Abstract
Arterial thromboembolic disease affects coronary vasculature and has an exhaustive list of etiologies. The aim of the present study was to investigate the effects of genetic variants in homocysteine pathway genes, homocysteine levels, and other modifiable and non-modifiable conventionally allotted risk factors for coronary artery disease. Study was retrospective case control study, comprised 404 participants (controls, n = 179, ischemic heart disease (IHD) patients, n = 89, and myocardial infarction (MI) cases, n = 136, respectively). Single nucleotide polymorphisms (SNPs); rs1801133, rs1801131 in methylenetetrahydrofolate reductase ‘MTHFR’ gene, rs1805087 in methyl tetrahydrofolate homocysteine methyltransferase ‘MTR’ gene, and rs662 in paroxanse1 ‘PON1’ gene, rs4646994, angiotensin converting enzyme ‘ACE’ insertion/deletion (I/D) polymorphism were resolved employing conventional, and by tetra primer allele refractory mutation system polymerase chain reaction (PCR). ANOVA association testing revealed that homocysteine, cholesterol, creatinine, triglyceride levels, age, family history of CAD, and polymorphisms in MTHFR and PON1 related to coronary artery disease. The post HOC analysis also maintained significance differences in the control, ischemic heart disease and case groups respectively. The regression analysis failed to maintain statistical significance for creatinine, triglycerides, age, and rs662 PON1 polymorphism, whereby, serum homocysteine, cholesterol, family history, and rs1801133/rs1801131 MTHFR SNPs maintained statistical significance. The results from the present study provide hint into interlaced nature of traditional and novel risk factors in the causation of arterial disease and an insight into their shared detrimental effects in affecting the coronary vasculature.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Coronary artery disease has multifactorial nature, [1] with many interactions amongst genetic variants and environmental factors [2, 3]. The rise in CAD cases worldwide has considerable burden on health care resources, associated with decline in quality of life. In the coming decade, CAD cases can rise from nearly 1600 per hundred thousand population to nearly 1800 per hundred thousand population [4]. A few of the implicative factors including dyslipidemia, gender, age, family history, diet, and lifestyle have gained significance as traditional risk factors of atherosclerosis and of arterial disease outcomes. Recent genetic studies in conjunction with genome wide association studies (GWAS) have included the novel risk factors; genetic polymorphisms, circulating biomarkers, gene—gene interactions, or gene—environment interactions as implicated with risk of vascular disease [2, 5, 6].
Atherothrombosis is at the heart of arterial diseases and its outcome is substantial burden on health care services with ensuing comparatively high morbidity and mortality. Age and gender differences (old age and males) in addition to hypertension and diabetes are strongly related with coronary artery disease, acute myocardial infarction (AMI), and fatal outcomes [7, 8]. High serum cholesterol levels and dyslipidemia have adverse relation with atherosclerotic plaque disease affecting the carotid and coronary vasculature [9–11]. Metabolic syndrome is associated with higher body mass index (BMI), dyslipidemia, hypertension, and angiographically documented coronary lesions [12]. Analogous to preceding factors, serum creatinine, triglycerides, high BMI, and family history of CAD have all been associated with deleterious outcomes in coronary vascular disease [12, 13].
Homocysteine as a novel biomarker, in association with traditional biomarkers, such as creatinine, blood sugar and lipid levels, is associated with acute myocardial infarction and coronary artery stenosis [14, 15]. Serum homocysteine along with cholesterol levels is associated with higher risk as well as greater progression of CAD [16, 17]. The SNPs in MTHFR gene, C677T polymorphism (rs1801133) and A1298C polymorphism (rs1801131) are independent risk factors for CAD, and these SNPs, furthermore are associated with hyperhomocystenemia, hypercholesterolemia, higher creatinine levels and higher risk of CAD [2, 9, 18, 19]. The A2756G polymorphism in MTR gene (rs1805087), the A192G polymorphism in PON1 gene, and the ACE insertion deletion polymorphism (rs4646994, gene/polymorphism not part of folate pathway) have additionally been implicated with vascular outcomes affecting coronary vessels [2, 20–22].
The genetic component, homocysteine levels, and the traditional/recognized serum parameters have confounded effect on coronary vasculature and resulting disease outcomes. No single entity has so far been designated the only role in the genesis of CAD (complex diseases). In the resident population, we studied the genetic links including the homocysteine pathway gene SNPs, serum homocysteine levels, and the well documented traditional risk predictors of CAD with a view to ascertain their grouping effects in the causation of CAD.
MATERIALS AND METHODS
Participants and Sample Characteristics
The design of study was in retrospective case control setting, all the study participants, namely control participants, MI/CAD patients, and the IHD patients were recruited from the District Headquarter Hospital and Benazir Bhutto Hospital, Rawalpindi. Patients with renal disease, cerebral vascular accidents, transient ischemic attacks, and with heart failure were exclude from the study.
Presentation Characteristics
Major presentation characteristics have been mentioned elsewhere [22], however there were some slight modification for disparate study groups in current study. The IHD cases were excluded in previous study but they have been included in current study. All the participants were hypertensive patients and were taking different antihypertensive prescriptions. The cases presented to hospital emergency within 3 to 4 hours of onset of symptoms. The major symptom for majority of the CAD patients was distinctive chest pain present for more than half an hour to one hour, discomfort with sense of anxiety, pressure, apprehension, and radiation of pain. All the participants were examined by clinicians from cardiology department. In the ER, tests for lung edema and repeat electrocardiography (ECG) was performed and cardiac enzymes were done for suspected patients with normal ECG (typical history, long duration pressing chest pain, continuous restlessness). The high risk patients, who had chest pain, ECG findings of ischemia (ST depression, T wave inversion) and negative cardiac markers were labelled IHD patients; whereas those with positive ST elevation on ECG (ST elevation MI, ‘STEMI’) or those with no ST change or ST depression but with positive CKMB or Troponin levels (non ST elevation MI, ‘NSTEMI’) were consequently labelled as cases. On presentations, vital signs of the participants were recorded including pulse rate, respiratory rate, systolic blood pressure (SBP), diastolic blood pressure (DBP) in mm Hg, (measured through mercury sphygmomanometer), and the readings of temperature (in degrees Celsius). All the vitals were recorded subsequently during the sampling as well. For BMI analysis, the height of individuals in meters and the weight in kilograms were recorded and the BMI computed by standard formula.
Collection of Blood Samples for Serum Analysis and Genetic Analysis
Samples were collected from all participants early in the morning after overnight fasting. For serum analysis, blood samples were collected in plain tubes, serum was extracted and stored at –20°C until analyzed. The covariates analyzed employing serum specimen included (a) cholesterol (mg/dL), (b) triglycerides (mg/dL), (c) creatinine (mg/dL), (d) fasting blood sugar (mg/dL), and (e) homocysteine (µmol/L). For homocysteine levels estimation, we employed enzyme-linked immunosorbent assay (ELISA) through EIA kit by DRG International Inc., USA. The routine serum parameters were analyzed through commercially available kits (AMP Diagnostics, AMEDA Labordiagnostik GmbH). For genetic analysis EDTA vacutainers were used for blood collection, DNA extracted from peripheral leukocytes through routine procedure and the extracted DNA samples were refrigerated at 4°C.
The Investigation of the Polymorphisms/SNPs for Study
We queried Santa Cruz Genome Browser Data base site genome.ucsc.edu/cgi-bin/hgGateway for the genomic sequences of SNP, flanking sequences and related details. For the primers design, the gene sequences were submitted to http://cedar.genetics.soton.ac.uk, and the primers generated by the website were reformed for optimum sequences and results. Tetra primer ARMS PCR employs 4 primers per reaction; pair of outer and pair of inner primers. The MTHFR rs1801133, rs1801131; MTR rs1805087; and PON1 rs662 were resolved using tetra primer technique while two outer primers only were only used to resolve the insertion deletion polymorphism (rs4646994) in ACE gene, respectively. The minute details of the primer sequences, amplified products, and restriction fragment length polymorphism (RFLP) validation was followed as is stated in detail elsewhere [2]
Statistical Analysis
The data was analyzed employing two different statistical programs and the software packages were used for the generation of figures as well. Since homocysteine levels were not normally distributed, the values were log transformed and all analysis were computed with log values for homocysteine. The one-way ANOVA test, and thereafter, Post Hoc ‘Tamhane’ test (assuming unequal variances) for multiple comparison was performed through the IBM SPSS statistics software version 25.0 (IBM Corp. Released 2017. IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY: IBM Corp.). The values with p < 0.05 were considered statistically significant. Thereafter, regression analysis for the SNPs, calculation of odds ratios (OR) and the 95% confidence intervals (95%CI) readings for the different parameters were carried out (to ascertain and validate the findings of the ANOVA analysis to find out disease (CAD) likelihood of the selected parameters. The regression analysis was carried out using the R 4.0.2 statistical package environment (R Core Team 2020. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
Baseline Data
The details of age for the three groups was, controls 63.75 ± 7.44 years (mean ± SD); IHD patients 61.85 ± 9.54 years; and MI cases 66.12 ± 8.57 years. Males numbered 108 (60.33%) in controls, 44 (49.43%) in IHD patients, and 89 (65.44%) in the MI cases. The values (mean ± SD) for systolic blood pressure (mm Hg) were 134.67 ± 13.99, 134.55 ± 17.40, 134.61 ± 18.93; and for the diastolic blood pressure (mm Hg) were 80.30 ± 10.02, 81.40 ± 10.83, and 79.52 ± 11.79 for the control, IHD, and MI groups respectively. The average values for the serum parameters were as follows; serum cholesterol (mg/dL) 178.42 ± 25.11, 182.18 ± 32.79, 189.16 ± 43.10; and serum creatinine 0.91 ± 0.36, 0.92 ± 0.36, 1.10 ± 0.41, respectively in the three groups. The values in controls, IHD, and MI cases for fasting blood sugar, and serum triglycerides (mean ± SD mg/dL) comprised 106.43 ± 35.94, 109.06 ± 31.46, 117.58 ± 52.46; and 117.86 ± 59.30, 108.94 ± 54.58, 150.63 ± 96.21, respectively. As already mentioned, the values for homocysteine were log transformed and result computed through log transformed data. The homocysteine (mean ± SD) was 1.170 ± 0.089 µmol/L in controls, 1.180 ± 0.081 µmol/L in IHD patients, and 1.218 ± 0.100 µmol/L in MI cases, respectively. The BMI readings were 31.21 ± 6.22, 32.31 ± 6.72, and 31.31 ± 5.17 in the groups respectively (Table 1).
ANOVA Analysis and Multiple Testing
The mentioned SNPs and factors were subsequently analyzed by ANOVA and Post Hoc test; the Post Hoc test was carried out assuming unequal variance of the data and Tamhane testing was used for the purpose. In accordance with one-way ANOVA analyses, serum homocysteine levels (p < 0.001), serum cholesterol levels (p = 0.021), serum creatinine levels (p < 0.001), serum triglyceride levels (p < 0.001), age (p = 0.001), family history of CAD (p = 0.001), rs1801133 MTHFR SNP (p = 0.008), rs1801131 MTHFR SNP (p < 0.001), and rs662 PON1 SNP (p < 0.001) exhibited statistical significance with CAD.
The multiple testing displayed group wise significance with CAD for the above documented parameters. Homocysteine levels displayed significance between IHD vs. MI group (p = 0.006) and the control vs. MI group (p < 0.001). Serum cholesterol was significant in control vs. MI (p = 0.031), whereas serum creatinine displayed noteworthy differences in IHD vs. MI cases (p = 0.004) and in control vs. cases (p < 0.001). There was also significant difference between IHD patient vs. MI cases for triglyceride (p < 0.001) and age (p = 0.002) and in controls vs. cases for triglycerides (p = 0.002) and age (p = 0.032). Similar to serum parameters, the family history of CAD also had significance between IHD and cases (p = 0.011) and controls vs. cases (p = 0.001), respectively. The Post Hoc analysis similarly displayed intragroup significance for the three significant polymorphisms. The rs1801133 polymorphisms was significant in control vs. MI (p = 0.010), the rs1801131 polymorphism exhibited significance in the IHD vs. MI (p = 0.008) and control vs. MI cases (p < 0.001). The last significant multiple comparison difference was in rs662 polymorphism, in the IHD vs MI (p = 0.005) and control vs MI cases (p < 0.001), respectively (Table 2).
Regression Analysis for Association of Factors with CAD
Regression analysis was carried out and accompanying computation performed for the odds ratio (OR) and 95% confidence intervals (95%CI) estimation. The rs662 PON1 polymorphism was insignificant with CAD following regression analysis (p < 0.001, OR 0.810, 95%CI 0.720–0.912). Limited number of parameters could not maintain significance through regression, such as serum creatinine, serum triglyceride levels, and age. The significant relation remained for the family history of CAD (p < 0.001, OR 1.341, 95%CI 1.138–1.580); for serum cholesterol levels (p = 0.015, OR 1.030, 95%CI 1.006–1.055); for serum homocysteine levels (p = 0.014, OR 1.845, 95%CI 1.110–2.836); for rs1801133 MTHFR SNP (p = 0.002, OR 1.270, 95%CI 1.093–1.478); and for rs1801131 MTHFR SNP (p < 0.001, OR 1.292, 95%CI 1.116– 1.495), respectively (Table 3).
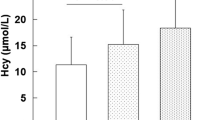
The differences in homocysteine levels µmol/L (log transformed), serum cholesterol levels mg/dL, serum creatinine levels mg/dL, and the serum triglycerides level mg/dL, between the controls, IHD, and MI cases have been plotted in Fig. 1. The frequency distribution of allelic variants between SNP rs662 in PON1, SNPs rs1801131 and rs1801133 in MTHFR, amongst the three studied groups, respectively, has been plotted in Fig. 2.
Graphical comparison of selected serum parameters in the studied samples. (а) Levels of serum homocysteine in studied groups. (b) Levels of serum cholesterol in studied groups. (с) Levels of serum creatinine in studied groups. (d) Levels of serum triglycerides in studied groups, respectively. (Studied groups—Controls, IHD patients, MI/CAD cases.)
Graphical representation of the allelic differences of three significant polymorphisms in homocysteine pathway genes, namely MTHFR and PON1 amid participants in study. (а) Differences between participants according to rs1801133 SNP. (b) Differences between participants according to rs1801131 SNP. (с) Differences between participants according to rs662 SNP, respectively. (Studied groups—Controls, IHD patients, MI/CAD cases.)
DISCUSSION
Complex diseases are characterized by numerous causes, both genetic and environmental. CAD is a leading cause of morbidity and mortality in developed and developing nations and the factor/factors causing disease outcome are a matter of extensive research. The present study looks into collaboration of traditional risk factors, newly elucidated genetic and serum related risk factors as the etiologic producers/predictors of CAD. We performed SNP analysis, ELISA for homocysteine, and used traditional methods for conventional risk factors estimation. ANOVA analysis and regression analysis was performed for statistical testing. Serum homocysteine, cholesterol levels, positive family history, and MTHFR SNPs reemerged as important risk predictors in the current study.
It is well documented that high cholesterol levels and dyslipidemia/hyperlipidemia, are related to carotid artery disease and CAD [16, 17, 23]. Reduction in total cholesterol level may be beneficial for CAD, [24] however some studies could not verify association between cholesterol and CAD [25, 26]. In the current study, there was modestly significant relation of cholesterol levels with CAD (p = 0.021; following ANOVA) with significant result in control versus MI case group (p = 0.031), and furthermore significance through regression analysis (p = 0.015, OR 1.030). Elevated serum creatinine is related to CAD and additionally, indirectly increases the risk for vascular disease outcomes [27–29]. We were able to repeat previous research findings regarding relation of creatinine to CAD (ANOVA results) but it failed to maintain significance following regression analysis. Another known traditional risk factor for CAD, the serum total triglyceride level, is also reported to have positive relation to atherosclerosis and arterial diseases [23, 30, 31]. In the current study, ANOVA analysis revealed association of triglyceride with CAD (p < 0.001) and significant intragroup differences, but failed to show significance following regression (p = 0.162, OR 1.001).
A major risk factor and the one with considerable importance in causation of CAD is family history of disease. Family history of coronary artery disease, and diabetes are significant, well established risk factors of CAD, moreover, they associate with incidence of early onset ailment [32–34]. The current study could not ascertain role of diabetes or elevated glucose levels, but family history of CAD was significantly associated with CAD. The family history had significance with disease outcome (p = 0.001), between IHD and CAD cases (p = 0.011), as well as between controls and cases (p = 0.001). The regression analysis also maintained significance of disease with family history (p < 0.001, OR 1.341) but not with diabetes mellitus. Elevated homocysteine level is novel, independent risk factor of atherosclerotic vascular disease affecting many vascular beds such as peripheral, carotid and coronary vasculature, [2, 17, 22, 35] the higher homocysteine levels being positively related to fatal outcomes [36]. The current study was consistent with previous literature, as both analyses maintained statistical significance of homocysteine with CAD. The elevated homocysteine levels were associated with more likelihood of disease in current study (p = 0.014, OR 1.845), and differences were more marked in between the ischemic group versus MI, as well as in controls versus MI cases, respectively.
There is inconsistency in data regarding SNP data in homocysteine pathway genes and the relation to CAD, as some studies favor and others negate their role [2, 22, 37–41]. The most studied SNP in MTHFR gene, the C677T (rs1801133, where alanine residue is changed to valine) is associated with elevated homocysteine levels and higher risk of CAD [2, 18, 19, 38]. The A1298C SNP (rs1801131) in MTHFR gene has disparate association with homocysteine levels and CAD, as some studies show positive [18, 22, 42] and others negative association for disease causation [38, 40]. The current study displayed strong relation of MTHFR SNPs with CAD; ANOVA analysis for rs1801133 had significance with CAD (p = 0.008) and in control vs. cases (p = 0.010). The rs1801131 SNP had even stronger relation with CAD (p < 0.001) and significant differences between IHD and MI cases (p = 0.008) and controls vs. cases (p = 0.001). The regression analysis, similarly showed positive correlation with disease likelihood for both rs1801133 MTHFR SNP (p = 0.002, OR 1.270) and rs1801131 MTHFR SNP (p < 0.001, OR 1.292). The MTR A2756G SNP (rs1805087) and PON1 SNP A192G (rs662) associate with hyperhomocysteinemia and CAD [43–47]. The former SNP could not attain significance in current study, however the latter polymorphism (rs662), despite displaying significance could not retain it in regression (p < 0.001, OR 0.801). The ACE insertion deletion polymorphism (rs4646994), is unrelated to homocysteine pathway, yet, it is involved in CAD both individually and in combination with homocysteine pathway genes [20, 22, 48, 49]. This affiliation warranted studying the polymorphism but the current study was unable to associate it with CAD.
The important predictors of CAD including the traditional risk factors such as cholesterol, and family history of CAD; the novel serum parameter homocysteine; and genetic polymorphisms in MTHFR gene, altogether are robustly allied to CAD in previous literature [17, 33, 40, 50, 51]. These parameters, in essence, achieved statistical significance with CAD in current study. The additive role of all the multiple studied factors has indispensable implication in coronary vascular disease.
LIMITATIONS OF STUDY
The limited number of samples and inclusion of hypertensive individuals as controls are important limitations in the study. The samples represented similar socioeconomic status but their recruitment/collection was limited to specific geographic strata of Pakistan, namely Rawalpindi and Islamabad region. Further studies incorporating considerably larger sample size and possibly patients from other regions of country may add to the risk stratification and validation of currently concluded results.
REFERENCES
Guven, M., Ismailoglu, Z., Batar, B., et al., The effect of genetic polymorphisms of TLR2 and TLR4 in Turkish patients with coronary artery disease, Gene, 2015, vol. 558, no. 1, pp. 99–102.
Masud, R. and Qureshi, I.Z., Tetra primer ARMS-PCR relates folate/homocysteine pathway genes and ACE gene polymorphism with coronary artery disease, Mol. Cell. Biochem., 2011, vol. 355, nos. 1—2, pp. 289–297.
Shabana, N.A., Ashiq, S., Ijaz, A., et al., Genetic risk score (GRS) constructed from polymorphisms in the PON1, IL-6, ITGB3, and ALDH2 genes is associated with the risk of coronary artery disease in Pakistani subjects, Lipids Health Dis., 2018, vol. 17, no. 1, p. 224.
Khan, M.A., Hashim, M.J., Mustafa, H., et al., Global epidemiology of ischemic heart disease: results from the global burden of disease study, Cureus, 2020, vol. 12, no. 7. e9349
Li, Y., Yan, H., Guo, J., et al., Down-regulated RGS5 by genetic variants impairs endothelial cell function and contributes to coronary artery disease, Cardiovasc. Res., 2019.
Ma, L., Chandel, N., Ermel, R., et al., Multiple independent mechanisms link gene polymorphisms in the region of ZEB2 with risk of coronary artery disease, Atherosclerosis, 2020, vol. 311, pp. 20–29.
Piatek, J., Kedziora, A., Kielbasa, R., et al., How to predict the risk of postoperative complications after coronary artery bypass grafting in patients under 50 and over 80 years old: a retrospective cross-sectional study, Kardiol. Pol., 2017, vol. 75, no. 10, pp. 975–982.
Tay, S.Y., Chang, P.Y., Lao, W.T., et al., The proper use of coronary calcium score and coronary computed tomography angiography for screening asymptomatic patients with cardiovascular risk factors, Sci. Rep., 2017, vol. 7, no. 1, p. 17653.
Charniot, J.C., Khani-Bittar, R., Albertini, J.P., et al., Interpretation of lipoprotein-associated phospholipase A2 levels is influenced by cardiac disease, comorbidities, extension of atherosclerosis and treatments, Int. J. Cardiol., 2013, vol. 168, no. 1, pp. 132–138.
Morito, N., Inoue, Y., Urata, M., et al., Increased carotid artery plaque score is an independent predictor of the presence and severity of coronary artery disease, J. Cardiol., 2008, vol. 51, no. 1, pp. 25–32.
Conkbayir, C., Fahrioglu Yamaci, R., Gencer, P., et al., Impact of genetic defects on coronary atherosclerosis among Turkish Cypriots, Heart Surg Forum, 2017, vol. 20, no. 5, pp. E223–E229.
Hu, R., Ma, C.S., Nie, S.P., et al., Effect of metabolic syndrome on prognosis and clinical characteristics of revascularization in patients with coronary artery disease, Chin. Med. J., 2006, vol. 119, no. 22, pp. 1871–1876.
Tasic, I., Kostic, S., Stojanovic, N.M., et al., Significance of asymptomatic hyperuricemia in patients after coronary events, Scand. J. Clin. Lab. Invest., 2018, vol. 78, no. 4, pp. 312–317.
Borowczyk, K., Piechocka, J., Glowacki, R., et al., Urinary excretion of homocysteine thiolactone and the risk of acute myocardial infarction in coronary artery disease patients: the WENBIT trial, J. Int. Med., 2019, vol. 285, no. 2, pp. 232–244.
Lee, T.H., Cheng, M.L., Shiao, M.S., and Lin, C.N., Metabolomics study in severe extracranial carotid artery stenosis, BMC Neurol., 2019, vol. 19, no. 1, p. 138.
Xie, J., Qi, J., Mao, H., et al., Coronary plaque tissue characterization in patients with premature coronary artery disease, Int. J. Cardiovasc. Imaging, 2020, vol. 36, no. 6, pp. 1003–1011.
Bosevski, M., Zlatanovikj, N., Petkoska, D., et al., Plasma homocysteine in patients with coronary and carotid artery disease: a case control study, Prilozi—Maked. Akad. Nauk. Umet., Odd. Biol. Med. Nauki, 2020, vol. 41, no. 1, pp. 15–22.
Falchi, A., Giovannoni, L., Piras, I.S., et al., Prevalence of genetic risk factors for coronary artery disease in Corsica Island (France), Exp. Mol. Pathol., 2005, vol. 79, no. 3, pp. 210–213.
Kerkeni, M., Addad, F., Chauffert, M., et al., Hyperhomocysteinaemia, methylenetetrahydrofolate reductase polymorphism and risk of coronary artery disease, Ann. Clin. Biochem., 2006, vol. 43, part 3, pp. 200–206.
Zak, I., Niemiec, P., Sarecka, B., et al., Carrier-state of D allele in ACE gene insertion/deletion polymorphism is associated with coronary artery disease, in contrast to the C677→ T transition in the MTHFR gene, Acta Biochim. Pol., 2003, vol. 50, no. 2, pp. 527–534.
Vinukonda, G., Shaik Mohammad, N., Md Nurul Jain, J., et al., Genetic and environmental influences on total plasma homocysteine and coronary artery disease (CAD) risk among South Indians, Clin. Chim. Acta, 2009, vol. 405, nos. 1–2, pp. 127–131.
Masud, R. and Baqai, H.Z., The communal relation of MTHFR, MTR, ACE gene polymorphisms and hyperhomocysteinemia as conceivable risk of coronary artery disease, Appl. Physiol. Nutr. Metab., 2017, vol. 42, no. 10, pp. 1009–1014.
Bansal, S.K. and Yadav, R., A study of the extended lipid profile including oxidized LDL, small dense LDL, lipoprotein (a) and apolipoproteins in the assessment of cardiovascular risk in hypothyroid patients, J. Clin. Diagn. Res., 2016, vol. 10, no. 6, pp. BC04–BC08.
Qunibi, W.Y., Dyslipidemia in dialysis patients, Semin. Dial., 2015, vol. 28, no. 4, pp. 345—353.
Abchee, A., Puzantian, H., Azar, S.T., et al., Predictors of coronary artery disease in the Lebanese population, Thromb. Res., 2006, vol. 117, no. 6, pp. 631–637.
Li, W., Li, X., Wang, M., et al., Association between red cell distribution width and the risk of heart events in patients with coronary artery disease, Exp. Ther. Med., 2015, vol. 9, no. 4, pp. 1508–1514.
Mann, J.F., Gerstein, H.C., Pogue, J., et al., Cardiovascular risk in patients with early renal insufficiency: implications for the use of ACE inhibitors, Am. J. Cardiovasc. Drugs, 2002, vol. 2, no. 3, pp. 157–162.
Huang, C.L., Wu, Y.W., Wu, C.C., et al., Association between serum adipocyte fatty-acid binding protein concentrations, left ventricular function and myocardial perfusion abnormalities in patients with coronary artery disease, Cardiovasc. Diabetol., 2013, vol. 12, p. 105.
Sahin, I., Karabulut, A., Gungor, B., et al., Correlation between the serum alkaline phosphatase level and the severity of coronary artery disease, Coron. Artery Dis., 2014, vol. 25, no. 4, pp. 349–352.
Bagheri, B., Zargari, M., Meshkini, F., et al., Uric acid and coronary artery disease, two sides of a single coin: a determinant of antioxidant system or a factor in metabolic syndrome, J. Clin. Diagn. Res., 2016, vol. 10, no. 2, pp. OC27–OC31.
Manochehri, M. and Moghadam, A.J., Studying the relation of postprandial triglyceride with coronary artery disease (CAD), Med. Arch., 2016, vol. 70, no. 4, pp. 261–264.
Shao, C., Wang, J., Tian, J., and Tang, Y.D., Coronary artery disease: from mechanism to clinical practice, Adv. Exp. Med. Biol., 2020, vol. 1177, pp. 1–36.
Brown, J.C., Gerhardt, T.E., Kwon, E., Risk factors for coronary artery disease, in Stat Pearls Treasure Island (FL), 2020.
Krittanawong, C., Kumar, A., Wang, Z., Narasimhan, B., et al., Coronary artery disease in the young in the US population-based cohort, Am. J. Cardiovasc. Dis., 2020, vol. 10, no. 3, pp. 189–194.
Cheng, M., Cheng, M., and Wei, Q., Association of myeloperoxidase, homocysteine and high-sensitivity C-reactive protein with the severity of coronary artery disease and their diagnostic and prognostic value, Exp. Ther. Med., 2020, vol. 20, no. 2, pp. 1532–1540.
Rallidis, L.S., Kosmas, N., Rallidi, T., et al., Homocysteine is an independent predictor of long-term cardiac mortality in patients with stable coronary artery disease in the era of statins, Coron. Artery Dis., 2020, vol. 31, no. 2, pp. 152–156.
Nakai, K., Habano, W., Fujita, T., et al., Highly multiplexed genotyping of coronary artery disease-associated SNPs using MALDI-TOF mass spectrometry, Hum. Mutat., 2020, vol. 20, no. 2, pp. 133–138.
Ghazouani, L., Abboud, N., Mtiraoui, N., et al., Homocysteine and methylenetetrahydrofolate reductase C677T and A1298C polymorphisms in Tunisian patients with severe coronary artery disease, J. Thromb. Thrombolysis, 2009, vol. 27, no. 2, pp. 191–197.
Bickel, C., Schnabel, R.B., Zengin, E., et al., Homocysteine concentration in coronary artery disease: influence of three common single nucleotide polymorphisms, Nutr. Metab. Cardiovasc. Dis., 2017, vol. 27, no. 2, pp. 168–175.
Long, Y., Zhao, X.T., Liu, C., et al., A case-control study of the association of the polymorphisms of MTHFR and APOE with risk factors and the severity of coronary artery disease, Cardiology, 2019, vol. 142, no. 3, pp. 149–157.
Raina, J.K., Sharma, M., Panjaliya, R.K., et al., Association of ESR1 (rs2234693 and rs9340799), CETP (rs708272), MTHFR (rs1801133 and rs2274976) and MS (rs185087) polymorphisms with coronary artery disease (CAD), BMC Cardiovasc. Disord., 2020, vol. 20, no. 1, p. 340.
Saad Shaukat, M.H., Toledo-Garcia, A., and Torosoff, M., Recurrent myocardial infarction despite normal C-reactive protein in a patient with Behcet’s disease and compound heterozygous methylenetetrahydrofolate reductase (MTHFR) mutations (C677T and A1298C), Cureus, 2019, vol. 11, no. 8, p. e5344.
Vijaya Lakshmi, S.V., Naushad, S.M., Rupasree, Y., et al., Interactions of 5'-UTR thymidylate synthase polymorphism with 677C → T methylene tetrahydrofolate reductase and 66A → G methyltetrahydrofolate homocysteine methyl-transferase reductase polymorphisms determine susceptibility to coronary artery disease, J. Atheroscler. Thromb., 2011, vol. 18, no. 1, pp. 56—64.
Hassan, M.A., Al-Attas, O.S., Hussain, T., et al., The Q192R polymorphism of the paraoxonase 1 gene is a risk factor for coronary artery disease in Saudi subjects, Mol. Cell Biochem., 2013, vol. 380, nos. 1–2, pp. 121–128.
Singh, P.R. and Lele, S.S., Folate gene polymorphisms MTR A2756G, MTRR A66G, and BHMT G742A and risk for coronary artery disease: a meta-analysis, Genet. Test Mol. Biomarkers, 2012, vol. 16, no. 6, pp. 471–475.
Huo, X., Guo, Y., Zhang, Y., et al., Paraoxonase 1 gene (Q192R) polymorphism confers susceptibility to coronary artery disease in type 2 diabetes patients: evidence from case—control studies, Drug Discov. Ther., 2019, vol. 13, no. 2, pp. 80–88.
Zeng, Q. and Zeng, J., A meta-analysis on relationship between paraoxonase 1 polymorphisms and atherosclerotic cardiovascular diseases, Life Sci., 2019, vol. 232, p. 116646.
Szperl, M., Dzielinska, Z., Roszczynko, M., et al., Genetic variants in hypertensive patients with coronary artery disease and coexisting atheromatous renal artery stenosis, Med. Sci. Monit., 2008, vol. 14, no. 12, pp. CR611–CR616.
Agirbasli, M., Guney, A.I., Ozturhan, H.S., et al., Multifactor dimensionality reduction analysis of MTHFR, PAI-1, ACE, PON1, and eNOS gene polymorphisms in patients with early onset coronary artery disease, Eur. J. Cardiovasc. Prev. Rehabil., 2011, vol. 18, no. 6, pp. 803–809.
Tripathi, R., Tewari, S., Singh, P.K., and Agarwal, S., Association of homocysteine and methylene tetrahydrofolate reductase (MTHFR C677T) gene polymorphism with coronary artery disease (CAD) in the population of North India, Genet. Mol. Biol., 2010, vol. 33, no. 2, pp. 224–228.
Rallidis, L.S., Gialeraki, A., Triantafyllis, A.S., et al., Characteristics and long-term prognosis of patients </=35 years of age with ST segment elevation myocardial infarction and normal or near normal coronary arteries, Am. J. Cardiol., 2017, vol. 120, no. 5, pp. 740–746.
ACKNOWLEDGMENTS
The authors are grateful to Prof Dr I.J, Kullo, Dr Keyue Ding, Dr Wasim, Dr Asad, and Dr Tariq for samples, for valuable comments, and help in manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest. The authors declare that they have no conflict of interests.
Statement of compliance with standards of research involving humans as subjects. The Pakistan Medical Research Council, Islamabad and Institutional Review Board CMH Kharian Medical College approved the study. All the participants provided written informed consent for use of data and samples for the current research project. The institutional ethical standards were rigorously exercised and all experiments were carried out under the Helsinki Declaration guidelines.
Rights and permissions
About this article
Cite this article
Masud, R., Khan, A.U., Baqai, H.Z. et al. The Correlation of MTHFR SNPs, Homocysteine, and Conventional Risk Predictors with Coronary Artery Disease. Russ J Genet 57, 1328–1336 (2021). https://doi.org/10.1134/S1022795421110077
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795421110077