it is to guard against the growing threat of antibiotic resistance by appropriately using medicines, especially those that are marked for "Watch" or "Reserve."
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Globally, antimicrobial resistance (AMR) is an increasing health concern. AMR is the ability of microorganisms to resist the effects of medicines that were previously used to treat such diseases [1]. Over 700,000 individuals worldwide die every year from illnesses linked to AMR; it is predicted that 10 million deaths will have occurred by 2050, costing the world $100 trillion [2]. In 2019, estimates of invasive infections, particularly antimicrobial-resistant invasive infections, accounted for 5.3 million deaths around the world annually [3]. A significant proportion of these deaths occurs in low- to middle-income countries (LMICs) [4]. While developing countries are battling an accelerated spread of AMR, developed countries are also experiencing the same trend [4].
AMR has increased as a result of the use of broad-spectrum antibiotics to treat invasive infections such as bloodstream [5, 6] and central nervous system (CNS) infections [6]. As a result, treating these infections is becoming more challenging, which increases treatment failures and death [7]. Over-the-counter medication access in developing countries such as Uganda is one of the drivers of AMR in such nations [8]. Indiscriminate usage of antimicrobials exerts increased selection pressure [9] on the bacterial population, resulting in accelerated emergence of AMR [10]. Antibiotic usage has led to the rise of widely multidrug-resistant germs, rendering even the most potent medications useless [11].
The unavailability of reliable data in developing countries such as Uganda makes it difficult to develop efficient methods to monitor and control AMR [12, 13]. Nonetheless, a few studies to date have reported [10, 14] trends of resistance in Uganda. A study that investigated antibiotic resistance in Uganda found that E. coli and K. pneumoniae were carried in the Gastrointestinal tract (GIT) of patients attending outpatient clinics in Kampala and two rural districts [15]. It further showed high rates of resistance to commonly used antibiotics such as ampicillin and cotrimoxazole (sulfamethoxazole and trimethoprim) and relatively low resistance rates to amoxicillin/clavulanate, chloramphenicol, ciprofloxacin, gentamicin, nitrofurantoin and ceftriaxone.
We described the organisms that are isolated from patients’ sterile site samples and their antibiotic resistance trends in Uganda to provide data to inform planning and AMR control interventions.
Methods
Study setting, design, and data source
We conducted a descriptive analysis of Uganda’s national AMR surveillance data, 2018–2021. National surveillance sites for AMR in human health comprise microbiology laboratories of regional referral hospitals, national referral hospitals, and selected institutions of learning. These sites analyze clinical samples (pathogen identification and antimicrobial susceptibility testing (AST)) for routine patient care using clinical and laboratory standards Institute (CLSI). The decision to perform a bacterial culture test is entirely at the discretion of the attending clinician. For quality control purposes, these sites also refer isolates with their relevant identifiers to the National Microbiology Reference Laboratory (NMRL) (at the National Health Laboratory and Diagnostic Services (NHLDS) department of the Ministry of Health) for reidentification and antimicrobial susceptibility retesting. NMRL is accredited by the College of American Pathologists (CAP) against International Organization for Standardization ISO 15189:2012. The accredited microbiology tests include microscopy, culture, identification, and antibiotic susceptibility testing The laboratory used both manual (phenotypic using biochemicals) and automated organism identification methods.The automated methods include Brucker Daltonics Microflex LT Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometer, and the Becton Dickson Phoenix™ automated microbiology system for automated identification and susceptibility testing system. Antimicrobial susceptibility of the isolates was determined by using Kirby-Bauer disk diffusion method according to Clinical and Laboratory Standard Institute (CLSI) guidelines. When necessary, the minimal inhibitory concentrations (MICs) are determined by BD phoenix and the E-Test (Bio Mérieux) according to CLSI guidelines The laboratory is the only microbiology reference laboratory for the whole country.
Inclusion and exclusion criteria
All isolates received from all the surveillance sites as patient samples were reanalyzed by the laboratory. However, only isolates with reidentification results matching primary sites were analyzed in this study.
Study variables
We extracted individual-level data from the NMRL database. Data were extracted as an excel sheet from this electronic database containing patient reports, organism identified, and sentinel sites. Information obtained on each case included patient age, patient sex, year of sample collection, specimen source, hospital in which the sample was drawn, isolated pathogen in the positive culture, and susceptibility results (defined as susceptible, intermediate, or resistant).
Data management and analysis
We stored data in password-protected computers and data was not shared with anyone outside the investigation team. The variables analyzed included age, sex, specimen type (i.e., blood and cerebrospinal fluid), organisms isolated, and antibiotics tested. For Gram positive organisms we analysed resistance to the following organisms clindamycin, erythromycin, gentamycin, oxacillin, penicillin G, cotrimoxazole, cefoxitin, vancomycin and ciprofloxacin. For Gram negative organisms, we analyzed for the resistance to amikacin, ampicillin, amoxicillin-clavulanate, cefotaxime, ceftriaxone, cefoxitin, cefepime, chloramphenicol, ciprofloxacin, gentamycin, imipenem, meropenem, piperacillin, piperacillin-tazobactam, cotrimoxazole and tetracycline. The outcome was the antibiotic resistance of the isolates to common antibiotics. Isolates were classified as either susceptible or resistant to an antimicrobial, and all isolates with intermediate reactions were classified as resistant. Data quality was checked using the completeness of data entries in the Laboratory Information System. Rates of resistance were calculated as a proportion of resistant organisms out of the total number of organisms that were tested for antimicrobial susceptibility per year. Resistance to each antibiotic was analyzed separately, and Microsoft Excel 2016 was used to plot the trends from 2018 to 2021. The chi-square test for trends was used to test the significance of antibiotic resistance trends over time. Epi-Info™ 7- (CDC, Atlanta, USA) statistical package was used for additional statistical analysis. A p value < 0.05 was considered statistically significant.
Ethical considerations
Our study utilized routinely collected aggregated program surveillance data that did not have any personal identifiers. We obtained permission to use the AMR surveillance data from the Division of Health Information, Ministry of Health which has the overall mandate to manage collected and stored health related information. Additionally, the U.S. Centers for Disease Control and Prevention (CDC) Center for Global Health determined our study as non-research whose primary intention was to address public health problems.
Results
Attributes of organisms isolated from sterile sample cultures, Uganda, 2018–2022
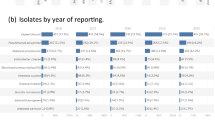
Of the 537 isolates analyzed, 188 (52.6%) were from children aged 0–5 years, 67 (18.7%) were from persons aged 6–18 years, 59 (16.2%) were from adults aged 19–45 years, and more than 66 years were from adults. Fifty-five percent of isolates were from females. Over time, 2018 had the highest number of isolates 151 (28.1%), while 2019 had the least number of isolates 93 (17%). Geographically, most isolates were from Mbarara Regional Referral Hospital 186 (48%), while the fewest came from Kiruddu National Referral Hospital and the General Military Hospital, each having 1 (0.3%). A total of 108 (20.1%) isolates were Staphylococcus aureus, followed by Salmonella species, with 101 (18.8%) (Table 1).
Trends of total antimicrobial resistance of gram-positive organisms to common antibiotics, Uganda, 2018–2021
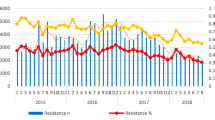
Increasing resistance to ciprofloxacin over the four-year study period was observed (26.2–44.9%; p < 0.01) (Fig. 1).
There is a general trend of increasing resistance among gram-positive organisms to most antibiotics tested in Uganda. Notably, there were significant increases in resistance to Clindamycin (25%-76%), Erythromycin (50%-76%), and Vancomycin (23–55%) over the years. Gentamycin showed fluctuating resistance rates, while resistance to Penicillin G remained consistently high. Cefoxitin susceptibility testing was only done for Staphylococcus aureus and resistance to it is a surrogate for methicillin resistance staphylococcus aureas(MRSA). See Table 2 below.
Trends of total antimicrobial resistance of gram-negative organisms to common antibiotics, Uganda 2018–2021
Overall, the percentage antibiotic resistance of gram-negative organisms increased over the years. Resistance to penicillins (piperacillin and ampicillin) was highest among gram-negative bacteria. Specifically, increases in resistance over time were noted for ceftriaxone (8.3–78.6%; p = 0.003), ciprofloxacin (17.1–42.6%; p-0.004), imipenem (5.7–29.7%; p-0.004), meropenem (5.3–18.4%; p-0.03), and tetracycline (28.6%-81.6%; p-0.02) (Table 3).
Trends of ciprofloxacin resistance by health region, 2018–2021
There was increasing resistance to ciprofloxacin across the health subregions. A significant decline was observed in 2021. There is an increasing resistance that is observed in the Elgon and Busoga subregions over time (Fig. 2).
Discussion
We described trends and patterns of antimicrobial resistance of organisms isolated from patients’ sterile site samples in Uganda during 2018–2021 with an emphasis on the antibiotics frequently utilized to treat common infections and the bacterial Gram classification. The rates of resistance to commonly-isolated antibiotics increased significantly over time. The most commonly encountered gram-positive organism was Staphylococcus aureus, while the most common gram-negative organisms were Salmonella typhii/paratyphii species and Escherichia coli. We found an increase in resistance of gram-negative organisms to ciprofloxacin, ceftriaxone, tetracycline, meropenem, and imipenem.
The increase in resistance to these antibiotic classes over time is consistent with a study by Mboowa et al. which found that also found an overall increase in resistance to antibiotics among surgical patients in a national referral hospital from 2014 to 2018 [16].
Over the study period, gram-negative bacteria had an increase in resistance to ciprofloxacin from 26 to 56%. This is similar to the findings from a systematic review that included studies assessing the prevalence of ciprofloxacin-resistant clinical isolates in Ethiopia. It found that 1 in 6 Proteus bacteria (mostly P. mirabilis) and about 1 in 7 Pseudomonas bacteria (mainly P. aeruginosa) were resistant to the ciprofloxacin [17]. A similar study in Ethiopia in 2015 observed that the rates of resistance to ciprofloxacin were high (20–70%) among bacterial organisms isolated from blood, which is a sterile site sample [18]. Currently the global recommendation on management of tuberculosis reserve ciprofloxacin as a second line option for drug resistant tuberculosis [19]. The above findings therefore pose a great threat in the management of drug resistant tuberculosis. Furthermore, as per the Uganda clinical guidelines, ciprofloxacin is the drug of choice for common conditions like enteric fever and a second line option in the management of urinary tract infections. This further complicates the treatment of these common conditions.
Gram- negative organisms had increasing resistance to ceftriaxone, ciprofloxacin, and tetracycline as well as last-resort antibiotics such as meropenem and imipenem. The growing resistance rates to carbapenems are worrisome and may lead to the spread of fatal infections, especially in hospitals [20].
Among gram-positive organisms still, particularly for Staphylococcus aureus, methicillin resistance fluctuated significantly, dropping from 60 to 45% over the study period. Cefoxitin resistance was utilized as a surrogate measure for oxacillin resistance, suggesting methicillin-resistant Staphylococcus aureus (MRSA) prevalence. This shift is relevant because MRSA is a major cause of healthcare-associated infections. A similar observation was seen by a study in Nigeria whuch also found varying MRSA frequencies, underlining the dynamic nature of MRSA resistance trends [21].
We also identified increase in resistance of gram-negative organisms to the antibiotics ceftriaxone (8–79%), tetracycline (28–81%), and imipenem (6–26%). Similar increase in resistance to third generation cephalosporins like ceftriaxone was found by a systematic review in Uganda, 2018 found that there was an overall increase in resistance to cefotaxime (46–49%) and cefuroxime, both of which are third-generation cephalosporins, for which resistance can be used to infer ceftriaxone resistance [22]. In contrast, in this same study, most organisms were susceptible to ciprofloxacin hence recommended in this study as a drug of choice for infections caused by gram-negative organisms. Another study in South Africa conducted over a 12-month from October 2011 to September 2012 at a tertiary facility in Cape Town, South Africa, Carroll et al. [23] found that for healthcare-associated Enterobacteriaceae bloodstream isolates, susceptibility rates were 58.5% to ceftriaxone and 70% to ciprofloxacin.
The observed increasing resistance across various classes of antibiotics for both gram-positive and gram-negative organisms signifies a declining range of effective treatment options. This may pose a direct threat to patient care, leading to prolonged illnesses, heightened severity of infections, and increased mortality rates. The study's identification of significant resistance spikes, particularly against commonly-used antibiotics such as ceftriaxone and ciprofloxacin, emphasizes the need for adherence to the WHO's Access, Watch, and Reserve (AWaRe) classification. Our findings highlight the need to strengthen antibiotic stewardship programs and public awareness campaigns on responsible antibiotic use to safeguard public health and ensure ongoing treatment efficacy.
Limitations
Our study only utilized secondary data from the national AMR surveillance system, which are limited in terms of variables available for analysis. Secondly, the study utilized resistance of only organisms obtained from sterile samples, and thus other organisms from non-sterile samples were excluded, resulting in an under-estimation of the rates of antibiotic resistance in Uganda. Patient-level studies using primary data to determine the factors rates and trends of antibiotic resistance from all patient samples may be beneficial in understanding the trends of antibiotic resistance in Uganda. An analysis with more samples would be more predictive of more significant findings. A longitudinal study of antibiotic prescriptions and its relationship to resistance would be helpful.
Conclusion
The study highlights a concerning increase in antibiotic resistance rates over four years, with significant increase in resistance observed across different classes of antibiotics for both gram-positive and gram-negative organisms. This increased antibiotic resistance, particularly to commonly used antibiotics like ceftriaxone and ciprofloxacin, makes adhering to the WHO's Access, Watch, and Reserve (AWaRe) category even more critical. It also emphasizes how important it is to guard against the growing threat of antibiotic resistance by appropriately using medicines, especially those that are marked for "Watch" or "Reserve."
Availability of data and materials
The datasets upon which our findings are based belong to the Uganda Public Health Fellowship Program. For confidentiality reasons, the datasets are not publicly available. The datasets can be availed upon reasonable request from the corresponding author with permission from the Uganda Public Health Fellowship Program.
Abbreviations
- AMR:
-
Antimicrobial resistance
- LIC:
-
Low-income country
- NMRL:
-
National microbiology reference laboratory
- RRH:
-
Regional Referral Hospital
- NRH:
-
National Referral Hospital
- IDI:
-
Infectious diseases institute
- MOH:
-
Ministry of Health
References
Thornber K, et al. Evaluating antimicrobial resistance in the global shrimp industry. Rev Aquac. 2020;12(2):966–86.
Mhondoro M, et al. Trends in antimicrobial resistance of bacterial pathogens in Harare, Zimbabwe, 2012–2017: a secondary dataset analysis. BMC Infect Dis. 2019;19(1):746.
Tamma PD, et al. Infectious diseases society of America 2023 guidance on the treatment of antimicrobial resistant gram-negative infections. Clin Infect Dis. 2023:ciad428.
Sulis G, Sayood S, Gandra S. Antimicrobial resistance in low-and middle-income countries: current status and future directions. Expert Rev Anti Infect Ther. 2022;20(2):147–60.
Akova M. Epidemiology of antimicrobial resistance in bloodstream infections. Virulence. 2016;7(3):252–66.
Chen T, et al. Acinetobacter baumannii strains isolated from cerebrospinal fluid (CSF) and bloodstream analysed by cgMLST: the dominance of clonal complex CC92 in CSF infections. Int J Antimicrob Agents. 2021;58(4):106404.
Javaid N, et al. Trends in antimicrobial resistance amongst pathogens isolated from blood and cerebrospinal fluid cultures in Pakistan (2011–2015): a retrospective cross-sectional study. PLoS ONE. 2021;16(4):e0250226.
Ayukekbong JA, Ntemgwa M, Atabe AN. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob Resist Infect Control. 2017;6(1):1–8.
Usha PT, Jose S, Nisha AR. Antimicrobial Drug Resistance-A global concern. Vet World. 2010;3(3).
Ferri M, et al. Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr. 2017;57(13):2857–76.
Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int. 2016;2016:2475067.
Grace D. Review of evidence on antimicrobial resistance and animal agriculture in developing countries. 2015.
Ampaire L, et al. A review of antimicrobial resistance in East Africa. Afr J Lab Med. 2016;5(1):1–6.
Omulo S, et al. A review of 40 years of enteric antimicrobial resistance research in Eastern Africa: what can be done better? Antimicrob Resist Infect Control. 2015;4:1–13.
Najjuka CF, et al. Antimicrobial susceptibility profiles of Escherichia coli and Klebsiella pneumoniae isolated from outpatients in urban and rural districts of Uganda. BMC Res Notes. 2016;9:1–14.
Llor C, Bjerrum L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther Adv Drug Saf. 2014;5(6):229–41.
Sisay M, et al. Resistance profile of clinically relevant bacterial isolates against fluoroquinolone in Ethiopia: a systematic review and meta-analysis. BMC Pharmacol Toxicol. 2018;19:1–14.
Ulhas AA. Prevalence and Characterization of carbapenem resistant organisms causing urinary tract infections among hospitalized patients and outcomes of these infections in a tertiary care center. Vellore: Christian Medical College; 2018.
World Health Organization. WHO treatment guidelines for isoniazid-resistant tuberculosis: supplement to the WHO treatment guidelines for drug-resistant tuberculosis. World Health Organization; 2018.
Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791.
Balami S, et al. Prevalence of Methicillin-Resistant Staphylococcus aureus (MRSA) in fattening lots in Maiduguri, Borno state, Nigeria. Quest J J Res Agric Anim Sci. 2016;4(2):23–7.
Ampaire L, Nduhura E, Wewedru I. Phenotypic prevalence of extended spectrum beta-lactamases among enterobacteriaceae isolated at Mulago National Referral Hospital: Uganda. BMC Res Notes. 2017;10(1):1–4.
Carroll M, et al. Five-year antimicrobial susceptibility trends among bacterial isolates from a tertiary health-care facility in Kigali, Rwanda. Am J Trop Med Hyg. 2016;95(6):1277.
Acknowledgements
The authors thank the staff of the Uganda Public Health Fellowship Program for the technical support and guidance offered during this study. The authors also extend their appreciation to the Ministry of Health, National Microbiology Reference Laboratory and National Health Laboratory and Diagnostic Centre for the technical support they offered during this study. Finally, we appreciate the microbiology laboratory staff of the different hospitals who ensure reporting of AMR data from their respective hospitals, as well as the team managing the AMR surveillance data at the National Coordination Centre; your hard work enabled the availability of data we used for this analysis.
Funding
The project was supported by the President’s Emergency Plan for AIDS (PEPFAR) through the United States Centers for Disease Control and Prevention Cooperative Agreement number GH001353-01 through Makerere University School of Public Health to the Uganda Public Health Fellowship Program, Ministry of Health. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the US Centers for Diseases Control and Prevention, the Department of Health and Human Services, Makerere University School of Public Health, or the Ministry of Health.
The funders had no role in the study design, data collection, data analysis and decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
SNK: participated in the conception, design, analysis, interpretation of the study and wrote the draft manuscript; RH, PEO, BS, ZK, BA, JFZ, HNN, RA, RN, IM and GN reviewed the report, reviewed the drafts of the manuscript for intellectual content and made multiple edits to the draft manuscript; RM, DK, BK POE, LB, ARA and SN reviewed the manuscript to ensure intellectual content and scientific integrity. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Because our study used routinely collected surveillance data reported by health facilities in the National AMR system which were also aggregated with no individual patient identifiers.However, we sought permission to use the data from the Uganda MoH. The US Centers for Disease Control and Prevention (CDC) also provided the non-research determination (NRD) for non-human subjects which is equivalent to a waiver. Data were only accessed by the study team.
Consent to publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Namubiru, S., Migisha, R., Okello, P.E. et al. Increasing trends of antibiotic resistance in Uganda: analysis of the national antimicrobial resistance surveillance data, 2018–2021. BMC Infect Dis 24, 930 (2024). https://doi.org/10.1186/s12879-024-09806-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-024-09806-y