Abstract
The paper briefly describes the evolution of the key enzyme of photosynthesis, RuBisCO. Before the emergence of the reaction of carbon dioxide assimilation via photosynthesis, this protein was involved in the methionine metabolism chain. Possibly, for this reason, the carboxylation reaction catalyzed by enzyme proceeds very slowly. In addition to carboxylation, RuBisCO can simultaneously oxidize ribulose bisphosphate, a substrate to which the fixed CO2 is attached. This, in turn, also reduces the effectiveness of photosynthesis. In this regard, the literature discusses various options for increasing plant productivity by creating new forms of RuBisCO or fundamentally different pathways of carbon dioxide assimilation. In this work, we propose a modification of the carboxylation reaction that makes it possible to avoid photorespiration and thus increase the efficiency of photosynthesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
According to the chronological estimates, the first prokaryotes emerged on Earth approximately 4 billion years ago. For hundreds of millions of years, the metabolic systems of living organisms were formed on the basis of CO2 fixation due to the energy of chemosynthesis [1]. By the beginning of assimilation of light energy to convert CO2 into carbohydrates approximately 3 billion years ago, they had a wide network of metabolic pathways into which it should have been inserted. To implement the key CO2 fixation reaction, the enzyme catalyzing enolation of 2,3-diketo-5-methylthiopenthyl phosphate (DK-MTP-1-P) and possibly involved in the carbon dioxide reduction in methanogenic bacteria, was recruited [2, 3]. However, these functions of the enzyme were discovered later than the attachment of carbon dioxide to ribulose 1,5-bisphosphate (RBP) during photosynthesis. For this reason, the enzyme was termed ribulose-1,5-bisphosphate carboxylase/oxygenase (generally accepted abbreviation, RuBisCO). To date, several types of this enzyme and RuBisCO-like proteins (RLPs) were found [4–6]. The second name, oxygenase, is due to the fact that, in addition to the assimilation of carbon dioxide, the enzyme catalyzes substrate oxidation (i.e., the process reverse to CO2 assimilation), which significantly reduces plant productivity [7]. The carboxylation reaction rate is very low—from 2 to 8 acts of catalysis per second [8]. During more than 3 billion years of evolution of photosynthesis, a considerable apparatus organizing the functioning of RuBisCO was created [9]. Therefore, the required level of carbon dioxide fixation is maintained by the large number of molecules of this enzyme—up to 50% of the soluble protein in leaves. Since the plant productivity directly depends on the CO2 assimilation efficiency, attempts to increase it [10, 11] or even to develop a fundamentally different pathway of CO2 fixation [12] have been made. Much attention is given to the possible improvement of the kinetic properties of RuBisCO in performing the carboxylation reaction [8]. However, despite the wide front of research (to date, RuBisCO and RBP genes have been sequenced in more than 2000 organisms), we do not have a complete picture of functioning of this enzyme. The biggest problem is associated with the implementation of RBP oxidation [8].
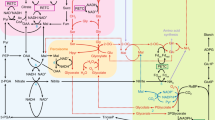
The sequence of reactions catalyzed by RuBisCO is shown in Fig. 1a. RBP carboxylation proceeds in five stages (tautomerization, carboxylation, hydration, bond cleavage, and stereospecific protonation (SP)). As a result, two molecules of 3-phosphoglyceric acid (3PGA) are produced, which can further be used both for the resynthesis of RBP and for the synthesis of fructose 6-phosphate and glucose at the stage of the Benson–Bassem–Calvin cycle. In the case of the oxygenase reaction, one molecule of 3PGA and one molecule of 2-phosphoglycolate (2PG) are produced. 3PGA can be used as that obtained in carboxylation, whereas 2PG requires special utilization as a toxic compound to chloroplasts [13]. It should be emphasized that, in living systems, the reactions that are catalyzed with the use of enolate and enamine-forming transitional forms are not always accompanied by oxidation of substrate. Therefore, photorespiration cannot be regarded as an inevitable consequence of the mechanism of tautomerization. The question arises as to why it is implemented in RuBisCO? Anything certain in this regard is still unknown. Perhaps, this is the result of a compromise between the chemical and metabolic imperatives, and only further research will shed light on this issue [8]. In general, the RuBisCO functioning mechanism is fairly sophisticated and comprises at least three intermediate and four transitional states with strict stereochemical constraints [14]. However, several important features of the catalytic process remain unclear: firstly, the sequence of proton dynamics; secondly, the origin of the oxygenase reaction [8]; and thirdly, the causes of the low carboxylation rate and the tendency of RuBisKO to multifunctionality [6, 8].
Scheme of reactions catalyzed by RuBisCO in photosynthetic organisms (a) and the RuBisCO-like proteins in Bacillus subtilis in the methionine metabolism chain (b). HK-MTP-1-P—2-hydroxy-3-keto-5-methylthiopenthyl 1-phosphate. Adapted from [13].
Note the remarkable fact that the double bonds formed during tautomerization under the control of RBP (evolutionarily older function of the future RuBisCO) in DK-MTP-1-P (B. subtilis) and under the control of RuBisCO in RBP during photosynthesis are oppositely directed (Figs. 1a, 1b). In the first case, the bond is formed between 1 and 2 carbon atoms; in the second, between 2 and 3 carbon atoms. It remains unclear why so different scenarios of enolization are realized. It should be noted that, in some live forms, the enzyme catalyzes reactions with both substrates (DK-MTP-1-P and RBP) [6, 13].
On this basis, we suggest to fundamentally change the mechanism of the RBP carboxylation reaction (Fig. 2). For this purpose, it is necessary to achieve the formation of a double bond between the 1 and 2 carbon atoms during RBP enolation, as it occurs in DK-MTP-1-P (B. subtilis). Further, it is necessary to perform CO2 attachment to the first carbon atom with the formation of 2,6-bis-phosphogluconic acid with its subsequent reduction to fructose or glucose. Thus, it is possible to avoid photorespiration and increase the yield of photosynthesis products. To assess the reality of the creation of this metabolic pathway, it is necessary to carry out quantum mechanical calculations, which should show the possibility of experimental solution of this problem. However, it cannot be ruled out that some simple photoautotrophs use just this pathway, since our knowledge about the mechanisms of photosynthesis is highly limited.
REFERENCES
Berg, I.A., Kockelkorn, D., Ramos, W.H., et al., Nat. Rev. Microbiol., 2010, vol. 8, pp. 447–460.
Ashida, H., Danchin, A., and Yokota, A., Res. Microbiol., 2005, vol. 156, nos. 5–6, pp. 611–618.
Ashida, H., Mizohata, E., and Yokota, A., Biochem. Soc. Trans., 2018, pp. 1–7.
Tabita, F.R., Satagopan, S., Hanson, T.E., et al., J. Exp. Bot., 2008, vol. 59, pp. 1515–1524.
Erb, T.J., Evans, B.S., Cho, K., et al., Nat. Chem. Biol., 2012, no. 8, pp. 926–932.
Dey, S., North, J.A., Sriram, J., et al., J. Biol. Chem., 2015, vol. 290, no. 52, pp. P. 30658–30668.
Ort, D.R., Merchant, S.S., Alric, J., et al., Proc. Natl. Acad. Sci. U. S. A., 2015, vol. 112, no. 28, pp. 8529–8536.
Bathellier, C., Tcherkez, G., Lorimer, G.H., and Farquhar, G.D., Plant, Cell Environ., 2018, vol. 41, pp. 705–716.
Bracher, A., Whitney, S.M., Hartl, F.U., and Hayer-Hartl, M., Review of Plant Biology, 2017, vol. 68, pp. 29–60.
Zhu, X.G., Long, S.P., and Ort, D.R., Annu. Rev. Plant Biol., 2010, vol. 61, pp. 235–261.
Weber, A.P.M. and Bar-Even, A., Plant Physiol., 2019, vol. 179, pp. 803–812.
Bar-Even, A., Noor, E., Lewis, N.E., and Milo, R., Proc. Natl. Acad. Sci. U. S. A., 2010, vol. 107, pp. 8889–8894.
Erb, T.J. and Zarzycki, J., Curr. Opin. Biotechnol., 2018, vol. 49, pp. 100–107.
Cleland, W.W., Andrews, T.J., Gutteridge, S., et al., Chem. Rev., 1998, vol. 98, pp. 549–562.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author declares that he has no conflict of interest. This article does not contain any studies involving animals or human participants performed by the author.
Additional information
Translated by M. Batrukova
Rights and permissions
About this article
Cite this article
Sokolov, V.A. On a Possible Way to Increase the Efficiency of Photosynthesis. Dokl Biochem Biophys 491, 98–100 (2020). https://doi.org/10.1134/S1607672920020131
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1607672920020131