Abstract
The preparatory pairing of homologous chromosomes is the obligatory step of meiosis. It occurs through formation of synaptonemal complexes (SC): the protein axes of two chromosomes are connected with the help of additional “central space proteins.” These proteins are sometimes species-specific and serve as the object of comparative studies. With the help of bioinformatics methods, we studied proteins structuring the SC central space in animals and fungi. We established that Ecm11 and Gmc2 had a low level of conservation even within the taxon of Ascomycetes. The SIX6OS1 protein of the mouse, as well as SYCE1–SYCE3 and TEX12 in animals, was moderately conserved only within the subphylum of vertebrates, despite these proteins (with the exception of SYCE3) occurring in invertebrates too. Thus, we have confirmed the thesis that, in addition to the common set of meiotic proteins, every evolutionary line of Eukaryotes has developed its own proteins for the formation of SC, the general structure of which is common between all eukaryotes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Meiosis is a special type of cell division, as a result of which haploid gametes are produced from diploid reproductive cells. Haploid chromosome sets are segregated by preparatory pairing of homologous chromosomes and subsequent exact separation of the homologs by the spindle apparatus. In most eukaryotes, special protein structures are formed for this process: synaptonemal complexes (SC) [1, 2]. Each SC consists of two lateral elements—preformed chromosome axes—and a central space (CS) between them. Generally (this varies between different organisms), a longitudinal central element (CE) is found in the middle of the central space. The CS is filled with transverse filaments that extend from each of the two lateral elements and connect like teeth of a zipper [3]. The CE is a line of contacts between the opposing teeth of the zipper. The SC was discovered by Moses in 1956 [4]; however, the first proteins in its composition were discovered much later. Proteins of the lateral elements and of the transverse filaments were identified mainly in the 20th century (see reviews [1, 5]). But there was another class of proteins predicted by Schmeckel and Daneholt back in 1995 [6]. These are the so-called pillars: proteins that hold the transverse filaments together and stabilize the SC structure. Researchers have started to identify such proteins only in the 21st century. These proteins include Corona and Corolla in the fruit fly, SYP-1–SYP-4 in the nematode [7, 8], and SYCE1–SYCE3 and TEX12 in the mouse [9–11]. Quite recently, the mouse protein SIX6OS1 [12] and the yeast proteins Ecm11 and Gmc2 were identified [13, 14]. The SYP proteins of the nematode (with the possible exception of SYP-2) are not pillars but instead form peculiar transverse SC filaments in the nematode. However, Corona and Corolla interact with each other and are true pillars [2, 7, 8]. The SYCE1–SYCE3 and TEX12 proteins are components of the CE of SC [15]. The SIX6OS protein is also a component of the CE of SC and interacts with SYCE1, strengthening the structure of the complex [12]. The Ecm11 and Gmc2 proteins interact with each other and assist in the connection of transverse SC filaments, which in yeast are formed by the Zip1 protein [13, 14].
The question of the conservation or unicity of SC proteins arose immediately after their discovery. It also immediately became clear that the proteins of the lateral elements and transverse filaments of SC are specific to individual evolutionary lines (branches) of Eukaryotes, although the structure of SC is generally much conserved [5, 16, 17]. After the discovery of new proteins of the central space of SC, it was found that the Corona, Corolla, and SYP proteins were genus-specific and therefore obviously not conserved. In contrast, SYCE2 and TEX12 were traced in the phylogenesis back to the ancestors of the present-day Eumetazoa and were experimentally found in Hydra. The SYCE1 protein was found in the ancestors of modern Bilateria, while SYCE3 was found in the ancestors of vertebrates [18, 19]. These studies mainly used bioinformatics methods, aligning the amino acid sequences of proteins using BLAST method. The SIX6OS1 protein was studied experimentally in mice, and the Ecm11 and Gmc2 proteins were studied in yeast Saccharomyces cerevisiae.
Our goal was to identify the level of conservation for the recently discovered proteins of the central space of SC using a complex of bioinformatics methods: Ecm11, Gmc2, and SIX6OS1. We aimed to answer the question of whether their conservation is comparable to that of the previously studied proteins (SYCE and TEX).
MATERIALS AND METHODS
The search for orthologs of synaptonemal complex proteins was performed in the databases UniProtKB/TrEMBL (http://www.uniprot.org/), NCBI (http://www.ncbi.nlm.nih.gov/guide/), and GeneCards (http://www.genecards.org/cgi-bin/). We chose experimentally tested proteins, either full-size forms or forms that were similar in length to other orthologs. Conservation of the proteins was estimated on the basis of four criteria: (1) presence or absence of identical functional domains and presence of additional domains in the studied orthologs; (2) spread of the values of isoelectric points (pI) in the studied orthologs; (3) presence of a similar or differing secondary structure (alpha-helical conformation); and (4) presence of common amino acid motifs. We detected the presence of functional domains using CDART (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?) and identified the set and sequence of conserved amino acid motifs using MEME (http://meme.nbcr.net/meme/tools/meme). We used COILS (http://www.ch.embnet.org/software/COILS_form.html) to determine the protein secondary structure (the probability of forming alpha-helical conformation). The isoelectric points of proteins (pI) were identified using the Compute pI/Mw tool (http://web.expasy.org/compute_pi/).
The following proteins were used in the analysis. For the SYCE1 protein, we used orthologs in human Homo sapiens (SYCE1_HUMAN), mouse Mus musculus (NP_001137237.1), rat Rattus norvegicus (NP_001020229.2), Chinese and golden hamsters Cricetulus griseus (A0A3L7HWV6_CRIGR) and Mesocricetus auratus (A0A1U7Q6L0_MESAU), prairie vole Microtus ochrogaster (XP_026637282.1), common shrew Sorex araneus (XP_012791351.1), rabbit Oryctolagus cuniculus (A0A5F9D5A2_RABIT), panda Ailuropoda melanoleuca (G1L8P2_AILME), elephant Loxodonta africana (G3TJY0_LOXAF), opossum Monodelphis domestica (D3JUJ3_MONDO), platypus Ornithorhynchus anatinus (F7BBC0_ORNAN), whale Delphinapterus leucas (A0A2Y9MS50_DELLE), dolphin Tursiops truncatus (A0A2U3V0Z1_TURTR), latimeria Latimeria chalumnae (H3ALY1_LATCH), fishes Salmo salar (A0A1S3M6G7_SALSA) and Danio rerio (B3DFT5_DANRE), alligator Alligator sinensis (A0A1U8DW85_ALLSI), lizards Anolis carolinensis (G1KEZ2_ANOCA) and Gekko japonicus (XP_015261632.1), frog Xenopus laevis (A0A1L8F471_XENLA), lancelet Branchiostoma floridae (XP_002592847.1), mollusk Lottia gigantea (XP_009044517.1), and annelid worm Capitella teleta (ELU12842.1).
For the SYCE3 protein, we used orthologs in Homo sapiens (SYCE3_HUMAN), Mus musculus (NP_001156352.1), Rattus norvegicus (NP_001128725.1), Cricetulus griseus (G3IM77_CRIGR), Mesocricetus auratus (A0A1U7Q7Y3_MESAU), Microtus ochrogaster (XP_026638266.1), Sorex araneus (XP_004610643.1), mole rat Nannospalax galili (XP_008833003.1), Oryctolagus cuniculus (G1U293_RABIT), Ailuropoda melanoleuca (D2H6D0_AILME), Loxodonta africana (G5E7G0_LOXAF), Monodelphis domestica (A0A5F8G5S8_MONDO), chicken Gallus gallus (NP_001265057.1), pigeon Columba livia (XP_021136937.1), Ornithorhynchus anatinus (XP_028935400.1), Delphinapterus leucas (A0A2Y9MKQ2_DELLE), Tursiops truncatus (A0A2U3V110_TURTR), Latimeria chalumnae (H3A163_LATCH), Danio rerio (NP_001129458.1), Alligator sinensis (A0A1U7SGC3_ALLSI), lizards Anolis carolinensis (R4GBT4_ANOCA) and Pogona vitticeps (XP_020645218.1), turtle Terrapene carolina triunguis (XP_024064554.2), and frog Xenopus tropicalis (XP_002939574.2).
For the TEX12 protein, we used orthologs in Homo sapiens (TEX12_HUMAN), Mus musculus (NP_079963.1), Rattus norvegicus (NP_001178035.1), Cricetulus griseus (A0A061I546_CRIGR), Mesocricetus auratus (A0A1U7Q596_MESAU), Microtus ochrogaster (XP_005347360.1), Sorex araneus (XP_004604751.1), Nannospalax galili (XP_017653573.1), Oryctolagus cuniculus (XP_008259599.1), Ailuropoda melanoleuca (G1LBH9_AILME), Loxodonta africana (G3SS29_LOXAF), Monodelphis domestica (F6UHS6_MONDO), Ornithorhynchus anatinus (XP_028931825.1), Delphinapterus leucas (A0A2Y9M6Z8_DELLE), Tursiops truncatus (A0A2U3V1E3_TURTR), Latimeria chalumnae (M3XKC5_LATCH), Danio rerio (A0A140LH85_DANRE), Alligator sinensis (A0A3Q0GUI2_ALLSI), Anolis carolinensis (XP_008121887.1), Xenopus laevis (A0A1L8FLU4_XENLA), Gallus gallus (A0A3Q2UGL4_CHICK, A0A3Q2U5T2_CHICK, XP_001233099.3), Branchiostoma floridae (EEN46167.1), and mollusk Hydra vulgaris (R4NDD8_HYDVU). The rest of the studied proteins are listed in Table 1.
RESULTS
The conservation of the SYCE1–SYCE3 and TEX12 proteins was studied by Fraune et al. [18, 19] using certain bioinformatics methods and experimentally, while in our work, we use a set of selected bioinformatics methods. Therefore, in order to compare the conservation of these proteins with that of SIX6OS1, Ecm11, and Gmc2, we first studied the SYCE and TEX proteins. We studied 24 SYCE1 protein orthologs, 31 SYCE2 orthologs, 25 TEX12 orthologs (in vertebrates and invertebrates), and 24 SYCE3 orthologs (in vertebrates). The SYCE3 protein (length from 73 to 179 amino acid residues, aa) has one functional domain (Synaptonemal_3 superfamily) and one common motif for all objects (most species also have common motifs at the ends of molecules). The protein is acidic; almost all the orthologs have an alpha-helical region. Thus, this protein is rather conserved according to all the characteristics, but it is present only in vertebrates. Almost all SYCE1 orthologs (length from 190 to 359 aa) also have one SYCE1 domain. We have identified additional domains in some species. The annelid worm Capitella teleta and the lancelet Branchiostoma floridae have other domains. The protein is acidic. Almost the entirety of the molecule is arranged in several fragments of a pronounced alpha helix. As for common conserved motifs, there is only one between all studied species, while vertebrates (with the exception of fish species Danio rerio) have four common motifs. The protein of annelid worm Capitella teleta has no common motifs with other proteins (perhaps it is not an ortholog). Thus, SYCE1 is moderately conserved, but only within the vertebrate subphylum. All orthologs of the TEX12 protein (length from 122 to 270 aa), with the exception of the protein of hydroid Hydra vulgaris, have one large TEX12 domain. The scatter of isoelectric points (pI) is wide (from 3.7 to 8.8). The secondary structure differs greatly (some proteins have alpha-helical regions, and others do not). Thus, the physicochemical properties of this protein are not conserved. One common conserved amino acid motif is present in all orthologs, with the exception of the protein in platypus Ornithorhynchus anatinus. Most vertebrates have three common motifs. Thus, despite the fact that vertebrates and invertebrates have orthologs of this protein, the protein is not very conserved.
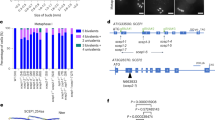
The last in this series is the SYCE2 protein (length from 113 to 327 aa, the objects of study are listed in Table 1). We detected no functional domains in the majority of orthologs. Human and a number of vertebrates have such domains, but those are not related to SYCE2. The protein is acidic. All orthologs have one common amino acid motif (highlighted by the light rectangle in Fig. 1); it has been changed in ascidian Ciona intestinalis and annelid worm Capitella teleta; most proteins have two common motifs in the second half of the molecule. Most orthologs have two fragments of the alpha-helical conformation, with varying degree of expression, and their location fully corresponds to the two main neighboring motifs: light gray and dark gray. In general, the secondary structure strictly corresponds to the identified motifs and is even more conserved than the primary structure, since the alpha-helical conformation can be identified even in the case where the motif has changed.
Conserved amino acid motifs in SYCE2 molecules of vertebrates and invertebrates: dolphin (Tt), whale (Dl), human (Hs), panda (Am), mouse (Mm), rabbit (Oc), elephant (La), turtle (Tct), alligator (As), latimeria (Lch), frog (Xl), chicken (Gg), lizard (Ac), opossum (Md), lancelet (Bf), platypus (Oa), sea anemone (Nv), mollusk (Lg), fish (Dr), ascidians (Ci), sea cucumber (Sj), and annelid worm (Ct). Identical motifs are marked by rectangles of identical color and size. N- and C-termini of protein molecules are shown.
Thus, the proteins of the central space of SC previously studied by Fraune et al. [18, 19] are not as conserved as expected. We compared the recently identified proteins of the central space of SC (SIX6OS1, Ecm11, and Gmc2) to the above-mentioned ones and established the following.
The SIX6OS1 protein (a component of the central element) was found mainly in vertebrates and only in a few invertebrates (Fig. 2, Table 1). We studied 26 orthologs with lengths ranging from 478 to 756 aa (including incomplete protein sequences). In most orthologs, almost the entire molecule, excluding the N-terminal fragment, consists of the S6OS1 domain. The sea urchin (Strongylocentrotus purpuratus) does not have the S6OS1 domain, but two other domains are present. The protein is acidic. The secondary structure is presented by one or more alpha-helical regions in the first half of the molecule.
Conserved motifs in SIX6OS1 molecules of vertebrates and invertebrates: human (Hs), rabbit (Oc), elephant (La), panda (Am), whale (Dl), dolphin (Tt), mouse (Mm), turtle (Tct), opossum (Md), chicken (Gg), lizard (Pv), platypus (Oa), latimeria (Lch), frog (Xt), oysters (Cgi), fish (Dr), gastropod (Lg), alligator (As), and sea urchin (Spu). Identical motifs are marked by rectangles of identical color and size. N- and C-termini of protein molecules are shown.
Almost all vertebrates have conserved sets of motifs (four motifs each) at the N- and C-termini of the molecule (Fig. 2). Invertebrates have only two common motifs at the N-terminus of the protein. The protein of the sea urchin has one common motif with the other objects, and even that motif is changed. The alligator has another motifs. It is possible that these two proteins are not orthologs (although they are annotated as putative SIX6OS1 proteins). Thus, the SIX6OS1 protein is fairly conserved only within the vertebrate subphylum, similar to the SYCE1, SYCE2, and TEX12 proteins.
The Ecm11 and Gmc2 proteins were previously studied in yeast Saccharomyces cerevisiae. We found orthologs of these proteins only in Ascomycete fungi (Table 1). We studied seven Ecm11 orthologs in representatives of different Ascomycete taxa. The length of the protein ranges from 302 to 997 aa. All studied proteins, with the exception of the protein in Pneumocystis murina, have the ECM11 domain at the C-terminus of the molecule. Some fungi have additional domains in the middle of the molecule. The isoelectric points of proteins (pI) vary over a wide range, as does the secondary structure of proteins. We also studied the set and arrangement of conserved motifs (Fig. 3). Very small common motifs (no longer than 50 aa) are shared by representatives of different Ascomycete taxa (motifs indicated by an asterisk and a lattice at the C‑terminus of the molecule). Thus, the Ecm11 orthologs are not conserved even within the same phylum of fungi (Ascomycetes). Other Eukaryotes do not have orthologs of these proteins.
Conserved amino acid motifs in Ecm11 proteins of seven species of Ascomycetes: Candida glabrata (Cgl), Saccharomyces cerevisiae (Sc), Cochliobolus sativus (Cs), Neurospora crassa (Nc), Emericella nidulans (En), Phialophora attae (Pa), and Pneumocystis murina (Pmu). Identical motifs are marked with identical symbols. N- and C-termini of protein molecules are shown. Notes are in text.
Another new protein, Gmc2, has been annotated in representatives of only one class of Ascomycetes, Saccharomycetes. We studied eight orthologs with lengths ranging from 168 to 228 aa. We found no functional domains in almost all of the orthologs. A common conserved motif was found at the C-terminus of the molecule (Fig. 4). Six of the eight orthologs have common motifs in the middle of the molecule.
Conserved amino acid motifs in Gmc2 protein molecules of fungi: Saccharomyces cerevisiae (Sc), Kluyveromyces marxianus (Km), Zygosaccharomyces parabailii (Zp), Kazachstania saulgeensis (Ks), Candida glabrata (Cgl), Hanseniaspora uvarum (Hu), Pichia kudriavzevii (Pk), and Lachancea quebecensis (Lq). Identical motifs are marked by rectangles of identical color and size. N- and C-termini of protein molecules are shown.
The isoelectric points of Gmc2, as with Ecm11, vary. The secondary structure (alpha helix) is present in all orthologs, but is expressed to varying degree. Thus, the level of conservation of the Gmc2 protein is as low as that of the Ecm11 protein.
DISCUSSION
As mentioned above, the proteins of the synaptonemal complex (SC) are specific to individual evolutionary lines (branches) of Eukaryotes, although the structure of the SC is quite conserved [5, 16, 17]. Our results support this conclusion. However, as a result of our study, we obtained a paradoxical data: the Ecm11 and Gmc2 proteins, present only in Ascomycete fungi, are less conserved even within this taxon than the SYCE and TEX proteins found in vertebrates and invertebrates. The degree of conservation of the SIX6OS1 protein is similar to the latter aforementioned proteins. Thus, we once again confirm the thesis that, along with the general set of meiotic proteins, each line of Eukaryotic evolution has developed its own proteins for the SC formation, which has a common structural plan in all Eukaryotes [5, 17, 20, 21]. To date, the answer to the question posed in the title of the unicity or universality of meiotic proteins is that there is a similarity of proteins within individual Eukaryotic lines.
In the early period of studying the structure of synaptonemal complexes, researchers were forced to limit themselves to studying their ultrastructure using electron microscopes. They accumulated many images of the ultrastructure of the central space and central elements of SC in plants, fungi, Invertebrates, and Vertebrates [22]. The table in this review contains information on the thickness and arrangement of the filaments forming the structure of the central space of SC in 52 species of organisms. These authors conditionally identified two types of ultrastructure of the central space: amorphous and lattice. Ascomycetes and mammals have the amorphous type: the arrangement of fine structures of the central space is less regular than in the lattice type, found in insects. The review [3] contains even more information that needs to be systematized. The width of the central space of SC in different organisms varies from 50 to 150 nm, and the thickness of its structural elements is in the range of 3 to 10 nm. With the advent of data on the structure of CS proteins, including the results obtained in the present study, significant work is required to reconcile the data of molecular (biochemical) and ultrastructural (electron microscopic) studies.
REFERENCES
Page, S.L. and Hawley, R.S., The genetics and molecular biology of the synaptonemal complex, Annu. Rev. Cell Dev. Biol., 2004, vol. 20, pp. 525—558. https://doi.org/10.1146/annurev.cellbio.19.111301.155141
Gao, J. and Colaiácovo, M.P., Zipping and unzipping: protein modifications regulating synaptonemal complex dynamics, Trends Genet., 2018, vol. 34, pp. 232—245. https://doi.org/10.1016/j.tig.2017.12.001
Zickler, D. and Kleckner, N., Meiotic chromosomes: integrating structure and function, Ann. Rev. Genet., 1999, vol. 33, pp. 603—754. https://doi.org/10.1146/annurev.genet.33.1.603
Moses, M.J., Chromosomal structures in crayfish spermatocytes, J. Biophys. Biochem. Cytol.,1956, vol. 2, pp. 215—219. https://doi.org/10.1083/jcb.2.2.215
Cahoon, C.K. and Hawley, R.S., Regulating the construction and demolition of the synaptonemal complex, Nat. Struct. Mol. Biol., 2016, vol. 23, pp. 369—377. https://doi.org/10.1038/nsmb.3208
Schmekel, K. and Daneholt, B., The central region of the synaptonemal complex revealed in three dimensions, Trends Cell Biol., 1995, vol. 5, pp. 239—242. https://doi.org/10.1016/s0962-8924(00)89017-0
Hawley, R.S., Solving a meiotic LEGO puzzle: transverse filaments and the assembly of the synaptonemal complex in Caenorhabditis elegans, Genetics, 2011, vol. 189, pp. 405—409. https://doi.org/10.1534/genetics.111.134197
Collins, K.A., Unruh, J.R., Slaughter, B.D., et al., Corolla is a novel protein that contributes to the architecture of the synaptonemal complex of Drosophila, Genetics, 2014, vol. 198, pp. 219—228. https://doi.org/10.1534/genetics.114.165290
Costa, Y., Speed, R., Ollinger, R., et al., Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis, J. Cell Sci., 2005, vol. 118, pp. 2755—2762. https://doi.org/10.1242/jcs.02402
Hamer, G., Gell, K., Kouznetsova, A., et al., Characterization of a novel meiosis-specific protein within the central element of the synaptonemal complex, J. Cell Sci., 2006, vol. 119, pp. 4025—4032. https://doi.org/10.1242/jcs.03182
Schramm, S., Fraune, J., Naumann, R., et al., A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility, PLoS Genet., 2011, vol. 7. e1002088. https://doi.org/10.1371/journal.pgen.1002088
Gόmez,-H. L., Felipe-Medina, N., Sáchez-Marti, M., et al., C14ORF39/SIX6OS1 is a constituent of the synaptonemal complex and is essential for mouse fertility, Nat. Commun., 2016, vol. 7, article number 13298. https://doi.org/10.1038/ncomms13298
Humphryes, N., Leung, W.-K., Argunhan, B., et al., The Ecm11–Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast, PLoS Genet., 2013, vol. 9. e1003194. https://doi.org/10.1371/journal.pgen.1003194
Voelkel-Meiman, K., Cheng, S.-Y., Morehouse, S.J., and MacQueen, A.J., Synaptonemal complex proteins of budding yeast define reciprocal roles in MutSγ-mediated crossover formation, Genetics, 2016, vol. 203, pp. 1091—1103. https://doi.org/10.1534/genetics.115.182923
Fraune, J., Schramm, S., Alsheimer, M., and Benavente, R., The mammalian synaptonemal complex: protein components, assembly and role in meiotic recombination, Exp. Cell Res., 2012, vol. 318, pp. 1340—1346. https://doi.org/10.1016/j.yexcr.2012.02.018
Bogdanov, Y.F., Grishaeva, T.M., and Dadashev, S.Y., Similarity of the domain structure of proteins as a basis for the conservation of meiosis, Int. Rev. Cytol., 2007, vol. 257, pp. 83—142. https://doi.org/10.1016/S0074-7696(07)57003-8
Grishaeva, T.M. and Bogdanov, Y.F., Conservation and variability of synaptonemal complex proteins in phylogenesis of eukaryotes, Int. J. Evol. Biol., 2014, vol. 2014, article number 856230. https://doi.org/10.1155/2014/856230
Fraune, J., Brochier-Armanet, C., Alsheimer, M., and Benavente, R., Phylogenies of central element proteins reveal the dynamic evolutionary history of the mammalian synaptonemal complex: ancient and recent components, Genetics, 2013, vol. 195, pp. 781—793. https://doi.org/10.1534/genetics.113.156679
Fraune, J., Brochier-Armanet, C., Alsheimer, M., et al., Evolutionary history of the mammalian synaptonemal complex, Chromosoma, 2016, vol. 125, pp. 355—360. https://doi.org/10.1007/s00412-016-0583-8
Ramesh, M.A., Malik, S.-B., and Logsdon, J.M., Jr., A phylogenomic inventory of meiotic genes: evidence for sex in giardia and an early eukaryotic origin of meiosis, Curr. Biol., 2005, vol. 15, pp. 185—191. https://doi.org/10.1016/j.cub.2005.01.003
Bogdanov, Y.F. and Grishaeva, T.M., Konservatizm, izmenchivost’ i evolyutsiya meioza (Conservatism, Variation and Evolution of Meiosis), Moscow: KMK, 2020, pp. 231—245, 260—277.
Westergaard, M. and von Wettstein, D., The synaptonemal complex, Ann. Rev. Genet., 1972, vol. 6, pp. 71—110. https://doi.org/10.1146/annurev.ge.06.120172.000443
Funding
The study was supported by the Russian Foundation for Basic Research, project no. 17-00-00430 (17-00-00429 KOMFI), and by the state assignment of the Vavilov Institute of General Genetics, Russian Academy of Sciences, topic no. 0112-2019-0002.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare they have no conflict of interest.
The present study does not contain any research using animals as subjects.
The present study does not contain any research involving people as subjects.
Additional information
Translated by A. Lisenkova
Rights and permissions
About this article
Cite this article
Grishaeva, T.M., Bogdanov, Y.F. Synaptonemal Complex Proteins: Unicity or Universality?. Russ J Genet 57, 912–919 (2021). https://doi.org/10.1134/S1022795421080068
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795421080068