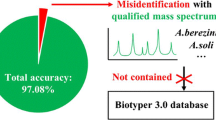
Abstract
Background
Nocardiosis, despite its rarity and underreporting, is significant due to its severe impact, characterized by high morbidity and mortality rates. The development of a precise, reliable, rapid, and straightforward technique for identifying the pathogenic agent in clinical specimens is crucial to reduce fatality rates and facilitate timely antimicrobial treatment. In this study, we aimed to identify Nocardia spp. in clinical isolates, using MALDI-TOF MS as the primary method, with molecular methods as the gold standard. Clinical Nocardia isolates were identified using 16S rRNA/hsp65/gyrB/secA1/rpoB gene sequencing. Identification performance of the Bruker MALDI Biotyper 3.1 (V09.0.0.0_8468) and MBT Compass 4.1 (V11.0.0.0_10833) for Nocardia identification was evaluated.
Results
Seventy-six Nocardia isolates were classified into 12 species through gene sequencing. The MALDI Biotyper 3.1 (V09.0.0.0_8468) achieved 100% genus-level accuracy and 84.2% species accuracy (64/76). The MBT Compass 4.1 with the BDAL Database (V11.0.0.0_10833) improved species identification to 98.7% (75/76). The updated database enhanced species level identification with scores > 1.7, increasing from 77.6% (59/76) to 94.7% (72/76), a significant improvement (P = 0.001). The new and simplified extraction increased the proportion of strains identified to the species level with scores > 1.7 from 62.0% (18/29) to 86.2% (25/29) (P = 0.016). An in-house library construction ensured accurate species identification for all isolates.
Conclusions
The Bruker mass spectrometer can accurately identify Nocardia species, albeit with some variations observed between different database versions. The MALDI Biotyper 3.1 (V09.0.0.0_8468) has limitations in identifying Nocardia brasiliensis, with some strains only identifiable to the genus level. MBT Compass 4.1 (V11.0.0.0_10833) effectively addresses this shortfall, improving species identification accuracy to 98.7%, and offering quick and reliable identification of Nocardia. Both database versions incorrectly identified the clinically less common Nocardia sputorum as Nocardia araoensis. For laboratories that have not upgraded their databases and are unable to achieve satisfactory identification results for Nocardia, employing the new and simplified extraction method can provide a degree of improvement in identification outcomes.
Similar content being viewed by others
Background
Nocardia species are aerobic opportunistic pathogens characterized by beaded and branched hyphae that are widely distributed in soil, dust, and decaying plants in nature as well as in freshwater, seawater, and animal excretions [1, 2]. Currently, 168 Nocardia spp. have been validly described from the environment samples (http://www.bacterio.CICT.FR/N/Nocardia.HTML). In recent years, new Nocardia species have been continually isolated from human specimens, and an increasing number of Nocardia species isolated from the environment, such as Nocardia huaxiensis and Nocardia takedensis, have been recently reported to infect humans. [3, 4] More than 40 valid Nocardia species are considered clinically relevant (https://www.cdc.gov/nocardiosis/health-care-workers/index.html). As an aerobic opportunistic pathogen, a significant proportion of the Nocardia species can cause infections in humans [5, 6]. According to data from the Centers for Disease Control and Prevention (CDC), there are approximately 500 to 1,000 new cases of Nocardia infection in the United States each year, with about 60% of these cases being associated with pre-existing immune compromise. Although the incidence rate data are limited, the number of cases may be rising due to an increase in individuals with severe immune compromise. (https://www.cdc.gov/nocardiosis/hcp/clinical-overview/index.html). It is important to note that due to the lack of systematic surveillance, the true prevalence of Nocardia infection is unknown. Despite being commonly classified as opportunistic, Nocardia species can also infect immunocompromised patients [7,8,9,10]. While pulmonary and disseminated nocardial diseases typically occur in immunocompromised patients, primary cutaneous nocardiosis is generally an infection found in immunocompetent individuals. Although any species of Nocardia species can cause primary cutaneous infections, N. brasiliensis is the predominant species, isolated from the majority (approximately 80%) of cases involving primary cutaneous or subcutaneous nocardiosis [1]. According to research reports, 33-42% of Nocardia infection cases do not exhibit significant immune deficiencies [7, 8].
As Nocardia has a slow growth rate and infections may show few symptoms, it can be challenging to detect them in clinical samples [11]. Therefore, laboratory diagnosis of Nocardia infection is of great significance in clinical practice. Identifying Nocardia isolates at the species level is crucial for providing the right level of care to patients because different Nocardia species display varying geographic prevalence, pathogenic characteristics, and patterns of susceptibility to antimicrobial agents [12].
Among the traditional microbiological methods for diagnosing Nocardia infections, the sensitivity of smear microscopy is low. It also has the drawback of not being able to identify species. Biochemical methods are inadequate for accurately differentiating among clinically significant species due to the vast number of identified Nocardia species.
Gene sequencing is the most precise molecular method for identifying Nocardia at the species level. The 16 S rRNA gene sequencing method is widely recognized as the most reliable technique, but it has limitations in distinguishing closely related species because it lacks adequate gene polymorphisms [1, 13]. Several additional housekeeping genes, including DNA gyrase subunit B (gyrB), ATPase secretory protein (secA1), heat shock protein gene (hsp65), and B-subunit of RNA polymerase (rpoB), were selected as supplementary genetic markers. When used in combination with 16 S rRNA, these genes establish a precise and highly specific method for the molecular identification of Nocardia [1, 14,15,16,17,18,19,20,21]. SecA1 is a housekeeping gene responsible for the production of the indispensable protein SecA1. Analysis of a specific 468-bp segment of this gene has proven to be effective in distinguishing between different Nocardia species [18]. The hsp65 gene, with a 441-bp sequence, is useful for identifying Nocardia species and offers more variability than the 16 S rRNA gene, allowing the discrimination of closely related species [19, 22]. The gyrB gene, which encodes the subunit B of DNA gyrase, has been utilized in the identification and phylogenetic analyses of Nocardia isolates [21, 23]. The B subunit of the RNA polymerase is encoded by rpoB, which has proven to be a valuable tool for identifying Nocardia isolates [21]. Although molecular methods have proven their ability to identify new species, distinguish within species, and accurately assign species, they have certain drawbacks such as relatively high costs and the need for staff to spend time analyzing the results.
Over the past couple of decades, the use of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) has completely transformed the way bacteria are identified in microbiology laboratories, enabling quick, effortless, low-cost, and reliable identification from culture growth. Early studies have shown that commercial databases are insufficient for accurate identification because they are inconsistent with sequencing methods (0–47.3%) [15, 24,25,26]. Recent studies have shown that VITEK ®-MS (MALDI-TOF), based on the VITEK ®-MS IVD database, performs well in the identification of Nocardia [27,28,29,30]. However, there have been limited studies evaluating the capability of Bruker mass spectrometry system to identify Nocardia, and prior research has indicated that Bruker mass spectrometry system may not be optimal for Nocardia identification, with only a range of 14.9–95.9% of the isolates being identifiable at the species level [25, 31,32,33,34]. Existing studies have utilized the manufacturer-recommended formic acid extraction method as a sample pretreatment, which is not only complex and time-consuming to execute, but also challenging to integrate into the daily workflow of clinical laboratories.
In this study, we examined the effectiveness of MALDI-TOF MS (Bruker Daltonics Inc., Billerica, MA, USA) for the identification of Nocardia species. Simultaneously, we aimed to explore a streamlined, rapid, and accurate workflow for identifying Nocardia strains that can be easily integrated into routine microbiology laboratory practices.
Methods
Nocardia isolates
This study analyzed 76 non-duplicate strains of Nocardia isolated from clinical specimens at Sichuan University West China Hospital from January 2016 to November 2022. All strains were stored at − 80℃ and revived by transferring onto blood agar plates and cultured at 35℃. The length of time for different strains to culture varied slightly; however, typical colony characteristics were observed in most strains within 48–72 h.
DNA extraction, PCR, 16S rRNA/hsp65/ gyrB/secA1/rpoB sequencing-based Nocardia Identification
Purified bacterial colonies were processed for DNA extraction using a TIAamp Bacteria DNA Kit (TIANGEN Biotech, Beijing, China), a centrifugal column-based kit, according to the manufacturer’s instructions. Sangon Biotech Co., Ltd. (Shanghai, China) synthesized the partial 16S rRNA, gyrB, secA1, hsp65, and rpoB.
Primers for 16S rRNA, gyrB, secA1, hsp65, and rpoB Table 1 lists the primers and PCR cycling conditions. After PCR amplification, the products were sent to Sangon Biotech Co. Ltd. (Shanghai, China) for bidirectional sequencing. The bidirectional sequence results were assembled using the DNASTAR/SeqMan software (7.1.0.44), and the sequences were cross-referenced with the type strains of Nocardia species present in the GenBank database using the BLAST program at NCBI for analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi? PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome).
According to the CLSI MM18, to classify two strains of aerobic actinomycetes as the same species, they must have a sequence similarity of at least 99.6%, with a minimum separation of 0.4% between different species. Furthermore, this document suggests that a sequence similarity of 99.0 to 99.5% is appropriate for identifying the genus. If a similarity of 99.0% in the 16 S rRNA gene sequence was detected between Nocardia species and multiple strains, species identification was performed by sequencing several other housekeeping genes: gyrB, secA1, hsp65, and rpoB.
MALDI-TOF MS identification
The MALDI biotyper microflex LT (Bruker Daltonics, Inc., Billerica, MA, USA) was used to analyze all Nocardia spp. isolates. To ensure calibration and quality control, a Bacterial Test Standard from the manufacturer was added each day. MALDI Biotyper 3.1 software (Bruker Daltonics) was used with the BDAL database (V09.0.0.0_8468) to identify Nocardia through extended direct transfer and a new, simplified extraction sample preparation. The MBT Compass 4.1 software (Bruker Daltonics) was used with the BDAL database (V11.0.0.0_10833) to identify Nocardia through extended direct transfer sample preparation. Two spot on the MALDI target plate was used for each strain, with the maximum score and the corresponding identification as determined by MALDI-TOF MS being documented. The experiment utilized two preprocessing methods, extended direct transfer and the new and simplified extraction sample preparation, to identify 29 Nocardia strains using the MALDI Biotyper 3.1 software with the BDAL database (V09.0.0.0_8468). Additionally, for the identification of 76 Nocardia strains, the extended direct transfer method was applied in conjunction with both the MALDI Biotyper 3.1 software using the BDAL database (V09.0.0.0_8468) and the MBT Compass 4.1 software along with the updated BDAL database (V11.0.0.0_10833).
The extended direct transfer method was carried out as follows. A small quantity of bacteria was deposited onto a MALDI target plate. 70% formic acid was poured over the samples, which were allowed to dry before the addition of 1 μl of a matrix solution known as α-cyano-4-hydroxycinnamic acid (α-HCCA).
A new and simplified extractionwas carried out as follows, 30 µL of 70% formic acid solution was added to a sterilized EP tube (Eppendorf tube), and the bacterial sample was scraped into the tube for thorough mixing. The mixture was left to stand for 10 min, and 1 µL of the mixture was applied onto MALDI target plate and once the drying process was completed, 1 µL of HCCA matrix was added, as instructed by the manufacturer. All the samples were identified using MALDI-TOF MS, and the highest score was recorded.
An analysis was conducted to compare the outcomes of the extended direct transfer method and the new and simplified extraction method. Strains identified at the species level with a confidence score exceeding 1.7 were classified as ‘a’. In contrast, those that failed to be identified at the species level and exhibited a confidence score below 1.7 were designated as ‘b’. The counts of strains for each method were recorded, and McNemar’s test was employed to ascertain whether there was a statistically significant disparity in the identification capacities of the two methods for Nocardia spp. A P-value < 0.05 was considered statistically significant. All analyses were performed using the Statistical Package for the Social Sciences version 24.0 software (IBM SPSS Inc., Chicago, IL, USA).
In-house library creation
According to the steps described in the “IVD MALDI Biotyper 2.3 User Manual for the fully automated rapid biomarker mass spectrometry detection system”, Nocardia species that could not be identified at the species level by the V09.0.0.0_8468 database version were added as MSP of the strains that produced identification results that were obtained using gene sequencing.
Before acquiring the spectral data, a freshly prepared bacterial test standard (Bruker Daltonics) was used to calibrate the instrument. Individual colonies were placed at eight different locations within the target plate, and readings were taken three times per position. Any isolate included as a reference MSP in the in-house library was required to have at least 20 indistinguishable spectra. The fingerprint spectra were manually collected, accumulated until the signal intensity reached over 10,000, and saved in a folder. The MALDI Biotyper software was used for library creation.
Results
Identification by genes sequencing
Sequencing of the 16S rRNA/hsp65/gyrB/secA1/rpoB genes allowed the identification of a combined total of 76 Nocardia isolates. Twelve different species of Nocardia spp. were identified. The detailed alignment results of all sequencing data are provided in Supplementary Table 1 (Sequencing of the 16 S rRNA, gyrB, hsp65, rpoB, and secA1 genes was conducted and compared against the GeneBank database using the BLAST software). The results of bidirectional sequencing were assembled and then uploaded to the BLAST software for sequence alignment (https://blast.ncbi.nlm.nih.gov/Blast.cgi? PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome). The result with the highest percent identity (Per.Ident) score was recorded. In cases where different identification outcomes were obtained and the difference in Per. Ident scores between them was less than 0.4%, a definitive identification could not be established, and both or multiple results were included in the final reference range. Results with a Per. Ident score below 95% were not considered for reference. For instance, the strain HX001 exhibited sequence alignment results for the 16 S rRNA gene with both Nocardia farcinica (Per. Ident: 100%) and Nocardia kroppenstedtii (Per. Ident: 100%). The gyrB gene sequencing alignment results included Nocardia farcinica (Per. Ident: 100%), Nocardia jejuensis (Per. Ident: 100%), Nocardia globerula (Per. Ident: 100%), and Nocardia exalbida (Per. Ident: 99.66%), none of which provided a definitive identification. The final identification was achieved by integrating the hsp65 gene result, identifying Nocardia farcinica (Per. Ident: 100%), the rpoB gene result as Nocardia farcinica (Per. Ident: 99.15%), and the secA gene result (Per.Ident: 99.38%), leading to the conclusive identification of the strain as Nocardia farcinica. The final identification was determined by integrating the test results of the 16 S rRNA gene with the other four genes. All isolated strains were unequivocally identified to the species level through sequencing. The data from this study was deposited in NCBI Sequence Read Archive under accession SRA data: PRJNA1091814. The numbers and composition ratios of the species are shown in Fig. 1.
Comparison of extended direct transfer and a new and simplified sample preparation using maldi biotyper 3.1 with BDAL database (V09.0.0.0_8468) for Nocardia identification
Using Extended direct transfer and a new and simplified sample preparation method, 29 Nocardia spp. isolates were identified using MALDI Biotyper 3.1 software was applied with the BDAL database (V09.0.0.0_8468). The specific identification results and scores are listed in Supplementary Table 2. We compiled the quantitative data on species and genus identifications by the two methods across different bacterial strains, including the distribution of confidence scores for species-level identifications, as shown in Table 2. Strains identified at the species level with a confidence score exceeding 1.7 were classified as ‘a’. In contrast, those that failed to be identified at the species level and/or exhibited a confidence score below 1.7 were designated as ‘b’. The statistical results are presented in Table 3.
The identification results for 29 Nocardia spp. isolates using the two methods showed that with the MALDI Biotyper 3.1 and BDAL Database (V09.0.0.0_8468), we achieved 100% genus identification accuracy. However species identification varied with the sample preparation method: it was 79.3% (23/29) using the extended direct transfer method, with six strains identified as Nocardia spp. and confirmed as Nocardia brasiliensis by sequencing (Table 2). Conversely, the new and simplified extraction method improved the species identification accuracy to 89.7% (26/29), with three strains identified as Nocardia spp. and confirmed as Nocardia brasiliensis by sequencing (Table 2). Compared to the extended direct transfer method, the new and simplified extraction method increased the proportion of strains identified at the species level with a confidence score > 1.7 from 62.0% (18/29) to 86.2% (25/29), an improvement of 24.2% (7/29). McNemar’s test revealed a statistically significant difference in the accuracy of Nocardia spp. preprocessing between the two experimental methods, with P = 0.016.
The strains HX007, HX018, and HX029 were identified as Nocardia spp. by MALDI-TOF MS. Sequencing identified these species as Nocardia brasiliensis. These strains were added as MSPs to an in-house library for future reference and retesting. The results confirmed their identification and the scores showed a significant improvement (Table 4).
Comparison of Nocardia spp. identification capabilities between maldi biotyper 3.1 with BDAL database (V09.0.0.0_8468) and MBT compass 4.1 with BDAL database (V11.0.0.0_10833) using extended direct transfer sample preparation
Using the Extended Direct Transfer Sample Preparation Methods, 76 Nocardia spp. isolates were identified using MALDI Biotyper 3.1 with BDAL Database (V09.0.0.0_8468) and MBT Compass 4.1 software with the BDAL database (V11.0.0.0_10833). The specific identification results and scores refer to (Table 5).
Strains identified at the species level with a confidence score exceeding 1.7 were classified as ‘a’. In contrast, those that failed to be identified at the species level and/or exhibited a confidence score below 1.7 were designated as ‘b’. The statistical results are presented in Table 6.
Compared to the sequencing outcomes, MALDI Biotyper 3.1 with the BDAL Database (V09.0.0.0_8468) achieved a genus identification accuracy of 100% and a species identification accuracy of 84.2% (64/76) for Nocardia spp. Among the results at the species level, 47.37% (36/76) had scores above 2.0, 30.26% (23/76) fell within the range of 1.7 to 1.99, and 6.58% (5/76) had scores below 1.7. 14.5% (11/76) of strains were identified as Nocardia spp., and according to the sequencing results, all eleven were identified as Nocardia brasiliensis (Table 5). However, one strain of Nocardia sputorum was erroneously identified as Nocardia araoensis.
Compared to the sequencing results, MBT Compass 4.1 with the BDAL Database (V11.0.0.0_10833) achieved a genus identification accuracy of 100% and 98.7% accuracy (75/76) in Nocardia species identification. At the species level, 64.47% (49/76) scored more than 2.0, 30.26% (23/76) scored between 1.7 and 1.99, and 3.95% (3/76) scored less than 1.7. One strain of Nocardia sputorum was misidentified as Nocardia araoensis.
Eleven isolates were identified as Nocardia sp. using the MALDI Biotyper 3.1 BDAL database (V09.0.0.0_8468) and were correctly identified as Nocardia brasiliensis using MBT Compass 4.1 software with the BDAL database (V11.0.0.0_10833). Comparing the identification capabilities of the two database versions for Nocardia, the MBT Compass 4.1 with the updated BDAL Database (V11.0.0.0_10833) showed an improvement over the MALDI Biotyper 3.1 BDAL database (V09.0.0.0_8468) in the proportion of Nocardia species accurately identified to the species level with a score > 1.7, increasing from 77.6% (59/76) to 94.7% (72/76). McNemar’s Test indicated a significant enhancement in identification capability after the database upgrade, with a P = 0.001, indicating a statistically significant difference between the two methods. Both databases incorrectly identified one strain of Nocardia sputorum as Nocardia araoensis. For the remaining strains, no species misidentifications occurred, even when the confidence scores were below 1.7.
Discussion
The MALDI-TOF MS identification technique has significantly transformed clinical bacteriology by offering a swift and dependable method for bacterial identification, which is crucial for clinicians in making informed treatment decisions. This technology holds particular relevance for the identification of Nocardia species, as it allows for the inference of antibiotic susceptibility profiles directly from the identification process [27].
The 16S rRNA sequence analysis is commonly used for bacterial identification. However, studies have shown that this sequence cannot effectively distinguish closely related Nocardia species [1, 13]. Consistent with previous reports [15], this study also revealed that 16S rRNA sequencing was unable to accurately distinguish between certain Nocardia species. For instance, the 16 S rRNA identification results for 16 strains of Nocardia farcinica, including HX001, HX004, HX006, etc., were identified as both Nocardia kroppenstedtii and Nocardia farcinica, with Per.Ident scores all exceeding 99.0% and the difference in scores being less than 0.4%. Similarly, the 16S rRNA identification results for three strains of Nocardia concava, numbered HX002, HX040, HX041, were identified as both Nocardia sp. and Nocardia concava, with Per.Ident scores all exceeding 99.0% and the difference in scores being less than 0.4%. Additionally, the 16S rRNA identification results for 11 strains of Nocardia brasiliensis, including HX003, HX005, HX007, etc., were identified as Nocardia brasiliensis, Nocardia sp., and Nocardia vulneris, with Per.Ident scores all exceeding 99.0% and the difference in scores being less than 0.4%. However, it is worth mentioning that sequencing a single gene locus is not sufficient to accurately identify a significant proportion of Nocardia species [20]. Nocardia species can be accurately identified by combining the 16S rRNA gene with the hsp65, gyrB, secA1, and rpoB genes.
In the present study, Nocardia farcinica (30%), Nocardia brasiliensis (24%), and Nocardia cyriacigeorgica (17%) were the three most common clinical Nocardia species. N. cyriacigeorgica and N. farcinica have been the most commonly reported clinical strains in previous studies [26, 27, 35], which is consistent with the results of the present study. However, in studies conducted by Xiao et al. [26] and Wang et al. [35], the proportion of Nocardia brasiliensis was low. In another study conducted by Lao et al. in Taiwan [36], Nocardia brasiliensis showed the highest clinical isolation rate. In this study, Nocardia brasiliensis ranked second in terms of the isolation rate, raising the question whether Nocardia species causing human infection vary from one geographical region to another.
The high costs and requirements of sequencing facilities hinder the implementation of sequencing-based methods in routine clinical laboratory operations, potentially impacting the efficiency and timeliness of result generation. The emergence of mass spectrometry technology has enabled the rapid and accurate clinical identification of Nocardia bacteria.
Previous studies have shown that accurate identification of Nocardia species requires the use of in-house libraries developed [24, 26, 33, 37], or additional tools and methods such as VITEK®- PICKMETM pen and a formic acid-based protein extraction [27] (with identification accuracies of 93.7–95.9%). With the expansion of commercial databases, the identification capabilities of Nocardia mass spectrometry have gradually improved. According to the research by M. Marín et al., the accuracy of identifying Nocardia species using MALDI-TOF MS with the updated BDAL database (Bruker Daltonics) containing 6,903 MSPs reached 94.5% [33]. Although this version of the database precedes the (V09.0.0.0_8468) version, our study revealed that the accuracy of identifying Nocardia species with the 8468 MSPs database was 84.2% at the species level (64 out of 76), indicating a noticeable discrepancy. The primary analysis for this discrepancy suggests that our study included a larger number of N. brasiliensis isolates, totaling 18, whereas M. Marín’s study had a smaller representation of this species with only 2 isolates. The insufficient diversity of strains in the earlier version (V09.0.0.0_8468) may have failed to uncover its limitations in identifying N. brasiliensis, highlighting a potential flaw in the database’s ability to accurately identify this species.
In the study by Gema Carrasco and colleagues, Nocardia species were categorized into three groups based on their prevalence in Spain [25]. The consistency of MALDI-TOF MS identification results compared to 16 S rRNA analysis for high and intermediate prevalence Nocardia species was 76% and 45%, respectively, indicating that the proteomic technique has room for improvement in accurately identifying Nocardia species. However, in our study, which included strains from the high and intermediate prevalence groups, excluding N. nova and N. carnea, we achieved a species identification accuracy of 98.7% (75/76) using the MBT Compass 4.1 (V11.0.0.0_10833). This suggests that the upgraded MALDI-TOF MS database has significantly enhanced its capability for identifying common Nocardia species.
Similar to the study by Melanie L. Yarbrough [34], our research has shown that the use of the Bruker mass spectrometry system with the direct on-target extraction method yields excellent identification results for Nocardia species. Yarbrough’s study compared three extraction methods: mycobacterial extraction, ethanol formic acid extraction, and direct on-target extraction, with the latter proving to be the superior approach. In line with their findings, our study found that for databases version V11.0.0.0_10833 and above, the direct on-target extraction, also known as Extended Direct Transfer, provides reliable identification results. In practical applications, due to the dry nature of Nocardia, which can be challenging to collect from agar plates, we can perform two spotting attempts on the target plate and record the highest score as the final result, obviating the need for more complex extraction methods. However, when using lower version databases or when Extended Direct Transfer does not yield high identification scores (> 1.7), the new and simplified extraction method can be employed. This method is less cumbersome than ethanol formic acid extraction and can enhance the reliability of the identification certain degree.
In this study, based on the species included in the BDAL database (V11.0.0.0_10833), the performance of Bruker MS using MBT Compass 4.1 software in identifying Nocardia through extended direct transfer sample preparation was highly effective. This method allowed for quick and reliable Nocardia identification in 98.7% (75/76) of the isolates using the same laboratory process as for conventional bacteria. The HX037 strain is Nocardia sputorum., which was newly discovered as a Nocardia species in 2023 by Hamada M et al., [38] was misidentified as Nocardia araoensis. The analysis revealed that the reason for this misidentification is the absence of the strain’s spectral profile in the current V11.0.0.0_10833 of the BDAL database. Among the identified results, 64.47% (49/76) had a score of 2.0 or higher, 30.26% (23/76) had a score ranging from 1.7 to 1.99, and 3.95% (3/76) had a score below 1.7. According to the manufacturer’s recommendations, a spectral score of ≥ 2.00 was considered species-level identification, a score of 1.700-1.999 indicated identification at the genus level, and a score of < 1.70 was deemed unreliable for identification purposes. In this study, 34.2% of the Nocardia strains had identification scores below 2.0. However, with the exception of a single case where the clinically less common Nocardia sputorum was misidentified, when the highest score among all identification results was in agreement with the molecular identification results, MBT Compass 4.1 with the BDAL Database (V11.0.0.0_10833) fulfilled the clinical requirement for the rapid and accurate identification of Nocardia species. Similar to the findings of Melanie L. et al. [34]. , for Nocardia spp., which is a challenging pathogen due to its difficult cell wall breakdown, adjusting the confidence threshold to 1.8 or 1.7 can yield satisfactory identification results with Bruker MS.
Database upgrades come at a certain cost, and unfortunately, not all laboratories have had the opportunity to undergo these important updates. Our research has shown that the extended direct transfer sample preparation method using Bruker MALDI-TOF MS (BDAL database v09.0.0.0_8468 database) has an identification accuracy of 100% for Nocardia at the genus level and 84.2% (64/76) at the species level. The main limitation of this database is the lack of diversity in MSP for Nocardia brasiliensis strains, resulting in some strains being identified only at the genus level. However, this issue was effectively resolved in the most recent iteration of the database (BDAL database V11.0.0.0_10833) with supplementary improvements. For laboratories still using the v09.0.0.0_8468 database and unable to identify Nocardia to the genus level or with no identification, a new and simplified extraction sample preparation method is recommended. Although this pre-processing method may be cumbersome, it is simpler than formic acid-based protein extraction and easier to implement in clinical practice. This new and simplified extraction sample preparation method has the potential to improve identification success rates. However, if a new and simplified extraction sample preparation method fails to yield reliable identification, conditional laboratories should use molecular methods as an initial step to accurately identify strains. If the laboratory encounters difficulties in sequencing using four or five genes, it should at least perform sequencing based on 16 S rRNA, along with gyrB and secA [15, 20] or sequence the full length of the 16 S rRNA gene(1500 bp), and then add any one or more of the gyrB, secA1, hsp65, and rpoB for identification [15, 39]. Additionally, the laboratory should incorporate MSP into its in-house library using a standardized procedure with the aim of enhancing the accuracy of Nocardia identification.
This study had certain limitations. Some rare Nocardia species such as N. wallacei, N. pseudobrasiliensis, N. veterana, N. carnea, N. elegans, and N. thailandica, which were rarely found in other studies [27, 33], were not detected at our research center. Therefore, it was not possible to evaluate the identification effectiveness of the current database for these rare Nocardia species.
Conclusion
In conclusion, we evaluated the ability of the Bruker MALDI-TOF MS to identify Nocardia spp. The Bruker MALDI-TOF MS provides fast and reliable results for identifying Nocardia in the BDAL database V11.0.0.0_10833. The identification accuracy of the most common Nocardia species in this region was 98.7%. However, for the BDAL database V09.0.0.0_8468, there was a deficiency in the identification efficiency for Nocardia brasiliensis, and it was necessary to add the MSP into the in-house library to accurately identify Nocardia species.
Data availability
The datasets supporting the conclusions of this article are included within the article (and its additional files).
Abbreviations
- MALDI-TOF MS:
-
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry
- MALDI:
-
Matrix-Assisted Laser Desorption/Ionization
- TOF:
-
Time-of-Flight
- MS:
-
Mass Spectrometry
- 16S rRNA:
-
16 S Ribosomal RNA
- hsp65:
-
Heat Shock Protein 65
- gyrB:
-
DNA Gyrase Subunit B
- secA1:
-
ATPase Secretory Protein
- rpoB:
-
RNA Polymerase B Subunit
- BDAL:
-
Bruker Daltonics Automated Library
- MBT Compass:
-
MALDI Biotyper Compass
- CDC:
-
Centers for Disease Control and Prevention
- NCBI:
-
National Center for Biotechnology Information
- BLAST:
-
Basic Local Alignment Search Tool
- CLSI:
-
Clinical and Laboratory Standards Institute
- SRA:
-
Sequence Read Archive
- MSP:
-
Matrix Spectra Profile
- EP Tube:
-
Eppendorf Tube
- HCCA:
-
α-Cyano-4-Hydroxycinnamic Acid
- SPSS:
-
Statistical Package for the Social Sciences
References
Brown-Elliott BA, Brown JM, Conville PS, Wallace RJ. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin Microbiol Rev. 2006;19:259–82. https://doi.org/10.1128/CMR.19.2.259-282.2006.
Wilson JW. Nocardiosis: updates and clinical overview. Inmayo Clinic Proceedings. 2012;87:403–7. https://doi.org/10.1016/j.mayocp.2011.11.016
Zhuang K, Liu Y, Dai Y, Xu J, Li W, Ming H, et al. Nocardia huaxiensis sp. nov., an actinomycete isolated from human skin. Int J Syst Evol Microbiol. 2021;71:004970. https://doi.org/10.1099/ijsem.0.004970.
Lotte R, Chevalier A, Dantas S, Degand N, Gaudart A, Rörhlich P-S, et al. Nocardia takedensis: a newly recognized pathogen responsible for skin and soft tissue infections. Ann Clin Microbiol Antimicrob. 2020;19:38. https://doi.org/10.1186/s12941-020-00379-7.
Hemmersbach-Miller M, Catania J, Saullo JL. Updates on Nocardia skin and soft tissue infections in solid organ transplantation. Curr Infect Dis Rep. 2019;21:27. https://doi.org/10.1007/s11908-019-0684-7.
Brown-Elliott BA, Conville P, Wallace RJ Jr. Current status of Nocardia taxonomy and recommended identification methods. Clin Microbiol Newsl. 2015;37:25–32. https://doi.org/10.1016/j.clinmicnews.2015.01.007.
Kim YK, Sung H, Jung J, Yu SN, Lee JY, Kim SH, et al. Impact of immune status on the clinical characteristics and treatment outcomes of nocardiosis. Diagn Microbiol Infect Dis. 2016;85:482–7. https://doi.org/10.1016/j.diagmicrobio.2016.05.004.
Gray D, Crawford S, Newton P. Nocardiosis in the Illawarra-Shoalhaven region from 2010 to 2019. Commun Dis Intell (2018). 2022;46. https://doi.org/10.33321/cdi.2022.46.7
Beaman BL, Beaman L. Nocardia species: host-parasite relationships. Clin Microbiol Rev. 1994;7:213–64. https://doi.org/10.1128/CMR.7.2.213.
Yi M, Wang L, Xu W, Sheng L, Jiang L, Yang F, et al. Species distribution and antibiotic susceptibility of Nocardia isolates from Yantai, China. Infect Drug Resist. 2019;12:3653–61. https://doi.org/10.2147/IDR.S232098.
Mehta HH, Shamoo Y. Pathogenic Nocardia: a diverse genus of emerging pathogens or just poorly recognized? PLoS Pathog. 2020;16:e1008280. https://doi.org/10.1371/journal.ppat.1008280.
Larruskain J, Idigoras P, Marimón JM, Pérez-Trallero E. Susceptibility of 186 Nocardia sp. isolates to 20 antimicrobial agents. Antimicrob Agents Chemother. 2011;55:2995–8. https://doi.org/10.1128/aac.01279-10.
Xiao M, Kong F, Sorrell TC, Cao Y, Lee OC, Liu Y, et al. Identification of pathogenic Nocardia species by reverse line blot hybridization targeting the 16S rRNA and 16S-23S rRNA gene spacer regions. J Clin Microbiol. 2010;48:503–11. https://doi.org/10.1128/JCM.01761-09.
Kong F, Chen SC, Chen X, Sintchenko V, Halliday C, Cai L, et al. Assignment of reference 5’-end 16S rDNA sequences and species-specific sequence polymorphisms improves species identification of Nocardia. Open Microbiol J. 2009;3:97. https://doi.org/10.2174/1874285800903010097.
Conville PS, Brown-Elliott BA, Smith T, Zelazny AM. The complexities of Nocardia taxonomy and identification. J Clin Microbiol. 2018;56:10–128. https://doi.org/10.1128/JCM.01419-17.
Wei M, Wang P, Yang C, Gu L. Molecular identification and phylogenetic relationships of clinical Nocardia isolates. Antonie Van Leeuwenhoek. 2019;112:1755–66. https://doi.org/10.1007/s10482-019-01296-2.
Carrasco G, Valdezate S, Garrido N, Medina-Pascual MJ, Villalón P, Sáez-Nieto JA. gyrB analysis as a tool for identifying Nocardia species and exploring their phylogeny. J Clin Microbiol. 2015;53:997–1001. https://doi.org/10.1128/JCM.03072-14.
Conville PS, Zelazny AM, Witebsky FG. Analysis of secA1 gene sequences for identification of Nocardia species. J Clin Microbiol. 2006;44:2760–6. https://doi.org/10.1128/jcm.00155-06.
Rodríguez-Nava V, Couble A, Devulder G, Flandrois JP, Boiron P, Laurent F. Use of PCR-restriction enzyme pattern analysis and sequencing database for hsp65 gene-based identification of Nocardia species. J Clin Microbiol. 2006;44:536–46. https://doi.org/10.1128/JCM.44.2.536-546.2006.
McTaggart LR, Richardson SE, Witkowska M, Zhang SX. Phylogeny and identification of Nocardia species on the basis of multilocus sequence analysis. J Clin Microbiol. 2010;48:4525–33. https://doi.org/10.1128/JCM.00883-10.
Carrasco G, Valdezate S, Garrido N, Villalon P, Medina-Pascual MJ, Saez-Nieto JA. Identification, typing, and phylogenetic relationships of the main clinical Nocardia species in Spain according to their gyrB and rpoB genes. J Clin Microbiol. 2013;51:3602–8. https://doi.org/10.1128/JCM.00515-13.
Kong F, Wang H, Zhang E, Sintchenko V, Xiao M, Sorrell TC, et al. secA1 gene sequence polymorphisms for species identification of Nocardia species and recognition of intraspecies genetic diversity. J Clin Microbiol. 2010;48:3928–34. https://doi.org/10.1128/JCM.01113-10.
Takeda K, Kang Y, Yazawa K, Gonoi T, Mikami Y. Phylogenetic studies of Nocardia species based on gyrB gene analyses. J Med Microbiol. 2010;59:165–71. https://doi.org/10.1099/jmm.0.011346-0.
Blosser SJ, Drake SK, Andrasko JL, Henderson CM, Kamboj K, Antonara S, et al. Multicenter matrix-assisted laser desorption ionization-time of flight mass spectrometry study for identification of clinically relevant Nocardia spp. J Clin Microbiol. 2016;54:1251–8. https://doi.org/10.1128/JCM.02942-15.
Carrasco G, de Dios Caballero J, Garrido N, Valdezate S, Cantón R, Sáez-Nieto JA. Shortcomings of the commercial MALDI-TOF MS database and use of MLSA as an arbiter in the identification of Nocardia species. Front Microbiol. 2016;7:542. https://doi.org/10.3389/fmicb.2016.00542.
Xiao M, Pang L, Chen SC, Fan X, Zhang L, Li HX, et al. Accurate identification of common pathogenic Nocardia species: evaluation of a multilocus sequence analysis platform and matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS ONE. 2016;11:e0147487. https://doi.org/10.1371/journal.pone.0147487.
Hodille E, Prudhomme C, Dumitrescu O, Benito Y, Dauwalder O, Lina G. Rapid, easy, and reliable identification of Nocardia sp. by MALDI-TOF mass spectrometry, VITEK((R))-MS IVD V3.2 database, using direct deposit. Int J Mol Sci. 2023;24:5469. https://doi.org/10.3390/ijms24065469.
Body BA, Beard MA, Slechta ES, Hanson KE, Barker AP, Babady NE, et al. Evaluation of the Vitek MS v3.0 matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of Mycobacterium and Nocardia species. J Clin Microbiol. 2018;56:10–128. https://doi.org/10.1128/JCM.00237-18.
Durand T, Vautrin F, Bergeron E, Girard V, Polsinelli S, Monnin V, et al. Assessment of VITEK(R) MS IVD database V3.0 for identification of Nocardia spp. using two culture media and comparing direct smear and protein extraction procedures. Eur J Clin Microbiol Infect Dis. 2020;39:559–67. https://doi.org/10.1007/s10096-019-03758-x.
Rodriguez-Temporal D, Zvezdanova ME, Benedi P, Marin M, Blazquez-Sanchez M, Ruiz-Serrano MJ, et al. Identification of Nocardia and non-tuberculous Mycobacterium species by MALDI-TOF MS using the VITEK MS coupled to IVD and RUO databases. Microb Biotechnol. 2023;16:778–83. https://doi.org/10.1111/1751-7915.14146.
Hsueh PR, Lee TF, Du SH, Teng SH, Liao CH, Sheng WH, et al. Bruker biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of Nocardia, Rhodococcus, Kocuria, Gordonia, Tsukamurella, and Listeria species. J Clin Microbiol. 2014;52:2371–9. https://doi.org/10.1128/JCM.00456-14.
Verroken A, Janssens M, Berhin C, Bogaerts P, Huang TD, Wauters G, et al. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Nocardia species. J Clin Microbiol. 2010;48:4015–21. https://doi.org/10.1128/JCM.01234-10.
Marin M, Ruiz A, Iglesias C, Quiroga L, Cercenado E, Martin-Rabadan P, et al. Identification of Nocardia species from clinical isolates using MALDI-TOF mass spectrometry. Clin Microbiol Infect. 2018;24:1342. https://doi.org/10.1016/j.cmi.2018.06.014. e5-1342 e8.
Yarbrough ML, Lainhart W, Burnham CA. Identification of Nocardia, Streptomyces, and Tsukamurella using MALDI-TOF MS with the Bruker Biotyper. Diagn Microbiol Infect Dis. 2017;89:92–7. https://doi.org/10.1016/j.diagmicrobio.2017.06.019.
Wang H, Zhu Y, Cui Q, Wu W, Li G, Chen D, et al. Epidemiology and antimicrobial resistance profiles of the Nocardia species in China, 2009 to 2021. Microbiol Spectr. 2022;10:e0156021. https://doi.org/10.1128/spectrum.01560-21.
Lao CK, Tseng MC, Chiu CH, Chen NY, Chen CH, Chung WH, et al. Clinical manifestations and antimicrobial susceptibility of Nocardia species at a tertiary hospital in Taiwan, 2011–2020. J Formos Med Assoc. 2022;121:2109–22. https://doi.org/10.1016/j.jfma.2022.06.011.
Buckwalter SP, Olson SL, Connelly BJ, Lucas BC, Rodning AA, Walchak RC, et al. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Mycobacterium species, Nocardia species, and other aerobic actinomycetes. J Clin Microbiol. 2016;54:376–84. https://doi.org/10.1128/JCM.02128-15.
Hamada M, Enomoto N, Yamashita T, Shimojima M, Tanno D, Ohana N, et al. Nocardia sputorum sp. nov., an actinobacterium isolated from clinical specimens in Japan. Int J Syst Evol Microbiol. 2023;73:005935. https://doi.org/10.1099/ijsem.0.005935.
Traxler RM, Bell ME, Lasker B, Headd B, Shieh W-J, McQuiston JR. Updated review on Nocardia species: 2006–2021. Clin Microbiol Rev. 2022;35:e0002721. https://doi.org/10.1128/cmr.00027-21.
Acknowledgements
We thank Prof Yongcheng Tong for providing technical support for this research and Editage (www.editage.cn) for English language editing.
Funding
The authors gratefully acknowledge the financial support from the Sichuan Science and Technology Program (No. 2022YFS0096), the 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (2019HXFH051).
Author information
Authors and Affiliations
Contributions
Y.L. led the conception and design of the study, performed experiments, created figures, analyzed data, and prepared the manuscript. S.Y.W., J.D., and N.W. also contributed to the experimental work. Z.K.W participated in the experimental sequencing of this study. N.W., Y.T., and Y.L.X. played key roles in data analysis. W.L. Z. and Q.F.L. conceived the study and performed experiments. Additionally, M. K. substantively revised the manuscript and approved the final version for submission.
Corresponding author
Ethics declarations
Ethical approval
This study has been approved by the Biomedical Research Ethics committee of West China Hospital of Sichuan University (approval number: 2021 (252)).
Consent for publication
Consent for publication is not applicable to this study as it does involve any individual person’s data.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, Y., Wu, SY., Deng, J. et al. Application of MALDI-TOF mass spectrometry for identification of Nocardia species. BMC Microbiol 24, 358 (2024). https://doi.org/10.1186/s12866-024-03483-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-024-03483-2