Abstract
To assess the biodiversity of symbiotic dinoflagellates (SD) in hydrocorals, we compared the molecular species compositions of four SD lipid classes such as diacylglyceryl-3-O-carboxyhydroxymethylcholine (DGCC), digalactosyldiacylglycerol (DGDG), monogalactosyldiacylglycerol (MGDG), and sulfoquinovosyldiacylglycerol (SQDG) in Millepora dichotoma and M. platyphylla collected in shallow waters of Vietnam. A statistical analysis showed significant differences in the SD lipid composition between the hydrocoral species. These differences were influenced rather by the lipid molecules of outer membrane (DGCC and DGDG) than by the lipids of chloroplast and thylakoid membranes in SD (MGDG and SQDG). As for the fatty acid (FA) composition of lipid molecules that allow discrimination of hydrocoral SDs, the medium-chain FA 18:4 was characteristic of M. dichotoma, while the long- and very-long-chain FAs (20:5, 22:6, and 28:8) were characteristic of M. platyphylla. Thus, two different Millepora species hosted different SD groups, which had similar thylakoid lipidomes, different lipid profiles of outer membrane, and different activities in biosynthesis of n-3 polyunsaturated FAs. The lipidomic approach has shown that the M. dichotoma population is heterogenic, and about 10% of its colonies can be infected with SDs that are common for M. platyphylla.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Investigations of coral reefs focus mainly on scleractinian corals, whereas hydrocorals of the genus Millepora are relatively poorly studied but important components of the reef ecosystem. In shallow tropical seas, Millepora species form conspicuous colonies on coral reefs and may be locally abundant reef-framework builders [1]. The Millepora species are hydrozoans (Hydrozoa), belonging to the phylum Cnidaria together with scleractinians (Anthozoa), which produce skeleton composed of calcium carbonate. Similarly to scleractinians, Millepora hydrocorals obtain their food partly from autotrophic sources, endocellular symbiotic dinoflagellates (SD) referred to as zooxanthellae that carry out photosynthesis [2]. Seasonal elevations in sea water temperature can induce a loss of SDs by host animal; this phenomenon, termed as “coral bleaching”, has resulted in large-scale mortality of coral colonies to date [3]. Different SD species show different thermal resistance [4], and, therefore, data on composition of SD communities in hydrozoan coral species are crucially important for predicting coral mortality risk and effective management of shallow-water ecosystems.
Several SD genera such as Symbiodinium, Breviolum, and Cladocopium (formerly Symbiodinium of clade A, B, and C, respectively) have been identified in Millepora hydrocorals using molecular markers (ITS-rDNA) [2, 5, 6]. The differences in the pattern of fatty acids (FA), lipid classes, and lipid molecular species between SDs isolated from different coral species were described earlier [4, 7, 8]. We supposed that lipid analyses can help us to estimate the SD biodiversity in Millepora hydrocorals.
Photosynthetic organisms, including SDs, contain specific lipid classes such as betaine lipid, diacylglyceryl-3-O-carboxyhydroxymethylcholine (DGCC), and three glycolipids: digalactosyldiacylglycerol (DGDG), monogalactosyldiacylglycerol (MGDG), and sulfoquinovosyldiacylglycerol (SQDG) [9]. These lipid classes are not synthesized in hydrocoral animals’ tissues and can be recognized in total lipids of holobiont without isolation of pure SD fractions [10]. The chemical structures of the molecular species of DGCC, DGDG, MGDG, and SQDG have recently been identified in the Millepora hydrocorals [11]. To study the SD biodiversity of hydrocorals, we compared the lipid molecular species profiles of SDs between two hydrocoral species, Millepora dichotoma and M. platyphylla, collected from the same place in shallow waters of Vietnam. The effect of each lipid class and molecular species on the differences between SDs was examined.
MATERIALS AND METHODS
Colonies of the hydrocorals Millepora dichotoma Forskål, 1775 and M. platyphylla Hemprich & Ehrenberg, 1834 (Cnidaria: Hydrozoa: Anthoathecata: Milleporidae) (84 colonies of each species) were collected at a depth of 2–4 m from the same place of a reef in Nha Trang Bay, South China Sea (12°17′ N, 109°14′ E). To obtain total lipids, polyps were washed out of each colony with pressure water and suspended. Then the suspension of polyps was extracted with a mixture of chloroform and methanol as described previously [12]. Total lipids of each colony were evaporated under reduced pressure, dissolved in chloroform, and stored at –80°C.
Total lipids were separated by high-performance liquid chromatography, and lipid molecular species were identified by high resolution tandem (ion trap-time-of-flight) mass spectrometry both in positive and negative modes with electrospray ionization as described previously [13]. MGDG molecular species were eluted between 2.5 and 4 min; DGCC, between 9.5 and 13.5 min; SQDG, between 9.8 and 12.2 min; DGDG between 11 and 14 min. Identification of each molecular species was performed manually based on the MS/MS fragmentation of molecular ions as described previously [8, 14]. The total abundance of each lipid class and the relative abundance of certain lipid molecular species within each lipid class were calculated according to Rosset et al. [4].
Differences in the mean percentage of each lipid molecular species (n = 84) between M. dichotoma and M. platyphylla were estimated by one-way analysis of variance (ANOVA). Raw data were used after being tested for homogeneity of variances (Levene’s test) and normality of distribution (Shapiro–Wilk’s test). To represent differences between SDs of two hydrocoral species, the variables (lipid molecular species contents) were included in principal components analyses (PCA). All statistical analyses were performed using STATISTICA 5.1 (StatSoft, Inc., USA). A probability level of P < 0.05 was considered statistically significant. Values are presented as the mean ± standard error.
RESULTS
Four lipid classes (DGCC, DGDG, MGDG, and SQDG), which are specific for SD, were found in total lipids of M. dichotoma and M. platyphylla. The presence of 14 molecules of DGCC, 13 molecules of DGDG, 11 molecules of MGDG, and 22 molecules of SQDG in the hydrocoral lipidome was confirmed by high-resolution tandem mass spectrometry [11]. The major molecular species (>5% of each lipid class) of SDs from both hydrocorals are listed in Table 1. We found significant differences (one-way ANOVA, P < 0.05) in the mean percentages of all DGCC molecules, most of DGDG and MGDG molecules, and two SQDG molecules between M. dichotoma and M. platyphylla (Table 1). There were no significant differences (P > 0.05) in the mean contents of DGDG 18:4/18:5, MGDG 16:2/18:2, SQDG 14:0/16:0, and SQDG 16:0/16:0.
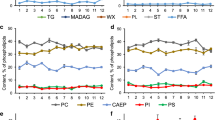
The effect of all molecular species of each lipid class on the SD diversity was assessed by PCA. The M. dichotoma and M. platyphylla specimens were well discriminated on the basis of all DGCC or DGDG molecular species (Figs. 1a, 1b). A particularly discrimination was observed, when all MGDG molecular species were used as variables (Fig. 1c). The two hydrocorals could not be discriminated on the basis of all SQDG molecular species (Fig. 1d).
Results of principal components analyses (PCA) using lipid molecular species composition (% of each lipid class) for the hydrocorals Millepora dichotoma (n = 84, black cycles) and M. platyphylla (n = 84, empty squares). Variables were the molecular species of (a) diacylglyceryl-3-O-carboxyhydroxymethylcholine (DGCC), (b) digalactosyldiacylglycerol (DGDG), (c) monogalactosyldiacylglycerol (MGDG), (d) sulfoquinovosyldiacylglycerol (SQDG), and (e) 22 selected lipid molecular species of all lipid classes (Table 1). The projection of 22 variables (f) are shown.
The major lipid molecules of SDs (Table 1) were used as variables for PCA to confirm the difference between SDs of the two hydrocorals. As shown in Fig. 1e, the M. dichotoma and M. platyphylla specimens were clearly separated into two regions of the two-dimensional space formed by PC 1 and PC 2, which together accounted for 43.68% of the total data variance. The separation of the M. dichotoma and M. platyphylla specimens was generated in PC 1, strongly influenced by DGCC, DGDG, and MGDG variables (Fig. 1f). The factor loading values on PC 1 were positive for DGCC 18:0/28:8 (0.79), DGDG 18:5/20:5 (0.70), DGDG 18:4/20:5 (0.73), DGDG 18:4/22:6 (0.74), DGDG 20:5/22:6 (0.56), and MGDG 18:4/18:5 (0.78) but negative for DGCC 16:0/18:4 (–0.82), DGCC 18:0/28:7 (–0.75), DGDG 18:4/18:4 (–0.78), and MGDG 18:4/18:4 (–0.84). Other variables generated the separation in PC 2 inside each region formed by the species (Figs. 1e, 1f). The factor loading values on PC 1 were consistent with our data shown in Table 1 and confirmed that SDs of M. dichotoma were reach in DGCC 16:0/18:4, DGCC 18:0/28:7, DGDG 18:4/18:4, and MGDG 18:4/18:4, but poor in DGCC 18:0/28:8, DGDG 18:5/20:5, DGDG 18:4/20:5, DGDG 18:4/22:6, and DGDG 20:5/22:6 vs. SDs of M. platyphylla.
The PCA results (Fig. 1f) showed that nine specimens of M. dichotoma got in the region of M. platyphylla. The percentages of some major molecular species of these unusual M. dichotoma specimens were similar to those of M. platyphylla specimens (Table 1). For example, nine M. dichotoma specimens contained 5.2% of DGCC 18:0/28:8, 3.4% of DGDG 18:5/20:5, 7.8% of DGDG 18:4/20:5, and 13.7% of DGDG 18:4/22:6. In the other 75 specimens of M. dichotoma, each of these four molecules constituted less than 1% of the corresponding lipid class.
DISCUSSION
The fatty acid and lipid class composition of marine and fresh-water dinoflagellates have been studied for more than 60 years [15, 16], but first studies on dinoflagellate lipidomes (chemical structures and contents of lipid molecular species) started only in 2003 [17]. Large data arrays on lipidomes of free-living and cultured dinoflagellates have been obtained [18], whereas information on the lipidome of symbiotic dinoflagellates (SD) living inside cells of marine cnidarians remains very limited [8, 19]. Until recently, all SDs from different cnidarian hosts were considered to belong to a single genus, Symbiodinium, which was then divided into several “clades” by a genetic analysis. In 2018, LaJeunesse et al. [5] suggested that the Symbiodinium clades are equivalent to the corresponding genera belonging to the family Symbiodiniaceae. The lipid composition is known to be genus-specific, and the differences in lipidome between two SD genera of reef-building corals have already been documented [4].
In our work, we found significant differences in the lipid molecular species profiles of SDs between two Millepora hydrocorals, which suggests that each hydrocoral species contains the species-specific SD group (most probably, a SD genus). The differences between two SD groups were mainly in DGCC and DGDG, whereas the effect of SQDG was insignificant. The plant lipid classes are non-uniformly distributed between lipid matrices of different biomembranes of photosynthetic cells. There are three membrane types in SD cells: outer membranes, membranes of chloroplast located inside SD cells, and membranes of thylakoids located in chloroplasts. The betaine lipid DGCC is synthesized in the endoplasmic reticulum but not in chloroplasts. DGCC is the predominant non plastidial polar lipid in dinoflagellates [20]. Glycolipids mainly perform a function of structural lipids of chloroplasts. MGDG are located exclusively in chloroplast membranes, but DGDG can also be found in extra-plastidic membranes. The proportion of SQDG in the thylakoid membranes is much higher. Hence, the major differences between the two SD groups of Millepora are determined rather by the extra-plastidial lipids (DGCC and DGDG) than by the lipids of plastid and thylakoid membranes (MGDG and SQDG).
The key lipid molecular species, which are responsible for the SD discrimination observed, are acylated by several specific polyunsaturated fatty acids (PUFA) of dinoflagellates such as, primarily, 18:4 and 18:5 [8]. The ratio 18:4/18:5 in the total FAs of M. dichotoma and M. platyphylla was 8.2 and 2.9, respectively. In contrast to M. platyphylla, the total FAs of M. dichotoma were rich in 18:4n-3 [11] and contained more lipid molecular species with 18:4n-3. The SD lipids contained very-long-chain FAs such as 28:7n-6 and 28:8n-3 [21]. The latter FA (28:8) can be synthesized from medium-chain acid 18:4n-3 via long-chain FAs 20:5n-3 and 22:6n-3. In the SDs of M. platyphylla, this biosynthetic pathway can decrease the level of 18:4 and increase the level of 28:8 and its intermediates. Therefore, most SD lipid molecules in M. dichotoma are built on 18:4, while the majority of SD lipid molecules in M. platyphylla are composed of long- and very-long-chain FAs. The different activity levels in the synthesis of n-3 PUFAs seem to explain the different lipidomes in the two SD groups.
It was surprising that the SD lipidome of some of M. dichotoma colonies (up to 10% of total FAs) was similar to that of M. platyphylla. We suppose that the population of M. dichotoma is heterogenic and some of M. dichotoma colonies can be infected with SDs that are common for M. platyphylla. The presence of different coral species with the same symbionts are very important for comparative studies of the role of host organism in the thermal tolerance of a coral holobiont under bleaching events.
Thus, the lipidomic approach has shown that different Millepora species host different SD groups, which have similar thylakoid lipidomes, different lipid profiles of outer membranes, and different biosynthetic activities of n-3 PUFAs. Obviously, further study should include analyses of genetic identity to determine the taxonomic position of hydrocoral SDs.
REFERENCES
Lewis, J.B., Biology and ecology of the hydrocoral Millepora on coral reefs, Adv. Mar. Biol., 2006, vol. 50, pp. 1–55.
Rodríguez, L., Lopez, C., Casado-Amezua, P., et al., Genetic relationships of the hydrocoral Millepora alcicornis and its symbionts within and between locations across the Atlantic, Coral Reefs, 2019, vol. 38, pp. 255–268. https://doi.org/10.1007/s00338-019-01772-1
Anthony, K.R.N., Connolly, S.R., and Hoegh-Guldberg, O., Bleaching, energetics, and coral mortality risk: Effects of temperature, light, and sediment regime, Limnol. Oceanog., 2007, vol. 52, pp. 716–726.
Rosset, S., Koster, G., Brandsma, J., et al., Lipidome analysis of Symbiodiniaceae reveals possible mechanisms of heat stress tolerance in reef coral symbionts, Coral Reefs, 2019, vol. 38, pp. 1241–1253. https://doi.org/10.1007/s00338-019-01865-x
LaJeunesse, T.C., Parkinson, J.E., Gabrielson, P.W., et al., Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts, Curr. Biol., 2018, vol. 28, pp. 2570–2580. https://doi.org/10.1016/j.cub.2018.07.008
Loh, W., Hidaka, M., Hirose, M., and Titlyanov, E.A., Genotypic diversity of symbiotic dinoflagellates associated with hermatypic coral from a fringing reef at Sesoko Island, Okinawa, Galaxea, 2002, vol. 4, pp. 1–9.
Tchernov, D., Gorbunov, M.Y., de Vargas, C., et al., Membrane lipids of symbiotic algae are diagnostic of sensitivity to thermal bleaching in corals, Proc. Nat. Acad. Sci. U.S.A., 2004, vol. 101, pp. 13531–13535. https://doi.org/10.1073/pnas.0402907101
Imbs, A.B., Rybin, V.G., Kharlamenko, V.I., et al., Polyunsaturated molecular species of galactolipids: Markers of zooxanthellae in a symbiotic association of the soft coral Capnella sp. (Anthozoa: Alcyonacea), Russ. J. Mar. Biol., 2015, vol. 41, pp. 461–467. https://doi.org/10.1134/S1063074015060048
Awai, K., Matsuoka, R., and Shioi, Y., Lipid and fatty acid compositions of Symbiodinium strains, Proc. 12th Inter. Coral Reef Symp., Cairns, 2012.
Sikorskaya, T.V., Ermolenko, E.V., and Imbs, A.B., Effect of experimental thermal stress on lipidomes of the soft coral Sinularia sp. and its symbiotic dinoflagellates, J. Exp. Mar. Biol. Ecol., 2020, vol. 524, artic. 151295, pp. 1–11. https://doi.org/10.1016/j.jembe.2019.151295.
Imbs, A.B., Ermolenko, E.V., Grigorchuk, V.P., and Dang, L.P.T., Seasonal variation in the lipidome of two species of Millepora hydrocorals from Vietnam coastal waters (the South China Sea), Coral Reefs, 2021, vol. 40, pp. 719–734. https://doi.org/10.1007/s00338-021-02073-2
Folch, J.F., Lees, M., and Sloane Stanley, G.H., A simple method for the isolation and purification of total lipids from animal tissue, J. Biol. Chem., 1957, vol. 226, pp. 497–509.
Imbs, A.B., Dang, L.P.T., and Nguyen, K.B., Comparative lipidomic analysis of phospholipids of hydrocorals and corals from tropical and cold-water regions, PLOS One, 2019, vol. 14, artic. e0215759, pp. 1–22. https://doi.org/10.1371/journal.pone.0215759.
Sikorskaya, T.V., Efimova, K.V., and Imbs, A.B., Lipidomes of phylogenetically different symbiotic dinoflagellates of corals, Phytochem., 2021, vol. 181, artic. 112579, pp. 1–9. https://doi.org/10.1016/j.phytochem.2020.112579.
Joseph, J.D., Identification of 3,6,9,12,15-octadecapentaenoic acid in laboratory-cultured photosynthetic dinoflagellates, Lipids, 1975, vol. 10, pp. 395–403.
Leblond, J.D. and Chapman, P.J., Lipid class distribution of highly unsaturated long chain fatty acids in marine dinoflagellates, J. Phycol., 2000, vol. 36, pp. 1103–1108. https://doi.org/10.1046/j.1529-8817.2000.00018.x
Gray, C.G., Lasiter, A.D., Li, C., and Leblond, J.D., Mono- and digalactosyldiacylglycerol composition of dinoflagellates. I. Peridinin-containing taxa, Eur. J. Phycol., 2009, vol. 44, pp. 191–197. https://doi.org/10.1080/09670260802419481
Leblond, J.D., McDaniel, S.L., Lowrie, S.D., et al., Mono- and digalactosyldiacylglycerol composition of dinoflagellates. VIII. Temperature effects and a perspective on the curious case of Karenia mikimotoi as a producer of the unusual, ‘green algal’ fatty acid hexadecatetraenoic acid 16:4(n-3), Eur. J. Phycol., 2019, vol. 54, pp. 78–90. https://doi.org/10.1080/09670262.2018.1519602
Garrett, T.A., Schmeitzel, J.L., Klein, J.A., et al., Comparative lipid profiling of the cnidarian Aiptasia pallida and its dinoflagellate symbiont, PLoS One, 2013, vol. 8, artic. e57975, рр. 1–17. https://doi.org/10.1371/journal.pone.0057975.
Leblond, J.D., Khadka, M., Duong, L., and Dahmen, J.L., Squishy lipids: Temperature effects on the betaine and galactolipid profiles of a C-18/C-18 peridinin-containing dinoflagellate, Symbiodinium microadriaticum (Dinophyceae), isolated from the mangrove jellyfish, Cassiopea xamachana, Phycol. Res., 2015, vol. 63, pp. 219–230. https://doi.org/10.1111/pre.12093
Rezanka, T., Lukavsky, J., Nedbalova, L., and Sigler, K., Lipidomic profile in three species of dinoflagellates (Amphidinium carterae, Cystodinium sp., and Peridinium aciculiferum) containing very long chain polyunsaturated fatty acids, Phytochem., 2017, vol. 139, pp. 88–97. https://doi.org/10.1016/j.phytochem.2017.04.912
ACKNOWLEDGMENTS
This work was supported by the Russian Foundation for Basic Research (grant no. 21-54-54002) and the Vietnam Academy of Science and Technology (grants no. QTRU01.10/21-22).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflict of interest.
Statement on the welfare of animals. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Imbs, A.B., Ermolenko, E.V., Grigorchuk, V.P. et al. A Lipidomic Approach to the Study of Biodiversity of Symbiotic Dinoflagellates in Millepora Hydrocorals from Vietnam Coral Reefs. Russ J Mar Biol 47, 312–317 (2021). https://doi.org/10.1134/S1063074021040064
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1063074021040064