Abstract
Two eukaryotic cell lines, A549 and A431, with stable expression of the nucleocapsid protein (N-protein) of the SARS-CoV-2 virus fused with the red fluorescent protein mRuby3 were obtained. Using microscopy, the volumes of the cytoplasm and nucleus were determined for these cells. Using quantitative immunoblotting techniques, the concentrations of the N-mRuby3 fusion protein in their cytoplasm were assessed. They were 19 and 9 μM for A549 and A431 cells, respectively. Using these concentrations, the initial rate of N-protein degradation in the studied cells was estimated from the decrease in cell fluorescence. In A549 and A431 cells, it was the same (84 nM per hour). The approach of quantitatively describing the degradation process can be applied to analyze the effectiveness of a wide class of antiviral drugs that cause degradation of viral proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
When developing new antiviral drugs (in particular, against the SARS-CoV-2 coronavirus), it is often necessary to obtain model cell systems to study this virus. For most viruses, there is a key protein or proteins without which the viral capsid cannot form. An example of such proteins is the nucleocapsid protein (N-protein) of the SARS-CoV-2 virus [1–3]. Usual interaction of antibody-like molecules with this protein can result in the disruption of the formation of the viral capsid, i.e., prevent the spread of viral infection [4]. To study the interaction of various drugs with this protein, it is most convenient to use cells with temporary or stable expression of this protein. In this work, two cell lines with stable expression of the N-protein of the SARS-CoV-2 virus were obtained as a model. In many delivery systems, various antibody-like molecules are commonly used [5]. For example, previously we showed that modular nanotransporters (MNTs) containing the NC2 monobody [6] to the N-protein of the SARS-CoV-2 virus are capable of binding to this protein, both in solution and in cells [7]. If these MNTs contain the DPETGEYL amino acid sequence, which is capable of high-affinity binding to the Keap1 ubiquitin ligase [8], then the simultaneous binding of MNTs to Keap1 and the N-protein should presumably lead to the degradation of the N-protein as a result of its ubiquitination [9]. In this study, we quantitatively described the process of N-protein degradation in cells under the influence of MNT. For this purpose, first of all, the intracellular concentration of N-protein in the obtained cell lines was assessed.
A549 and A431 cells, with stable expression of N-protein fused to the fluorescent protein mRuby3 (N-mRuby3), were obtained by lentiviral transfection of cells using plasmids pMD2.G, psPAX2, and pHAGE N-mRuby3 (from SARS-CoV-2) IRES puro (Addgene). Recombinant lentiviral particles were prepared by cotransfection of HEK293T cells in a 25-cm2 flask with 2.3 μg pMD2.G, 4.3 μg psPAX2, and 10.2 μg pHAGE N-mRuby3 (from SARS-CoV-2) IRES puro using Calcium Phosphate Transfection Reagent (Thermo Fisher Scientific). The virus suspension was collected 72 h after transfection. Lentivirus was concentrated using a Lenti-X concentrator (TakaraBio) according to the protocol provided by the company. Human lung adenocarcinoma A549 cells and human epidermoid carcinoma A431 cells were seeded into 24-well plates (1 × 104 cells per well) 1 day before viral infection. To perform lentiviral transfection, lentiviral particles were added to a culture medium containing 10 μg/mL polybrene (Sigma-Aldrich). Stable cell lines were selected on medium containing 1 μg/mL pyromycin (Acros Organics). Transfection of cells with the N-mRuby3 fusion protein was determined visually using confocal microscopy in the mRuby3 channel using an LSM 510 META NLO multiphoton scanning microscope (Carl Zeiss, Germany) with NA 0.4 lens (magnification 20×), as well as using Western blot analysis with antibodies to the N-protein (PA5-116894, Invitrogen) (Fig. 1a).
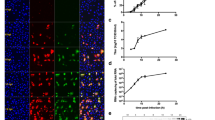
(a) Western blot with antibodies to the N-protein of the SARS-CoV-2 virus for lysates of N-mRuby3 transfected A549 cells (1) and A431 cells (2) and recombinant N-protein concentrations: 200 (3), 600 (4), and 1000 nM (5). M—protein standards. (b) Calibration curve of dependence of Western-blot total band intensity on N-protein concentration.
MNT expression was studied in the E. coli strain Ros(DE3)pLysS. Autoiinduction of MNT expression was performed by incubation at 18°C for 48 h. MNTs were isolated from the soluble fraction [10] and then purified by affinity chromatography on HisTrap FF column (Cytiva). Denaturing electrophoresis in polyacrylamide gel showed a sufficient degree of purity of the obtained MNTs (86.6%).
As a result of analysis of 69–87 images of cells detached from the substrate, which was performed as described in [11], the volumes of cells and their nuclei were determined: cell volumes, 1814 ± 87 and 1450 ± 37 µm3; nucleus volumes, 374 ± 28 and 160 ± 7 µm3 for A549 and A431 cells, respectively. The N-mRuby3 fusion protein was located predominantly in the cytoplasm of A549 and A431 cells. The cytoplasm volume was 1439 ± 91 and 1289 ± 38 µm3 for A549 and A431 cells, respectively. These values were subsequently used to calculate the concentration of N-mRuby3 in the cytoplasm.
The obtained A549 and A431 cells were collected from the substrate using trypsin treatment and subsequent centrifugation. The cells were then suspended in 100 μL of phosphate buffer (pH 8.0) containing cOmplete (EDTA free protease inhibitor cocktail, 11873580001, www.sigmaaldrich.com) and 10 mM EDTA. The number of cells in the studied samples was measured using a MACSQuant Analyzer flow cytometer (Miltenyi Biotec GmbH, France). Cells were lysed by three cycles of freezing in liquid nitrogen and thawing at 37°C. When cells are lysed, N-protein is released and distributed throughout the entire volume of the sample. The ratio of the volume of the sample to the total volume of the cytoplasm of all cells contained in the sample before lysis (the so-called dilution factor) was 126–280 and 197–371 for A549 and A431 cells, respectively (for three or four samples with different concentrations of cells).
The concentration of the N-mRuby3 protein in A549 and A431 cell lysates was measured using N-protein calibration solutions of known concentrations. The N-protein used for these purposes was obtained and purified as described in [7]; its purity was 98.9%. The obtained lysates of A549 and A431 cells were loaded onto polyacrylamide gel together with N-protein of known concentration. Electrophoresis was followed by immunoblotting. For this purpose, samples were transferred to a 0.22-μm nitrocellulose membrane and stained with antibodies to N-protein and secondary goat-antirabbit+Peroxidase antibodies (G21234, ThermoFisher Scientific) (Fig. 1a). Using the known N-protein concentration, calibration curves were constructed (Fig. 1b) and used to calculate the concentration of N-protein in the studied samples. Knowing the dilution factor of the N-mRuby3 protein during cell lysis and the concentration of N-protein in the lysates, we calculated the N-mRuby3 concentration in the cytoplasm: 19.0±1.3 and 9.0 ± 1.8 µM for A549 and A431 cells, respectively.
The differences in the N-mRuby3 concentrations in A549 and A431 cells are clearly seen under a fluorescence microscope: the fluorescence intensity of A549 cells is noticeably higher than that of A431 cells. If we calculate the average fluorescence intensity of mRuby3 in the area of cells and subtract from it the average fluorescence intensity of the noise signal (for areas without cells), then the ratio of the mRuby3 fluorescence intensity for A549 cells to that for A431 cells is 1.6 ± 0.2. This value is close to the ratio of N‑mRuby3 concentrations we determined for these two cell lines (2.1 ± 0.4). Thus, the difference in fluorescence of A549 and A431 cells reflects the difference in the concentrations of the N-mRuby3 protein in these cells.
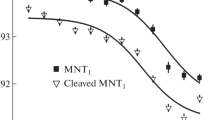
The process of degradation of the N-mRuby3 fusion protein in A431 and A549 cells was monitored using flow cytometry with a CytoFLEX S instrument (Beckman Coulter) and a MACSQuant Analyzer (Miltenyi Biotec, France) in the fluorescence channel 564–606 nm; fluorescence was excited using a laser with a wavelength of 561 nm. Cells transiently expressing the N-mRuby3 protein were incubated with 500 nM MNT for a specified time, washed, removed from the substrate, and analyzed with the flow cytometer. Table 1 shows the fluorescence values of A431 and A549 cells at different times of incubation with MNT. Assuming that the decrease in the fluorescence intensity is caused by the degradation of the N-mRuby3 protein and knowing the concentrations of the N-protein in the cell lines studied, the dependence of the concentration of the N-protein that undergoes degradation on the duration of incubation with MNT can be calculated (Fig. 2). It was found that, for A431 and A549 cells, these dependences give the same initial rate of N-protein degradation, equal to 84 ± 7 nM per hour (Fig. 2). It should be noted that the actual rate of N-protein degradation may be higher, because the fluorescent protein mRuby3, similarly to other fluorescent proteins, may not be degraded completely [12]. This means that, even after degradation, residual mRuby3 fluorescence can be observed.
Thus, in this work, cell lines A431 and A549 with stable expression of the N-protein fused with the fluorescent protein mRuby3 were obtained. MNT added to these cells caused N-protein degradation. The initial rate of N-protein degradation in cells was determined and shown to be the same for A431 and A549 cells. The proposed MNTs can become the basis for a future antiviral drug, and the approach to quantitative analysis of the degradation process can be used to analyze the efficacy of a wide class of drugs aimed at suppressing the activity of various viruses.
REFERENCES
Surjit, M. and Lal, S.K., Infect. Genet. Evol., 2008, vol. 8, pp. 397–405.
Wu, C. and Zheng, M., Preprints, no. 2020020247, 2020.
Prajapat, M., Sarma, P., Shekhar, N., et al., Indian J. Pharmacol., 2020, vol. 52, p. 56.
Liao, H.-I., Olson, C.A., Hwang, S., et al., J. Biol. Chem., 2009, vol. 284, pp. 17512–17520.
Shipunova, V.O. and Deyev, S.M., Acta Nat., 2022, vol. 14, no. 1 (52), pp. 54–72.
Du, Y., Zhang, T., Meng, X., et al., Preprints, 2020. https://doi.org/10.21203/rs.3.rs-25828/v1
Khramtsov, Y.V., Ulasov, A.V., Lupanova, T.N., et al., Dokl. Biochem. Biophys., 2023, vol. 510, pp. 87–90.
Lu, M., Liu, T., Jiao, Q., et al., Eur. J. Med. Chem., 2018, vol. 146, pp. 251–259.
Fulcher, L.J., Hutchinson, L.D., Macartney, T.J., et al., Open Biol., 2017, vol. 7, p. 170066.
Slastnikova, T.A., Rosenkranz, A.A., Khramtsov, Y.V., et al., Drug Des. Dev. Ther., 2017, vol. 11, pp. 1315–1334.
Khramtsov, Y.V., Ulasov, A.V., Rosenkranz, A.A., et al., Pharmaceutics, 2023, vol. 15, p. 324.
Klionsky, D.J., Abdel-Aziz, A.K., Abdelfatah, S., et al., Autophagy, 2021, vol. 17, no. 1, pp. 1–382.
ACKNOWLEDGMENTS
The authors are grateful to E.S. Bunin for assistance in performing Western blot analysis and A.S. Saburov for help in isolating proteins. The experiments were performed using the equipment of the Core Facility of the Institute of Gene Biology, Russian Academy of Sciences.
Funding
The study was supported by the Russian Science Foundation (project no. 21-14-00130).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
CONFLICT OF INTEREST
The authors of this work declare that they have no conflicts of interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This work does not contain any studies involving human and animal subjects.
Additional information
Translated by M. Batrukova
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khramtsov, Y.V., Ulasov, A.V., Lupanova, T.N. et al. Quantitative Description of the N-Protein of the SARS-CoV-2 Virus Degradation in Cells Stably Expressing It under the Influence of New Modular Nanotransporters. Dokl Biochem Biophys 513 (Suppl 1), S63–S66 (2023). https://doi.org/10.1134/S1607672923700709
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1607672923700709