Abstract—The current stage of genetic certification of horses of cultural and local breeds based on microsatellite analysis makes it possible to quite effectively carry out identification and genetic examination of the origin of breeding animals, as well as solve the problem of assessing and preserving genetic resources. With a reduction in the number of breeding stock to 200–300 mares observed in a number of breeds, the threat of a decrease in the genetic diversity of populations and the accumulation of genetic load increases, which necessitates the need to study and monitor the genetic structure of horse breeds. In this regard, our comparative genetic analysis of polymorphism of 17 microsatellite loci in 20 541 horses of 29 cultural and local breeds allows us to certify the basic part of the genetic resources of the horse breeding of the Russian Federation. including riding, trotter, draft, and local breeds. During the genetic population analysis of the studied breeds, basic parameters were assessed: the total number of allele variants (Na), the effective number of alleles (Ae), the average number of alleles per locus (MNA), the level of observed (Ho) and expected heterozygosity (He), as well as the coefficient of intrapopulation inbreeding Fis. Phylogenetic relationships of breeds were assessed using the R and R Studio software packages. Among horse breeds of different specializations, the highest values of all indicators of genetic diversity (Ae, Ho, He, and Na) were determined in aboriginal populations. In the allele pool of local horse breeds, there were rare alleles ASB2T, HMS7S, HMS6J, HMS6H, HMS2T, HMS1O, HTG7L, HTG6L, HTG6H, VHL20S, ASB17Z, ASB17X, ASB17U, LEX3S, LEX3R, and CA425E, which were absent in horses of stud breeds. Among the riding horse breeds created in Russia, the Budennovskaya, Donskaya, and Kabardian horse breeds stood out due to the presence of rare alleles. Alleles ASB2G, ASB2F, HMS2F, HTG7Q, and ASB23O were found in trotter horses, which were not identified in the genetic structure of other breeds. The phylogenetic analysis showed the division of horse breeds into two clear subclusters, the first of which included only stud breeds. The second cluster united all the native breeds, as well as the Orlov Trotter and a group of draft breeds, which were used for many years as improvers of the local horse population. The analysis of the genetic structure of domestic horse breeds revealed a fairly high reserve of diversity even in small populations, which is an indispensable condition for successful selection in horse breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
In past centuries, the development of horse breeding largely determined the volume of agricultural production and was of particular importance for Russian livestock farming. The horse was not only a symbol of the power and prestige of the country, but also saved people in difficult times of history. However, with the development of mechanization in agriculture, the army, and industry, the approach to the use of horses changed. By the middle of the 20th century. the number of horses declined sharply, but then stabilized and even began to grow in many countries. Now horses play an important role in tourism, sports, and the hippodrome business, as well as in food production (milk, meat, kumiss, etc.). Horses of factory breeds are distinguished by the highest cost in the modern world market of breeding resources of farm animals. Therefore, a key condition for effective selection of breeding animals is the undeniable authenticity of the origin of each animal that is involved in the reproduction process. Objective and accurate identification of horses and certification of livestock are very urgent tasks. To solve this problem, molecular genetic methods are used, which in turn make it possible to in-depth study the identity and polymorphism of the genetic structures of populations in order to preserve them and effectively manage the selection process in all areas of improving horse breeds [1–6].
Currently, DNA technologies are widely used in diverse genetic and genomic studies, in monitoring the origin of animals, to study phylogenetic relationships and microevolution of breeds, as well as to improve the genotypic assessment of animals at the individual and population levels [5–13]. As genetic markers, microsatellites are interesting because they are subject to a higher level of mutation than the rest of the genome [1, 4, 14–16].
The register of breeding achievements of the Russian Federation includes 44 breeds of horses, which are unique in their versatility in use and high adaptive qualities and are a valuable genetic resource for the world horse breeding. Already the first studies on the genetic structure of factory and local horse breeds of domestic selection using microsatellite markers [6, 9, 17] showed a high level of allelic variability in the studied populations. Subsequent studies confirmed the presence of genetic specificity in many breeds of horses, especially those with a limited breeding area [1–3, 18–20].
The goal of our research is a comparative assessment of the genetic diversity of microsatellite loci in the genomes of horses of 29 breeds and populations bred in the Russian Federation, as well as the study of their phylogenetic relationships.
MATERIALS AND METHODS
The material for the research was biosamples of hair, blood, and sperm from 20 541 horses of 29 factory and local breeds bred on the territory of the Russian Federation. The studies included horses of the following riding breeds: Akhal-Teke (n = 1040), Budennovskaya (n = 93), Arabian (n = 2971), Donskaya (n = 21), Hanoverian (n = 33), Trakehner (n = 93), Kabardian (n = 289), Thoroughbred (n = 9600), four trotting breeds: Orlov Trotter (n = 4177), French Trotter (n = 381), Russian Trotter (n = 975), American standardbred (n = 434) and four heavy draft breeds: Russian Heavy Draft (n = 71), Vladimirskaya (n = 233), Soviet Heavy Draft (n = 51), Percheron (n = 57), as well as Shetland Pony (n = 46). Local breeds and populations were represented by Khakasskaya (n = 25) and Yakutskaya (n = 24), Buryatskaya (n = 20), Bashkirskaya (n = 100), Zabaikalskaya (n = 24), Vyatskaya (n = 219), Mezenskaya (n = 97), Priobskaya (n = 25), Pechorskaya (n = 17), Novoaltaiskaya (n = 150), Mugalzhar (n = 94), Tuvinian (n = 569), and Altaiskaya horses (n = 39).
Testing of the entire horse population for 17 microsatellite DNA loci: VHL20, AHT4, HMS2, HMS3, HMS1, AHT5, HTG7, HTG6, HTG4, HTG10, HMS7, HMS6, ASB23, ASB2, ASB17, LEX3 and CA425 was carried out in the genetics laboratory of the All-Russian Research Institute of Horse Breeding, using protocols and equipment recommended by the International Society for the Study of Animal Genetics (ISAG). Research period 2009–2021.
Isolation of DNA from different types of biomaterial (hair, blood, sperm, etc.) was carried out using ExtraGene DNA Prep 200 reagents (Isogen Laboratory LLC, Russia). Amplification of the resulting DNA was carried out using 17 complex primer sets for horse genotyping StockMarks® for Horses manufactured in the United States and COrDIS S550 (GORDIZ LLC, Russia) on a Termocycler 2730 DNA amplifier (Germany). Separation and detection of amplification products was carried out by capillary electrophoresis on an automatic 4-capillary genetic analyzer AB 3130 (United States). The sizes of amplified DNA fragments were determined using the Ge-neMapper™ V4.0 program. To designate alleles, we used the international nomenclature adopted by ISAG when conducting comparative tests on equine DNA testing (HCT), in which our laboratory regularly took part in obtaining I degree certificates.
When assessing population genetic parameters, we took into account the frequencies of genotypes, alleles, and the level of polymorphism (Ae), as well as the degree of expected (Ne) and observed (No) heterozygosity, which were calculated using the algorithm of PLINK 1.9 programs [21], MS Excel 2010. Assessment of genetic similarity and genetic distances was carried out using the Statistics 12 program (https://statsoft-statistica.ru/). To calculate genetic distances and construct a phylogenetic tree, we used the R program package and R Studio 1.3.1093 [22].
RESULTS
In the course of studies of the variability of 17 DNA microsatellite loci in horses of eight riding breeds, 169 alleles were discovered, with variations from 4 to 13. The richest spectrum of alleles was recorded in horses of the Kabardian and Akhal-Teke breeds, each of which had 147 and 126 alleles, respectively, while only 96 alleles were tested in horses of Donskaya breed. The tested Arabian horses had 104 alleles at 17 STR loci, including CA425F and CA425P, which were unique to this breed and were not found among representatives of other riding breeds, while ASB2C was found only in Arabian and Kabardian horses.
In horses of the ancient Akhal-Teke breed, 126 alleles were identified, the most typical of which are HMS1M, 0.581; AHT4H, 0.544; ASB23J, 0.427; HMS7J, 0.461; HMS6O, 0.489; HTG7O, 0.665; HTG6G, 0.526; HTG10O, 0.503, and CA425N, 0.499. Unique alleles were discovered in the genotypes of horses of the Kabardian and Akhal-Teke breeds ASB2J and HTG4Q unlike other riding breeds. HTG10Q was found in horses of the Budennovskaya, Akhal-Teke and Kabardian breeds, and the allele ASB23R, only for Don, Akhal-Teke, and Kabardian horses.
In the genotypes of Thoroughbred horses, the most common alleles were AHT4O, AHT5K, HMS1J, HMS2L, HMS3I, HMS6P, HTG4K, HTG4M, HTG6J, HTG7O, and CA425N, while the average number of alleles per locus was 5.88, with fluctuations from 4 to 9.
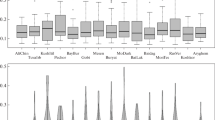
Figure 1 shows the variability and frequencies of alleles at the locus AHT4, demonstrating significant differences between horse breeds.
When analyzing the polymorphism of the locus AHT4 in horses of riding breeds it was found that a widespread allele AHT4H is typical for all breeds, allele AHT4J is characteristic of the Arabian and Akhal-Teke, and the allele AHT4O for purebred riding and other half-bred breeds.
Horses of the Hanoverian and Trakehner breeds with a total number of 100 and 105 alleles, respectively, had a relatively low level of genetic diversity. The genetic similarity of the Trakehner breed with purebred riding horses reached 0.922, which is of course natural, since stallions of the purebred riding breed have always been used in breeding the Trakehner horse.
One of the most informative panel STR loci is LEX3, located on the X chromosome, as it characterizes the diversity and connections of populations along the maternal line. A study of this locus in riding breeds showed the presence of 11 alleles out of 14 registered ISAGs. In this case, the maximum allele frequency LEX3M was established in the Arabian breed, 0.366; LEX3P, in Thoroughbred, 0.330; and LEX3K, 0.108, in horses of the Kabardian breed. Rare alleles were discovered in Kabardian horses LEX3Q and LEX3G, which suggests the presence of original female lines in this population (Fig. 2).
In horses of domestic breeds, the entire range of standard alleles registered by the International Society of Animal Genetics (ISAG) was determined. At loci ASB2, ASB17, CA425, HMS2, HMS6, HMS7, HMS1O, HTG6, HTG7, LEX3, and VHL20 16 new alleles not found in Western European horses were identified (Table 1).
Additional alleles were found in horses: ASB2T, Novoaltaiskaya; ASB17U, Bashkirskaya, Tuvinian; ASB17X, Mezenskaya, Tuvinian; ASB17Z, Tuvinian; HMS1O, Altaiskaya; HMS2T, Altaiskaya; HMS6, Tuvinian; HMS6J, American Trotter, Mezenskaya; HMS7S, Priobskaya; HTG6H, Buryatskaya; HTG6L, Buryatskaya, Vyatskaya, Tuvinian; HTG7L, Mugalzhar; LEX3R, French Trotter, Mezenskaya; LEX3S, Mezenskaya; and VHL20S, Novoaltaiskaya, Mugalzhar, Tuvinian.
An analysis of the genetic and population parameters of riding horse breeds indicates that the highest level of genetic diversity belongs to the Kabardian breed, while the Arabian horses had a low level of polymorphism, 3.18. The highest value of the degree of actual heterozygosity Ho, 0.727 was observed in the Trakehner breed, while its minimum value, 0.631 was found in horses of the Arabian breed (Table 2).
The obtained data on genetic similarity between riding horse breeds indicate the close similarity (0.961) between Budyonnovskaya and Thoroughbred breeds, while the relationship between the Budyonnovskaya and Akhal-Teke breeds was minimal (0.732). Analysis of genetic distances also confirmed these results, showing the greatest difference between Akhal-Teke and Thoroughbred breeds (0.495), and the smallest between Budennovskaya and Thoroughbred breeds (0.194) (Fig. 3a). The data we obtained allow us to better understand the genetic nature of various horse breeds and contribute to their more efficient breeding.
Genetic identification and clustering of horses of different breeds according to Nei [24].
When conducting a comparative analysis of polymorphism of 17 STR loci of four trotting breeds, 157 alleles were discovered, while the highest number of alleles belonged to the Russian Trotter breed with a wide range of alleles.
Orlov Trotters demonstrated the presence of 128 alleles with variants from four alleles at the locus HMS3 and up to 12 at locus ASB17. The genetic structure of this trotting breed showed a high frequency of occurrence of alleles HMS6P, 0.476; HMS3O, 0.553; HMS1M, 0.577; HTG7O, 0.553; HTG6O, 0.700; and HTG4M, 0.572. However, rare allelic variants HTG7I, AHT5P, and CA425G noted only in horses of Orlov Trotter breed.
Rare alleles were discovered in standard delirious breed trotters ASB2J, ASB2F, HMS6J, HMS2F, HTG7Q, HTG7P, and ASB23O, which were not found in horses of other trotting breeds.
In the French Trotter, alleles had a high frequency of occurrence AHT4O, 0.571; HMS3P, 0.514; HMS1J, 0.507; HTG10I, 0.536; HTG7O, 0.627; HTG6J, 0.629 and LEX3M, 0.358 and only in horses of this trotting breed were alleles found ASB17U, ASB17V, ASB2G, HMS2N, HTG6L, and LEX3R.
Among the trotting breeds of horses, the French Trotter stood out for its maximum levels of polymorphism Ae (3.81) and degree of heterozygosity He (0.72), while standardbred horses had the lowest level of genetic diversity (Ae (3.457), Ho (0.663), and He (0.679) with a positive value of Fis (0.016).
The greatest genetic differences and the lowest coefficient of genetic similarity (0.577) were noted between Orlov and Standardbred trotters. Russian Trotter, which has been constantly improved by Standardbred, had a very high coefficient of genetic similarity (0.978) with this American breed.
Based on the characteristics of horses of four trotting breeds based on STR loci, we constructed a dendrogram of their phylogenetic relationships (Fig. 3b), which demonstrated that Orlov Trotter is the most genetically isolated and forms a separate branch. Horses of prize breeds (Russian Trotter, French and Standardbred) form a common cluster, which confirms their close genetic connection.
When testing horses of four draft breeds at microsatellite DNA loci, a fairly wide range of alleles was identified (143). The greatest variability of alleles was confirmed in horses of Russian and Soviet Draft breeds (Table 2). The number of alleles in the studied STR loci varied from 4 to 12, with an average value of 6.41–7.12 per locus. The maximum number of alleles was identified at the locus ASB17 (9–12), the least variability was noted in the loci HTG6 (4–6), HTG7 (4) and HMS1 (4–6).
In horses of Vladimirskaya breed, 112 alleles were identified, two of which, HMS1Q and HTG4Q, were not found in other heavy draft breeds. The genetic structure of Vladimirskaya horse breed showed a relatively high frequency of occurrence of alleles HMS1M (0.699), HTG10R (0.488), HTG7O (0.529), HTG6O (0.433), AHT4L (0.507), and LEX3L (0.588).
Alleles have been identified in horses of Russian Draft breed AHT4M, HMS7G, HMS2O, HMS6Q, HMS7P, and HMS2P, which are absent in the allele pool of other breeds of this group. A feature for these medium-sized horses was the maximum frequency of occurrence of alleles, such as HMS3P, HMS1M, HTG10M, HTG7O, HTG6O, HTG4M, and CA425N.
In the gene pool of horses of Soviet Draft breed, two special rare alleles were discovered, HMS1I and HMS6N. Analysis of the genetic structure of this breed revealed a high concentration of alleles HMS2H, 0.406; HMS1M, 0.422; HTG10M, 0.422; HTG6O, 0.833; HTG4M, 0.559; ASB17M, 0.400; and CA425N, 0.500.
In the genotypes of horses of Percheron breed, mainly represented by animals brought from France, 109 alleles were discovered, among which VHL20K, ASB23Н, and ASB23Q were absent in horses of domestic breeds.
It is interesting to note that horses of Vladimirskaya and Soviet Draft breeds had the largest number of alleles at the locus LEX3. Overall, the four draft breeds differed significantly in allelic structure at this locus. Horses of Vladimirskaya breed had a high frequency of occurrence of alleles LEX3H, 0.203 and LEX3L, 0.588, while for Soviet and Russian Heavy Draft trucks the dominance of ancestors with LEX3Р on the X chromosome (0.284 and 0.310, respectively).
In terms of genetic diversity (Ae 3.982 and Ho 0.723) among heavy draft horses, Soviet Draft breed had the highest level (Table 2). Even in conditions of reduction in the number of breeding queens in this breed to 200 head, the population retains a fairly high genetic resource and heterozygosity, which confirms the negative value of the coefficient Fis (–0.002). Horses of Russian Draft breed had a low degree of Ho (0.676) in combination with intrapopulation inbreeding (Fis, 0.036).
The highest coefficient of genetic similarity (0.941) was found between Russian and Soviet Draft horses. At the same time, the relationship between Vladimirskaya and Soviet Heavy Draft breeds turned out to be minimal (0.680). The tree of the phylogenetic relationship (Fig. 3c) clearly demonstrated the similarity between the studied draft breeds, which are divided into two branches of continental (Percheron and Belgian Drafts) and English breeds (Clydesdale and Shire).
The results of the analysis of horses of 14 aboriginal breeds revealed the widest range of alleles of 17 panel (n = 195), including a number of unique alleles (Table 1). The largest number of allelic variants was identified in the Tuvinian horse (n = 170), and the smallest occurs for Buryatskaya breed (n = 117). Local horse breeds differed in basic population parameters, including Na, Ae, Ho, He and Fis, as well as the presence of unique alleles not included in the standardized nomenclature (Table 1), including HMS7S, HMS6J, HMS6H, HMS2T, HMS1O, ASB2T, HTG7L, HTG6L, HTG6H, ASB17Z, ASB17X, ASB17U, VHL20S, LEX3S, LEX3R, and CA425E.
The allele pool of horses of Mezenskaya breed, which is bred in the Arkhangelsk region, included five unique alleles, HMS6J, ASB17Y, ASB17X, LEX3S, and LEX3R. The alleles in this breed of horses had the highest concentration HMS7L, 0.670; HMS3M, 0.426; AHT4O, 0.407; HTG7O, 0.407; HTG7K, 0.418; HTG6O, 0.789; HTG4M, 0.443 and LEX3M, 0.528.
Vyatskaya horse breed was distinguished by the presence of two unique alleles HTG6L and AHT5P; typical for this breed were HMS7L (0.449), HMS2H (0.448), HMS1M (0.481), AHT5J (0.412), HTG7 (0.548), HTG6O (0.708), and HTG4M (0.680).
In the genetic structure of the Bashkir horses, there is a high frequency of occurrence of certain alleles (HTG10O, 0.439; HTG6O, 0.510; HTG4M, 0.590, and HMS7L, 0.490) against the background of rare alleles ASB17S and ASB17U.
Tuvinian horse breed, bred in Siberia, was distinguished by a high level of genetic diversity (Na, 170; Ae, 5.197; Ho, 0.782). The genetic structure of this breed has a high concentration of alleles HTG6O (0.545), HTG4M (0.630), and HMS7L (0.419) and the presence of very rare alleles HMS6H, HMS3L, VHL20K, ASB23N, ASB17Z, and LEX3J.
The LEX3 locus polymorphism in horses of local breeds has 12 alleles, with variants LEX3F, LEX3L, and LEX3M were identified in all studied populations. LEX3G occasionally found in horses of Western and Southern Siberia (Novoaltaiskaya and Tuvinian). The Altai horses were distinguished by the presence of rare alleles LEX3J and LEX3I and only in the genotypes of Mezenskaya horses were rare alleles identified LEX3S (0.042) and LEX3R (0.014), which are absent in representatives of other breeds.
Indicators of the level of polymorphism and degree of heterozygosity of aboriginal populations as a whole were slightly higher than those of factory horse breeds, with negative values Fis, which indicates a genetic balance of heterozygotes in the studied populations (Table 2).
Relatively high coefficients of genetic similarity were found between horses of steppe breeds: Bashkirskaya and Tuvinian (0.945), Khakasskaya and Bashkirskaya (0.915) and Zabaikalskaya and Bashkirskaya (0.903) breeds. On the dendrogram reflecting the genetic distances between native horse breeds (Fig. 3d) it has been demonstrated that horses of Khakasskaya, Zabaikalskaya, Tuvinian, and Bashkirskaya breeds are combined into a separate cluster of steppe breeds. The most distant branch was the Shetland ponies of foreign origin.
Phylogenetic analysis of 29 horse breeds of different specializations showed the presence of two clear subclusters (Fig. 4). The first cluster united horses of factory breeds, while the second included all aboriginal populations, as well as Orlov Trotter and draft breeds, which were used as improvers of the local horse population for a long time.
DISCUSSION
Our country has unique genetic resources for horse breeding, the study of the biological characteristics of which is the basis for programs for their conservation and improvement. As a result of studies of the molecular genetic characteristics of 29 horse breeds of different specializations, we identified a fairly high level of polymorphism of microsatellite loci in almost all studied populations (Ae = 3.2–5.2) and their high information content as genetic markers is shown.
When analyzing the polymorphism of 17 microsatellite loci in horses of domestic breeds, all standard alleles that are found in European horses were found [23, 25]. In addition, new alleles were identified that could have been preserved in the centers of domestication of ancient horses in the territory, and also appeared as a result of genomic mutations or the introduction of genes with horses of nomads from different regions of Asia. Additional confirmation of this hypothesis is the study of the genetic characteristics of local Chinese horses, in which a wide range of alleles at STR loci were identified, as well as the presence of common mtDNA haplogroups of eastern origin.
A comparative analysis of the genetic structure of horses of different specializations revealed significant differences in parameters: the total number of allele variants, the effective number of alleles (Ae) and the number of alleles per locus. According to the average indicators of all studied populations, the highest values of Ae, Ho, He, and Na were identified in horses of native breeds. Among the riding breeds, Kabardian horses stood out most due to the presence of alleles AHT4R, AHG4Q, LEX3Q, and CA425P, which were found only in horses of local breeds. This may be due to the fact that this breed, which are bred in the foothills of the North Caucasus, was formed with the participation of horses of nomadic tribes and improved by Persian, Turkmen, and Karabakh horses (the Circassian horse).
A feature of the allele pool of trotting breeds are the alleles ASB2G, ASB2F, HMS2F, HTG7Q, and ASB23O, which are not identified in other horses. The high coefficient of genetic similarity between Russian and Standardbred Trotters (0.978) clearly reflects the results of the breeding strategy to increase the blood quality and agility of prize breeds. The differences between the American Standardbred and Orlov Trotter breeds in terms of genetic markers are due to both the allele pool of the original breeds used in the formation process and the direction of breeding work. In the Orlov Trotting breed, a high level of genetic diversity was preserved due to the fact that selection was systematically carried out in the breed for a set of economically useful traits. In addition to the high degree of genetic variability, a characteristic feature of local breeds was the presence of a number of unique alleles ASB2T, HMS7S, HMS6J, HMS6H, HMS2T, HMS1O, HTG7L, HTG6L, HTG6H, VHL20S, ASB17Z, ASB17X, ASB17U, LEX3S, LEX3R, and CA425E, which were not found in horses of domestic breeds and in the studied European populations [23].
Unique alleles have been identified in horses of a number of breeds, including Altaiskaya, HMS2T; Bashkirskaya, ASB17U; Buryatskaya, HTG6H; HTG6L; Vyatskaya, HTG6L and AHT5P; Mezenskaya ASB17X, ASB17Y, HMS6J, LEX3R, and LEX3S; Tuvan, VHL20S, HMS6H, HTG6L, ASB17U, ASB17X, and ASB17Z. Among the factory breeds of horses, Donskaya and Budennovskaya stood out due to the presence of unique alleles, ASB17T, as well as the Orlov Trotter, AHT5P.
Summarizing the results of the cluster analysis, we can say that there are clear genetically determined levels of fundamental evolutionary connections between modern representatives of the overwhelming number of breeds bred on the territory of the Russian Federation. The dendrogram of genetic distances between the studied horse breeds clearly reflects their specialization by type of economic use: riding, trotting, draft, and local. At the same time, the cluster of aboriginal breeds included individual factory breeds, such as Orlov Trotter and heavy draft horses, which over the past centuries have been used as improvers of local livestock.
The closest genetic relationship was established between the Thoroughbred and Budennovskaya breeds, as well as American Standardbred and Russian Trotter, with a significant constant influence of the improving breed. It is obvious that the formation of the genetic structure of breeds is influenced by many factors, including the introduction of genes, selection vectors, and genetic-population processes in populations. The results of our research indicate that all domestic horse breeds included in the Register of Breeding Achievements of the Russian Federation have a unique genetic profile and differ from other cultivated breeds. Modern indigenous breeds of horses, even with a common origin from Mongolian roots, have their own characteristic genetic structure with the presence of unique alleles, despite periodic crossing with factory breeds of riding, trotting, and draft breeds.
Our results confirm the published data of foreign scientists [12, 26] that the area of horse domestication occupied a significant part of modern Russia, which, due to its geographical location, was a historical crossroads of the routes of many nomadic peoples of Eurasia, which contributed to the intensive process of the formation of new breeds of horses. Russia has an impressive diversity of horse breeds with unique gene pools, which are well adapted to the harshest climatic conditions, are characterized by universal performance and high productive qualities and are of significant interest to the world horse breeding. Domestic horse breeds have a unique genetic profile with the presence of unique alleles, which must be taken into account when monitoring the origin and assessing population diversity, as well as when conducting genetic monitoring and planning programs for the conservation and breeding of horses of factory and local breeds.
REFERENCES
Vdovina, N.V. and Yur’eva, I.B., Monitoring for the genetic structure of Mezenskaya breed horses in terms of DNA microsatellites, Vavilovskii Zh. Genet. Sel., 2021, vol. 25, no. 2, pp. 202—207. https://doi.org/10.18699/VJ21.024
Blohina, N.V., Khrabrova, L.A., Zaitsev, A.M., and Gavrilicheva, I.S., Assessment of genetic diversity of microsatellite loci in draft horses, Genet. Razvedenie Zhivotn., 2018, no. 2, pp. 39—44. https://doi.org/10.31043/2410-2733-2018-2-39-44
Gavrilicheva, I.S., Genetic and population characteristics of the Russian trotting horse breed based on DNA microsatellite loci, AgroZooTekhnika, 2019, vol. 2, no. 3, p. 2. https://doi.org/10.15838/alt.2019.2.3.2
Glazko, V.I., Kosovskii, G.Yu., Glazko, T.T., and Fedorova, L.M., DNA markers and the “microsatellite code” (a review), S-kh. Biol., 2023, vol. 58, no. 2, pp. 223—248.
Dolmatova, I.Yu., Niyatshin, F.I., and Urazbakhtin, R.F., Population genetic characteristics of Bashkir breed horses based on DNA microsatellites, Konevod. Konnyi Sport., 2017, no. 4, pp. 17—19.
Kalashnikov, V.V., Khrabrova, L.A., Zaitsev, A.M., et al., Study of polymorphism of satellite DNA of improved and local horse breeds, Russ. Agric. Sci., 2011, vol. 36, no. 6, pp. 460—462. https://doi.org/10.3103/S1068367410060194
Kalashnikova, L.A., Novikov, A.A., and Semak, M.S., Progress in genetic examination of pedigree stock in animal husbandry, Zootekhniya, 2022, no. 11, pp. 25—28.
Marzanov, N.S., Nasibov, M.G., Marzanova, L.K., et al., Geneticheskie markery v teorii i praktike razvedeniya ovets (Genetic Markers in the Theory and Practice of Sheep Breeding), Moskow: Pioner, 2010.
Khrabrova, L.A., Theoretical and practical aspects of genetic monitoring in horse breeding, Doctoral (Agric.) Dissertation, Divovo: All-Russian Research Institute of Horse Breeding, 2011.
Khrabrova, L.A., Strategiya ispol’zovaniya geneticheskikh markerov i genomnoi selektsii v konevodstve (Strategy for the Use of Genetic Markers and Genomic Selection in Horse Breeding), Divovo, 2015.
Ernst, L.K. and Zinov’eva, N.A., Biologicheskie problemy zhivotnovodstva v XXI veke (Biological Issues in Livestock Farming in the 21st Century), Moscow: Ross. Akad. S-kh. Nauk, 2008.
Lippold, S., Matzke, N.J., and Reissmann, M., Whole mitochondrial genome sequencing of domestic horses reveals incorporation of extensive wild horse diversity during domestication, BMC Evol. Biol., 2011, vol. 11. https://doi.org/10.1186/1471-2148-11-328
Stock, K.F., Jönsson, L, Ricard, A., and Mark, T., Genomic applications in horse breeding, Anim. Front., 2016, vol. 6, no. 1, pp. 45—52. https://doi.org/10.2527/af.2016-0007
Atroshchenko, M., Dementieva, N., Shcherbakov, Yu., et al., The genetic diversity of horse native breeds in Russia, Genes, 2023, vol. 14. https://doi.org/10.3390/genes14122148
Jarne, P. and Lagoda, P.J.L., Microsatellites, from molecules to populations and back, Trends Ecol. Evol., 1996, vol. 11, no. 10, pp. 424—429. https://doi.org/10.1016/0169-5347(96)10049-5
Khaudov, A.D., Duduev, A.S., Kokov, Z.A., et al., Diversity of Kabardian horses and their genetic relationships with selected breeds in the Russian Federation based on 17 microsatellite loci, IOP Conf. Ser.: Earth Environ. Sci., 2019, vol. 341, p. 12072. https://doi.org/10.1088/1755-1315/341/1/012072
Khrabrova, L.A., Characterization of Genetic Horse Breeding Resources in Russia, Saarbrucken: Lambert, 2015. https://www.abebooks.com/9783659808814/ Characterization-genetic-horse-breeding-resources-3659808814/plp.
Khrabrova, L.A., Blohina, N.V., Belousova, N.F., and Cothran, E.G., Estimation of the genealogical structure of Vyatka horse breed (Equus ferus caballus) using DNA analysis, Russ. J. Genet., 2022, vol. 58, no. 4. pp. 462—466. https://doi.org/10.1134/S1022795422040068
Blohina, N.V., Khrabrova, L.A., and Gavrilicheva, I.S., Application of modern technologies in identifying distinctive features in the subpopulation of Novoaltaiskaya horses, IOP Conf. Ser.: Earth Environ. Sci., 2021, vol. 624, no. 1, p. 12019.
Zaitcev, A.M., Gavrilicheva, I.S., and Blohina, N.V., Assessment of the population structure of horses of the Priobskaya breed based on modern technologies, IOP Conf. Ser.: Earth Environ. Sci., 2021, vol. 624. https://doi.org/10.1088/1755-1315/624/1/012032
Chang, C.C., Chow, C.C., Tellier, L.C.A.M., et al., Second-generation PLINK: rising to the challenge of larger and richer datasets, GigaScience, 2015, vol. 4, no. 1, p. s13742-015-0047-8. https://doi.org/10.1186/s13742-015-0047-8
Francis, R.M., Pophelper: an R package and web app to analyse and visualize population structure, Mol. Ecol. Resour., 2017, vol. 17, pp. 27—32.
Van de Goor, L.H.P., Panneman, H., and van Haeringen, W.A., A proposal for standardization in forensic equine DNA typing: allele nomenclature for equine-specific STR loci, Anim. Genet., 2010, vol. 41, no. 2, pp. 122—127.
Nei, M., Molecular Evolutionary Genetics, New York: Columbia Univ. Press, 1987.
Van de Goor, L.H., van Haeringen, W.A., and Lenstra, J.A., Population studies of 17 equine STR for forensic and phylogenetical analysis, Anim. Genet., 2011, vol. 42, no. 6, pp. 627—633. https://doi.org/10.1111/j.1365-2052.2011.02194.x
Librado, P., Khan, N., Fages, A., et al., The origins and spread of domestic horses from the Western Eurasian steppes, Nature, 2021, vol. 598, pp. 634—640. https://doi.org/10.1038/s41586-021-04018-9
ACKNOWLEDGMENTS
The authors express deep gratitude and appreciation to their colleagues of the laboratory of genetics and the selection department of the Federal State Budgetary Institution All-Russian Research Institute for Horse Breeding, as well as breed registrars and horse breeding specialists from various farms.
Funding
The work was carried out with financial support from the Ministry of Science and Higher Education of the Russian Federation, grant no. 075-15-2021-1037 (internal no. 15.BRK.21.0001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the All-Russian Research Institute of Horse Breeding Ethics Committee–Federal State Budgetary Scientific Institution protocol no. 35 of December 14, 2023.
All applicable international, national and/or institutional guidelines for the care and use of animals were followed.
CONFLICT OF INTEREST
The authors of this work declare that they have no conflicts of interest.
Additional information
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Blohina, N.V., Khrabrova, L.A. Analysis of the Genetic Structures of 29 Horse Breeds of Russian Selection by STR Markers. Russ J Genet 60, 1046–1055 (2024). https://doi.org/10.1134/S1022795424700510
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795424700510