Abstract
This study was aimed to investigate the expression levels of transcription factor (TF) genes (MYF6, MYOD1, MYF5, MYOG) and proteins associated with muscle growth in the longissimus dorsi (LD) and gluteal (GL) muscles in sheep. For this aim, two fat-tailed sheep breeds (Akkaraman (n = 10), Awassi (n = 10)) and two thin-tailed sheep breeds (Kivircik (n = 10) and Karayaka (n = 10)) from Türkiye’s native sheep breeds were examined. The expression level was analyzed for MYF6, MYOD1, MYF5, and MYOG genes and proteins that RNAs and proteins were isolated from fresh tissues. As a result of the statistical analysis, in the LD tissue, respectively, MYOG and MYF5 genes in the Karayaka sheep breed; MYOD1 gene in Akkaraman sheep breed; MYF5 gene in Awassi sheep breed were found to be significant (P < 0.05). In GL tissue, respectively, MYOG and MYF6 genes in Akkaraman sheep breed; MYOD1 gene in Karayaka sheep breed; MYF6 gene in Akkaraman and Awassi sheep breed were significant (P < 0.05). The MYOG (fold change 6.87) and MYOD1 (fold change 15.41) genes were upregulated in the GL muscle of the fat-tailed Akkaraman sheep breed. In addition, in the thin-tailed Karayaka sheep breed, down-regulation of MYOD1 (fold change –0.22) gene in LD muscle and up-regulation of MYOD1 (fold change 6.67) gene in GL muscle was found. As a result, it can be considered that MYOG and MYOD1 genes as potential candidate genes in molecular selection studies for Akkaraman sheep breed in terms of muscle development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Improving meat yield characteristics such as rapid growth, high carcass yield, increasing carcass and meat quality are among the most important topics in breeding studies regarding the livestock animals raised for meat yield. In recent years, there has been an increasing interest in the use of genetic markers in the studies conducted for this purpose. Studies in sheep, which includes 25% of the known mammalian livestock animal breeds, are relatively limited [1]. Sheep offer great opportunities in meeting the future demand for food of animal origin and responding to changing market needs because they are more flexible livestock animals compared to cattle due to their adaptability to very different geographies [2].
In addition to being resistant to bad environmental conditions and having the capacity to make high use of low-yielding pastures, fat-tailed sheep breeds store excess calories as tail fat, which also has commercial value, in the season when grass is abundant. Due to these characteristics, fat-tailed sheep breeds have adapted well to Anatolian geography, where steppe climate conditions prevail in general, compared to thin-tailed breeds Ertugrul et al. (Ertugrul et al., 2009). For this reason, 87% of domestic sheep population in Türkiye consists of fat-tailed sheep breeds [4]. With narrower breeding areas, there are also thin-tailed domestic sheep breeds such as Kivircik and Karayaka, which store fat in body cavities and between muscle fibers instead of the tail, and therefore have higher quality meat compared to fat-tailed sheep breeds [3, 5].
The Akkaraman breed, one of the fat-tailed sheep breeds in Türkiye, is mainly bred in the Central Anatolia Region, where sheep breeding is most intense, but this breed is also common in the transition regions close to this area. Therefore, it is the sheep breed with the largest population size in Türkiye [3]. Another fat-tailed breed bred in Türkiye, the Awassi breed, is bred in the Southeastern Anatolia Region in Türkiye, Iraq, Syria, Jordan, Israel, and Palestine [6]. The Kivircik breed, one of the thin-tailed domestic sheep breeds in Türkiye, is bred in the Marmara Region and North Aegean region of Türkiye and is superior to other domestic sheep breeds in terms of meat quality [7]. Another thin-tailed sheep breed is the Karayaka breed, which, like the Kivircik breed, is better compared to the fat-tailed breeds in terms of meat quality and taste and is bred in the inner parts of the Black Sea Region [3].
One of the important challenges in molecular biology in understanding gene regulation is to reveal genetic information that control gene regulation, which is the basis of all biological processes and phenotype [8]. Transcription factors, an important family of gene regulatory proteins, play an important role in the growth, development, and evolution of higher organisms [9]. Therefore, investigating the transcriptome profile of transcription regulatory proteins, especially in different muscle tissues, provide useful information to improve meat production and quality in, especially sheep.
The myogenic regulatory factor (MRF) gene family, which includes myogenin (MYOG), myoblast detection protein-1 (MYOD1), myogenic factor 5 (MYF5) and myogenic factor 6 (MYF6), has important roles in the muscle growth and development [10, 11]. During this process, MYOD1, MYF5 and MRF6 control the formation of myogenic cells [12], while MYOG is required for terminal differentiation of skeletal muscle [10].
Skeletal muscle growth and development is a complex process supported by transcriptome regulation involving regulatory networks and signaling pathways [13]. To date, MRFs [14] in livestock animals were associated with growth hormone [15], insulin-like growth factors [16] and myostatin genes [17] were reported to be associated with muscle growth and development. The transcriptome analyzes to be performed in different species and different muscle groups will contribute to the understanding of the effects of the candidate genes responsible for muscle development or the previously determined candidate genes in different muscle groups.
The aim of the study was to investigate MYOG, MYOD1, MYF5 and MYF6 genes and protein expression levels in longissimus dorsi (LD) and gluteal (GL) muscles in two fat-tailed sheep breeds (Akkaraman and Awassi) and two thin-tailed sheep breeds (Kivircik and Karayaka) from among the sheep breeds in Türkiye.
MATERIALS AND METHODS
Animal Material
The animal material of the study consisted of 5‑month-old four different sheep breed (n = 10 Kivircik lambs, n = 10 Karayaka lambs, n = 10 Akkaraman lambs and n = 10 Awassi lambs). Fresh LD and GL samples were taken on the day of slaughter and frozen in liquid nitrogen. It was stored at –80°C until protein and RNA isolation.
RNA Isolation and RT-qPCR
LD and GL tissue samples were homogenized with the help of liquid nitrogen and take in sterile 1.5 mL plastic tubes. RNA isolation was performed with trizol (Roche, Mannheim, Germany) from LD and GL tissue samples [18]. The concentration of the RNAs were determined with a nanodrop device (Synergy H1 Hybrid Multi-Mode Microplate Reader, BioTek, USA).
DNase (Invitrogen DNA-free DNA removal kit, Invitrogen, Thermo Fisher Scientific, Carlsbad, US) was applied to RNA samples to prevent DNA contamination. DNase-treated RNA samples were checked on a 1% agarose gel. c-DNA synthesis was performed with these RNA samples by First Strand c-DNA Synthesis Kit (Roche Ltd., Mannheim, Germany) and gene expression analysis was determined by RT-qPCR [19]. The Ct values of target genes designed with Primer 3 software [20] were normalized with housekeeping gene [21] and normalized data was applied to the 2–ΔΔCt methods for statistical analyses [22].
Western Blot Analysis
Protein isolation was performed on ice with Cell lysis buffer (Cell Signaling Technology, USA) on the muscle tissue samples taken fresh from the slaughterhouse on the study day and frozen in liquid nitrogen. The concentration of isolated proteins was determined by the Lowry method [23]. Marker and protein samples, whose concentration was adjusted according to protein size, were loaded into 8–12% SDS-polyacrylamide (SDS-PAGE) gel as 60 µg protein and electrophoresis was carried out in vertical electrophoresis (MiniProtean Tetra System, BioRad, Hercules, CA, USA) system in running buffer at 70 V for two hours. After the electrophoresis, the gel was transferred to methanol-activated Polyvinylidene difluoride (PVDF) membranes (Immobilon-PSQ Transfer membrane, Millipore) and the transfer process was applied at 4 C for one hour at 100 V to the membranes placed in the transfer tank. The membrane obtained as a result of the transfer process was incubated for one hour in an orbital shaker for one hour in blotting solution with 5% skim milk powder (Tris buffer saline-Tween 20 (TBS-T): 10 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% Tween-20). The membranes were kept in the primary antibody solution (1 : 500 dilution ratio) overnight at 4°C for each protein (MYOD1 (LSC130926 (LSBio, Lynnwood, Washington, USA)), MYOG (LS-C113013 (LSBio, Lynnwood, Washington, USA)), MYF5 (LSBIO LS-C29704 (LSBio, Lynnwood, Washington, USA)), MYF6 (LS-C191948 (LSBio, Lynnwood, Washington, USA)) and B-Actin (4967S (Cell Signaling Technology, Danvers, Massachusetts, USA))) planned to be studied after blotting. The following day, the membrane was incubated with (HRP)-Conjugated secondary antibodies for one hour at room temperature. To control the primary antibody incubation, chemiluminescence dye was applied to the membrane and incubated for five minutes at room temperature in the dark, imaging was done in the BioRad (ChemiDoc™ XRS + System with Image Lab™) system, the band sizes were evaluated, and the data were normalized with the β-Actin.
Statistical Analyzes
Histogram, q-q plots, and Kolmogorov–Smirnov test were used to test the conformity of the study data to normal distribution. The expression differences of the genes by tissues between the breeds were tested with One-Way ANOVA. Statistical significance control of the difference in expression levels of genes between the tissues within the breed was tested with the Paired Sample t-test. Windows 14.01 SPSS software was used in the statistical analysis of the data and the significance level was accepted as P < 0.05.
RESULTS
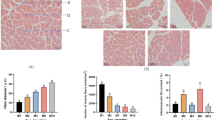
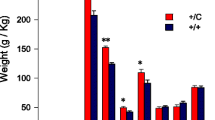
MYOG gene expression level was upregulated (P < 0.05) in Karayaka breed in LD muscle, while the expression levels of MYOD1 was downregulated in Akkaraman breed and MYF5 genes were downregulated in Awassi and Karayaka breeds (P < 0.05) (Fig. 1).
The expression of MYOG, MYF5 and MYF6 genes were upregulated in Akkaraman breed, MYOD1 genes in Karayaka breed and MYF6 genes in Awassi breed (P < 0.05) (Fig. 2).
Longissimus dorsi tissues showed that the expression levels of MYOG, MYOD1, MYF5 and MYF6 genes and the expression levels of MYOG, MYOD1, MYF5 and MYF6 proteins in these muscles changed (increased and/or decreased) harmoniously in all breeds (Fig. 3).
Gluteal tissues showed that the expression levels of MYOG, MYOD1, MYF5 and MYF6 genes and the expression levels of MYOG, MYOD1, MYF5 and MYF6 proteins in these muscles changed (increased and/or decreased) harmoniously in all breeds (Fig. 4).
DISCUSSION
This study aimed to present the gene and protein expression profiles of the four members of the myogenic regulatory gene family MYOG, MYOD1, MYF5 and MYF6 in the LD and GL muscles of the fat-tailed and thin-tailed native Turkish sheep breeds and to explore the potential of using the obtained data in breeding studies to increase the meat yield in sheep.
Siqin et al. [24] reported that the MYOG and MYF6 gene expression levels in the LD muscle were stable during postnatal development in the fat-tailed Wuzhumuqin sheep breed. A study investigating these effects on muscle development in short-tailed Hu sheep found that the expression level of the MYOG gene was downregulated in the LD muscle, whereas the expression level was upregulated in the gastrocnemius and soleus muscles, reporting the dependence of the expression level of the MYOG gene on the muscle groups as one of the reasons for this difference [25]. In support of this view, this study found that the MYOG gene and protein expression level was statistically significantly (P < 0.05) upregulated in the LD muscle in the thin-tailed Karayaka sheep breed, while the expression level of the same gene and protein was downregulated in the GL muscle (P < 0.05). On the other hand, it was determined that the MYOG gene expression level was upregulated in GL muscle tissue in Akkaraman breed, a fat-tailed breed. This supports the knowledge that the activity of the MYOG gene, which plays a key role in regulating the development of muscle cells during the formation of skeletal muscle, changes specifically for the muscle group [26].
The increase in the number of satellite cells responsible for muscle cell development differs from breed to breed, species to species, and muscle group to muscle group [27–29]. MYOD1 gene is involved in satellite cell increase in different ways [30, 31]. It was determined that the expression level of MYOD1 gene in pigs varies according to age, breed and muscle type, and it was reported that this may be related to the number of satellite cells in different breeds and different muscle groups [32]. The cited study suggested that new candidate genes related to meat yield could be revealed by investigating MYOD1 gene and closely related MYF5 and MYF6 gene expression levels in different pig breeds or at different developmental stages of pigs [32]. This situation can be considered to be the case for all livestock animals.
Sun et al. [13] investigated the transcriptome profile of MYOD1 and MYOG genes with the aim of improving sheep meat yield and quality in thin-tail Merino and Han sheep breeds and reported that the gene expression levels of these genes were downregulated in muscle tissue. The present study presented that the MYOG gene expression was statistically significantly higher (P < 0.05) in the LD muscle in the Karayaka breed, one of the sheep breeds that was examined in this study. On the other hand, MYOG and MYOD1 gene expression levels were found to be upregulated in the GL muscle in the fat-tailed Akkaraman breed. It was observed in the present study that the expression level of MYOD1 gene decreased statistically significantly in the LD tissue of the fat-tailed Akkaraman breed samples (P < 0.05). This difference may be due to the nutritional difference and the geographical region where the animals are breed. These expression differences obtained in this study may be due to the different tail phenotype. Again, Sun et al. [13] reported that MYF6 gene expression was upregulated in Merino and Han sheep which they investigated. Similarly, in this study, it was also observed that MYF6 gene and protein expression levels were statistically significantly higher in GL muscle tissue of Akkaraman and Awassi breeds (P < 0.05).
Ropka-Molik et al. [29] reported that the expression level of MYF6 in body muscles in different pig breeds did not change significantly. In this study, the expression level of MYF6 gene was found to increase statistically significantly in fat-tailed Akkaraman and Awassi breeds (p < 0.05). This situation might be due to difference in species. Fan et al. [33] investigated the expression level of MYF6 gene and protein in the skeletal muscle tissue of Duroc breed pigs with high meat fat ratio and Pietrain breed pigs with low meat fat ratio; and reported that MYF6 gene and protein expression levels were statistically high in the Pietrain breed. In the present study, the fact that the expression level of MYF6 gene and protein was upregulated in Akkaraman and Awassi breeds, the two fat-tailed breeds with high body fat accumulation in the tail, suggested that there may be a relationship between the expression level of this gene and fat metabolism. Studies on the expression patterns of major myogenic regulatory factors in pigs, an important source of red meat, may clarify the genetic basis of the postnatal myogenesis process and its effects on high or low muscle levels in different breeds of pigs [32]. This case may be valid for all livestock animals.
According to the results obtained in the study, it was observed that the gene and protein expression profiles of MYOG, MYOD1, MYF5 and MYF6 genes in both fat-tailed and thin-tailed sheep breeds were showed parallel increases and/or decreases in LD and GL muscles. With this the potential of using MYOG, MYOD1 gene and protein expressions as markers in selection studies to improve meat yield in different muscle groups in Akkaraman sheep breed.
REFERENCES
Sheriff, O. and Alemayehu K., Genetic diversity studies using microsatellite markers and their contribution in supporting sustainable sheep breeding programs: a review, Cogent Food Agric., 2018, vol. 4, no. 1, p. 1459062. https://doi.org/10.1080/23311932.2018.1459062
Wollny, C.B.A., The need to conserve farm animal genetic resources in Africa: should policy makers be concerned?, Ecol. Econ., 2003, vol. 45, no. 3, pp. 341—351. https://doi.org/10.1016/S0921-8009(03)00089-2
Ertugrul, M., Dellal, G., Soysal, I.M., et al., Turkiye Yerli Koyun Irklarinin Korunmasi, Bursa Uludag Üniv., Ziraat Fak. Derg., 2009, vol. 23, no. 2, pp. 97—119.
Balcioğlu, M.S., Șahin, E., Karabağ, K., et al., Determination of DNA fingerprinting of Turkish fat-tailed sheep breeds by RAPD-PCR method, JAS, 2010, vol. 16, no. 1, pp. 55—61.
Kaymakçı, M., Özder, M., Karaca, O., et al., Türkiye Koyun Islahı Stratejisi, Bursa Uludag Üniv., Ziraat Fak. Derg., 2009, vol. 23, no. 2, pp. 67—77. https://doi.org/10.20479/uuzfd.29112
Galal, S., Gürsoy, O., and Shaat, I., Awassi sheep as a genetic resource and efforts for their genetic improvement—a review, Small Rumin. Res., 2008, vol. 79, nos. 2—3, pp. 99—108. https://doi.org/10.1016/j.smallrumres.2008.07.018
Öner, Y., Üstüner, H., Orman, A., et al., Genetic diversity of Kıvırcık sheep breed reared in different regions and its relationship with other sheep breeds in Turkey, Ital. J. Anim. Sci., 2014, vol. 13, no. 3, p. 3382. https://doi.org/10.4081/ijas.2014.3382
Hobert, O., Gene regulation by transcription factors and microRNAs, Science, 2008, vol. 319, no. 5871, pp. 1785—1786.
Chen, K. and Rajewsky, N., The evolution of gene regulation by transcription factors and microRNAs, Nat. Rev. Genet., 2007, vol. 8, no. 2, pp. 93—103. https://doi.org/10.1038/nrg1990
Cheng, S., Wang, X., Wang, Q., et al., Comparative analysis of Longissimus dorsi tissue from two sheep groups identifies differentially expressed genes related to growth, development and meat quality, Genomics, 2020, vol. 112, no. 5, pp. 3322—3330. https://doi.org/10.1016/j.ygeno.2020.06.011
Rihan, W., Yueying, Y., Qinghui, F., et al. Expression of myogenic regulatory factor genes in skeletal muscle satellite cells from Wuzhumuqin sheep in vitro, Small Rumin. Res., 2020, vol. 193, p. 106251. https://doi.org/10.1016/j.smallrumres.2020.106251
Hernández-Hernández, J.M., García-González, E.G., Brun, C.E., et al., The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration, Semin. Cell Dev. Biol., 2017, pp. 10—18. https://doi.org/10.1016/j.semcdb.2017.11.010
Sun, L., Bai, M., Xiang, L., et al., Comparative transcriptome profiling of longissimus muscle tissues from Qianhua Mutton Merino and Small Tail Han sheep, Sci. Rep., 2016, vol. 6, no. 1, pp. 1—13. https://doi.org/10.1038/srep33586
Bryson-Richardson, R.J. and Currie, P.D., The genetics of vertebrate myogenesis, Nat. Rev. Genet., 2008, vol. 9, no. 8, pp. 632—646. https://doi.org/10.1038/nrg2369
Gent, J., van den Eijnden, M., van Kerkhof, P., et al., Dimerization and signal transduction of the growth hormone receptor, J. Mol. Endocrinol., 2003, vol. 17, no. 5, pp. 967—975. https://doi.org/10.1210/me.2002-0261
Duan, C., Ren, H., and Gao, S., General and comparative endocrinology insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins: roles in skeletal muscle growth and differentiation, Gen. Comp. Endocrinol., 2010, vol. 167, pp. 344—351. https://doi.org/10.1016/j.ygcen.2010.04.009
Abe, S., Soejima, M., Iwanuma, O., et al., Expression of myostatin and follistatin in Mdx mice, an animal model for muscular dystrophy, Zool. Sci., 2009, vol. 26, no. 5, pp. 315—320. https://doi.org/10.2108/zsj.26.315
Harwood, A.J., Basic DNA and RNA Protocols, Totowa, NJ: Humana Press, 1996.
Daldaban, F., Karaca Bekdik, İ., Aslan, Ö., et al., Investigation of TLR1-9 genes and miR-155 expression in dogs infected with canine distemper, Comp. Immunol. Microbiol. Infect. Dis., 2021, vol. 79. https://doi.org/10.1016/j.cimid.2021.101711
Rozen, S. and Skaletsky, H., Primer3 on the WWW for general users and for biologist programmers, Methods Mol. Biol., 2000, vol. 132, no. 1, pp. 365—386. https://doi.org/10.1385/1-59259-192-2:365
Dervishi, E., Serrano, C., Joy, M., et al., The effect of feeding system in the expression of genes related with fat metabolism in semitendinous muscle in sheep, Meat Sci., 2011, vol. 89, no. 1, pp. 91—97. https://doi.org/10.1016/j.meatsci.2011.04.003
Schmittgen, T.D. and Livak, K.J., Analyzing real-time PCR data by the comparative C T method, Nat. Protoc., 2008, vol. 3, no. 6, p. 1101. https://doi.org/10.1038/nprot.2008.73
Lowry, O.H., Rosebrough, N.J., Farr, A.L., et al., Protein measurement with the Folin phenol reagent, J. Biol. Chem., 1951, vol. 193, pp. 265—275. https://doi.org/10.1016/S0021-9258(19)52451-6
Siqin, Q., Nishiumi, T., Yamada, T., et al., Relationships among muscle fiber type composition, fiber diameter and MRF gene expression in different skeletal muscles of naturally grazing Wuzhumuqin sheep during postnatal development, Anim. Sci. J., 2017, vol. 88, no. 12, pp. 2033—2043. https://doi.org/10.1111/asj.12848
Lv, X.Y., Sun, W., Su, R., et al., Correlation between sheep YAP1 temporal and spatial expression trends and MSTN and MyoG gene expression, Genet. Mol. Res., 2015, vol. 14, no. 2, pp. 3244—3256. https://doi.org/10.4238/2015.A
Faralli, H. and Dilworth, F.J., Turning on myogenin in muscle: a paradigm for understanding mechanisms of tissue-specific gene expression, Comp. Funct. Genom., 2012, vol. 2012. https://doi.org/10.1155/2012/836374
Campion, D.R., Richardson, R.L., Reagan, J.O., et al., Changes in the satellite cell population during postnatal growth of pig skeletal muscle, J. Anim. Sci., 1981, vol. 52, no. 5, pp. 1014—1018. https://doi.org/10.2527/jas1981.5251014x
Gibson, M.C. and Schultz, E., Age-related differences in absolute numbers of skeletal muscle satellite cells, Muscle Nerve, 1983, vol. 6, no. 8, pp. 574—580. https://doi.org/10.1002/mus.880060807
Ropka-Molik, K., Żukowski, K., Eckert, R., et al., Whole transcriptome analysis of the porcine muscle tissue of breeds differing in muscularity and meat quality traits, Livest. Sci., 2015, vol. 182, pp. 93—100. https://doi.org/10.1016/j.livsci.2015.10.024
Zhang, C., Wang, G., Wang, J., et al., Analysis of differential gene expression and novel transcript units of ovine muscle transcriptomes, PLoS One, 2014, vol. 9, no. 2, p. e89817. https://doi.org/10.1371/journal.pone.0089817
Zhang, W., Tong, H., Zhang, Z., et al., Transcription factor EGR1 promotes differentiation of bovine skeletal muscle satellite cells by regulating MyoG gene expression, J. Cell Physiol., 2018, vol. 233, no. 1, pp. 350—362. https://doi.org/10.1002/jcp.25883
Ropka-Molik, K., Eckert, R., and Piórkowska, K., The expression pattern of myogenic regulatory factors MyoD, Myf6 and Pax7 in postnatal porcine skeletal muscles, Gene Expr. Patterns., 2011, vol. 11, nos. 1—2, pp. 79—83. https://doi.org/10.1016/j.gep.2010.09.005
Fan, H., Cinar, M.U., Phatsara, C., et al., Molecular mechanism underlying the differential MYF6 expression in postnatal skeletal muscle of Duroc and Pietrain breeds, Gene, 2011, vol. 486, nos. 1—2, pp. 8—14. https://doi.org/10.1016/j.gene.2011.06.031
Funding
This study was supported by Erciyes University Scientific Research Projects (ERÜ BAP project no. TCD-2015-5653).
Author information
Authors and Affiliations
Contributions
KA designed the study and supervised the experiments. DB contributed to animal material. FD contributed to gene expression and western blot analyses. MHS and MUÇ analyzed statistical data. The manuscript was prepared by FD and edited by KA and BA. All authors contributed to the manuscript and approved the submitted version.
Corresponding author
Ethics declarations
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Erciyes University (09.04.2014/14-78).
CONFLICT OF INTEREST
The authors of this work declare that they have no conflicts of interest.
Additional information
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Arslan, K., Daldaban, F., Bayram, D. et al. Investigation of Expression Levels of Transcription Factor Genes in Native Sheep Breeds of Türkiye. Russ J Genet 60, 932–938 (2024). https://doi.org/10.1134/S1022795424700364
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795424700364