Abstract
The K-ratio between diadducts (D) and monoadducts (M) induced by 8-methoxypsoralen (8-MOP) in the DNA packed in the head of bacteriophage λ and in the DNA of pBR322 plasmid under UV irradiation (λ ≥ 320 nm) was measured. The probabilities of UVR excision repair of 8-MOP monoadducts (P) and of SOS repair of 8-MOP diadducts and monoadducts (S and Sm) were determined. P was measured using an angular derivative of angelicin. It was demonstrated that P = 0.86. S and Smwere determined using bacteriophage λ11 treated with 8-MOP + UV (λ ≥ 320 nm) or 8-MOP + UV (λ ≥ 380 nm) and inoculated either onto bacteria pre-irradiated with short-wave UV light (λ = 254 nm) or onto bacteria with constitutive synthesis of the SOS regulon genes, as well as onto bacteria containing the pKM101 plasmid. In bacteriophage λ11, the level of W-reactivation (α) and the frequency of W-mutagenesis of clear mutations (m) were determined. It was demonstrated that 8-MOP diadducts (“crosslinks”) were repaired by the bacterial SOS system with the probability S = 0.28–0.29, and 8-MOP monoadducts were repaired by the bacterial SOS system with the probability Sm = 0.41 only with the participation of the MucA’2B enzyme, the genes for which were located in the conjugative plasmid pKM101.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Photosensitization of the bacteria and bacteriophages with 8-methoxypsoralen (8-MOP) plus UV (λ ≥ 320 nm) causes the formation of two types of DNA photoproducts, i.e., monoadducts (one 8-MOP molecule is covalently linked to a pyrimidine base) and diadductsor interstrand “crosslinks” (one 8-MOP molecule is covalently linked to two pyrimidine bases from complementary strands) [1–4].
In the analysis of the 8-MOP effect on DNA, the value of K, which characterizes the ratio between monoadducts and diadducts (M = K × D, where M is the number of monoadducts and D is the number of diadducts), is of critical importance. The K value depends on a number of factors. First, K depends on the UV wavelength used for illumination of 8-MOP solution. For example, when UV with wavelength longer than 380 nm is used, 8-MOP monoadducts are formed in DNA almost exclusively, since the secondary reaction that crosslinks a monoadduct with the complementary strand requires a much shorter UV wavelength (320–350 nm) [5]. Second, K depends on the DNA state. For example, DNA packed in the bacteriophage head is characterized by a lower K value compared to free DNA [6].
It is also necessary to take into account the possibility of repair of monoadducts and diadducts in the bacterial cell. Since 8-MOP monoadducts and diadducts block replication carried out by DNA polymerase III, it is reasonable to suppose that, in the case of inoculation of non-UV-irradiated E. coli ΔuvrΔrecA with 8-MOP treated plasmids and phage, each DNA monoadduct and diadduct is lethal because of the absence of excision repair (UVR) and post-replication repair (RecA) systems in these bacteria. In E. coli uvr+recA+, 8-MOP monoadducts are repaired by the enzymes of the UVR excision repair pathway with the same efficiency P (P is the probability of excision repair of monoadducts and cyclobutane pyrimidine dimers) as cyclobutane pyrimidine dimers [7]. However, 8-MOP monoadducts in the phage and plasmid DNA cannot be repaired by the post-replication recombination repair system (RecA). This is caused by the need for a second copy of the homologous genome (which is intact at this locus) to be present in the cell, since usually phages and plasmids infect the cell in one copy (the multiplicity of infection used in the experiment is much lower than one).
Diadducts (“crosslinks”) in the DNA of phages and plasmids are 100% lethal in the case of inoculation of non-UV-irradiated E. coli ΔuvrΔrecA and E. coli Δuvr strains, because they block replication and are not repaired. The crosslink repair requires excision of the linker arm on one of the complementary strands, which is carried out by the UVR excision repair enzymes, and then filling of the formed “gap.” 8-MOP diadducts in the DNA of phages and plasmids are also 100% lethal in the case of inoculation of non-irradiated uvr+recA+ bacteria, i.e., in the case of the absence of active SOS repair system in the cell, because despite the excision of linker arm by UVR enzymes, the system of post-replication RecA-recombination repair is unable to fill the gap owing to the absence of a second copy of the phage or plasmid genome in the cell. These findings were experimentally confirmed in [8]. In the DNA of phage λ inoculated onto non-irradiated E. coli uvr+recA+, the formation of one diadduct in DNA corresponded to approximately one lethal hit, while 8-MOP monoadducts were repaired with high efficiency by the UVR enzymes.
UV irradiation of the bacteria induces the SOS response system of DNA repair, the regulation of which is determined by two proteins, the LexA repressor protein and RecA protein [9, 10]. In E. coli, the SOS regulon contains the umuC and umuD genes [11], encoding the subunits of PolV (UmuD’2C), which is able to bypass the replication block caused by the lethal DNA defect, preferentially incorporating a noncomplementary nucleotide opposite the damaged one [12–14]. As a result of the process, which was named “translesion synthesis,” the survival rate of bacteria is increased and mutations are induced [15]. It was demonstrated that, in addition to PolV polymerase, an important role in this process is played by the activated RecA* protein (an asterisk denotes the activated state of the protein) [16, 17]. The RecA activation occurs as a result of binding to single-stranded DNA, which is formed in considerable amounts in UV-irradiated bacteria during replication and its blockage by lethal defects. The SOS repair proteins are capable of repairing and causing mutations not only in the bacterial chromosome but also in plasmids and DNA of bacteriophages. They are especially effective in temperate bacteriophages (for example, phage λ), are less effective in conditionally lethal bacteriophages (for example, phages T1, T3, T7 [18]), and are unable to repair lytic bacteriophages, such as T2 and T4, which kill the cell and inhibit the main intracellular processes almost at the moment of adsorption [19]. In honor of the discoverer of the effect, J. Weigle (1953), the phenomenon of the increase in survival rate and the frequency of mutations in phages and plasmids inoculated onto bacteria with pre-induced SOS system is called W‑reactivation and W-mutagenesis [9].
In the present study, the K-ratio between diadducts (D) and monoadducts (M) in the DNA packed in the head of bacteriophage λ and in plasmid DNA was measured. The probabilities of excision repair of monoadducts (P) and of SOS repair of 8-MOS diadducts and monoadducts (S and Sm) (according to [20]) were determined. The measurement of P was carried out using an angular derivative of angelic in, which owing to its structure is capable of forming monoadducts and is not capable of forming diadducts [20]. S and Sm were determined using bacteriophage λ11 treated with 8-MOS + UV (λ ≥ 320 nm) or 8-MOP + UV (λ ≥ 380 nm) and inoculated ontobacteria pre-irradiated with shortwave UV light (λ = 254 nm) or onto bacteria with constitutive synthesis of the SOSregulon genes, as well as onto bacteria containing the pKM101 plasmid.
MATERIALS AND METHODS
Bacterial Strains, Bacteriophages, and Plasmids
The Escherichia coli K12 strains: AB1157 F--- thr-1 leu-6 proA2 his-4 thi-1 argE3 lacY1 galK2 ara14 xyl-5 mtl-1 tsx-33 rpsL31 supE44; AB1886 uvrA6; AB2480 uvrA6 recA13; other markers are as in AB1157 (the strains were obtained from Prof. P. Howard-Flanders, USA); Escherichia coli DM1187 recA441 lexA51 sfiA11 arg+; other markers are as in AB1157 (the strains were obtained from Prof. V.A. Lantsov, St. Petersburg State University). The TK603 strains arg+ilvuvrA6; other markers as in AB1157; GW514 = TK603 (pKM101 mucAB+) were obtained from Prof. G.C. Walker (USA).
Bacteriophage λ11 (λv2v3) was obtained from R. Devoret (France). The pBR322 plasmid.
Bacterial Growth Media and Conditions
LB medium: 1% tryptone, 0.5% yeast extract, 0.5% NaCl; 0.7 and 1.8% LB agar were used as top and bottom agar for Petri dishes. The bacteria were grown in LB or LB plus 100 μg/mL ampicillin with constant shaking until middle exponential phase at 30°С. The transformation of bacterial cells treated with Ca2+ ions was carried out according to [21].
Buffers and Reagents
Tris buffer: 0.05 M Tris-HCl, 0.01 M NaCl, pH 7.5; TM buffer: 0.05 M Tris-HCl, 0.01 M NaCl, 0.001 M MgSO4, pH 7. 8.8-Methoxypsoralen (8-MOP) was purchased from Sigma. Angelicin was obtained from Prof. G. Rodighiero (Italy).
Irradiation of Bacteria, Bacteriophage, and Plasmids
DNA solution in Tris buffer and phage suspension in TM buffer containing 40 μg/mL 8-MOP or 200 μg/mL angelicin were irradiated at 4°С in Pyrex cuvettes. An SVD-120A high-pressure mercury lamp with a UFS-6 light filter served as a light source of λ ≥ 320 nm. An SVD-120A lamp with a ZhS-4 light filter was used as a light source of λ ≥ 380 nm. The distance between the lamp and the specimen was 20 cm. The bacterial suspension in TM buffer was irradiated with shortwave UV light (254 nm), the source of which was a BUV-15 low-pressure mercury lamp. The dose of UV light was measured with a UVD-4 dosimeter with a magnesium photocell.
W-Reactivation and W-Mutagenesis
Bacteria in the exponential phase were washed twice with TM buffer, concentrated five times in the same buffer, and irradiated at 20°С with a BUV-15 lamp (254 nm) with different doses. Bacteriophage λ adsorption on unirradiated and UV-irradiated cells was performed for 15 min in TM buffer at 37°C. Inoculation of phage–bacterium complex was carried out using the double-layer agar technique with the addition of the AB2480 indicator culture. Clear mutants were scored using bacteriophage λ11, which is capable of forming two one-step mutations, clear and vir. The spontaneous background of clear mutations for different phage preparations was 8 × 10–4–1 × 10–3. In bacteriophage treated with 8-MOP + UV (λ ≥ 320 nm), the frequency of clear mutations among survivors was determined. The level of W-mutagenesis was determined as the ratio of the mutation frequency upon inoculation of UV-irradiated (254 nm) cells with bacteriophage to the mutation frequency upon inoculation of unirradiated cells. The multiplicity of infection did not exceed 0.01.
To measure W-reactivation in the pBR322 plasmid, cells were treated with Ca2+ ions and then irradiated with different UV doses (254 nm). The level of W‑reactivation (α) for phage and plasmid was determined according to the formula α = NnnNii/NinNni, where Nnn is the titer of unirradiated phage and plasmid on unirradiated bacteria, Nii is the titer of irradiated phage and plasmid on irradiated bacteria, Nin is the titer of irradiated phage and plasmid on unirradiated bacteria, and Nni is the titer of unirradiated phage and plasmid on irradiated bacteria.
RESULTS
Inoculation of Unirradiated Bacteria with Phages and Plasmids
In the present study, P (the probability of UVR excision repair) was measured using an angular derivative of angelicin, which, owing to its structure, is capable of forming only monoadducts and is not capable of forming diadducts. The curves of inactivation kinetics for the pBR322 plasmid treated with angelicin + UV (λ ≥ 320 nm) during transformation of unirradiated E. coli AB1157uvr+recA+ and AB2480ΔuvrΔrecA strains were obtained (Fig. 1), which were described by the following formulas:
N/N0 = e–M, for strain AB2480, since each monoadduct is lethal;
N/N0 = e–(1 – P)M, for strain AB1157, since only each unrepaired monoadduct is lethal.
From the obtained slope ratio of the inactivation curves, σΑΒ1157/σΑΒ2480 = 0.14, we obtain M(1 – P)/M = (1 – P) = 0.14, and the probability of monoadduct excision repair P = 0.86.
To measure the value of K (M = KD), the kinetic curves of 8-MOP + UV (λ ≥ 320 nm) inactivation of pBR322 plasmid and bacteriophage λ were determined upon their inoculation onto unirradiated AB2480 and AB1157 bacteria (Fig. 2):
The kinetic curves of inactivaition for bacteriophage λ (a) and pBR322 plasmid (b) treated with 8-MOP + UV (λ ≥ 320 nm) upon inoculation or transformation of E. coli K12 strains: (1) AB1157 uvr+recA+; (2) AB2480 uvrA6 recA13. On the y axis, the survival rate, N/N0; on the x axis, the preparation exposure time (min).
N/N0 = e–D(1 + K), for strain AB2480, since each monoadduct and diadduct is lethal;
N/N0 = e–D[1+K(1 – P)], for strain AB1157, because each diadduct is lethal, and monoadduct is lethal with the probability (1 – P). From the obtained slope ratio equal to σAB2480/σΑΒ1157 = 1.7 for the bacteriophage and 3.3 for the plasmid and taking into account the previously obtained value of P = 0.86, the values of K are determined: σAB2480/σΑΒ1157 = (1 + K)/[1 + (1 – P)K]. K = 0.92 for bacteriophage λ; i.e., on average, one monoadduct is formed per one diadduct; and K = 4.3 for plasmid DNA; i.e., on average, about 4.5 monoadducts are formed per diadduct.
Inoculation of Phages and Plasmids onto UV-Irradiated AB1157 and AB1886 Bacteria, as well as onto Unirradiated DM1187 Bacteria
First, for the AB1157 and AB1886 uvrA6 bacteria, optimum doses of UV irradiation (254 nm) at which the SOS repair system is maximally induced in cells were determined. Figures 3 and 4 show data on the increase in the survival of phage λ1l (hereinafter denoted as λ) (Fig. 3) and pBR322 plasmid (Fig. 4) irradiated with either shortwave UV light (254 nm), inducing cyclobutane pyrimidine dimers as the main replication-blocking lethal DNA defects, or UV light with λ ≥ 320 nm in the presence of 8-MOP. In bacteriophage λ1l, the frequency of “clear” mutations was also determined. The UV dose for irradiation of phage and plasmid preparations was chosen so that the level of object inactivation upon inoculation onto unirradiated bacteria was approximately 10–3. The AB1157 and AB1886 uvrA6 bacteria were irradiated with different doses of shortwave UV light (254 nm). The level of W‑reactivation α for phages and plasmids was determined according to the formula α = NnnNii/NinNni (see Materials and Methods).
W-reactivation and W-mutagenesis of bacteriophage λ11 irradiated with UV (254 nm) (a) and treated with 8-MOP + UV (λ ≥ 320 nm) (b) upon infection of the E. coli K12 strains AB1157 uvr+recA+ (1, 2) and AB1886 uvrA6 (3, 4). Bacteria were irradiated with different doses of UV light (254 nm). (1 and 3) The curves of W-reactivation; (2 and 4) the curves of W-mutagenesis. On the x axis, UV doses (254 nm) on bacteria, in J/m2. On the y axis, α-level of W-reactivation and m, the frequency of clear mutations among the survivors. The UV (254 nm) dose on phage upon inoculation on to AB1157 was 140 J/m2; the phage survival on unirradiated AB1157 bacteria was 1.2 × 10–3. The UV (254 nm) dose on phage upon inoculation on to AB1886 was 20 J/m2; phage survival on unirradiated AB1886 bacteria was 1.1 × 10–3. The exposure time of phage with 8-MOP + UV (λ ≥ 320 nm) upon inoculation onto AB1157 was 55 min; the phage survival on unirradiated culture was 1.7 × 10–3 and on AB1886 was 45 min; the phage survival on unirradiated AB1886 bacteria was 1.4 × 10–3. The frequency of phage clear mutations on unirradiated AB1886 and AB1157 bacteria was 1.0 × 10–3.
W-reactivation of the pBR322 plasmid irradiated with UV (254 nm) and treated with 8-MOP + UV (λ ≥ 320 nm) upon infection of E. coli K12 strains AB1157 uvr+recA+ and AB1886 uvrA6. The UV doses (254 nm) of plasmid irradiation were 50 J/m2 (AB1886) and 210 J/m2 (AB1157); the survival on unirradiated cells was 1.5 × 10–3and 1.3 × 10–3, respectively. The plasmid exposure times to UV (λ ≥ 320 nm) + MOP were; 50 min (AB1886 uvrA6) and 90 min (AB1157); the plasmid survival rates on unirradiated bacteria were 1.2 × 10–3 (AB1886) and 1.4 × 10–3 (AB1157). Bacteria were irradiated with different doses of UV (254 nm). On the x axis, the doses of UV (254 nm) on bacteriain J/m2; on the y axis, α-level of W-reactivation.
As can be seen from Figs. 3 and 4, the survival of the phage and plasmids increases, and at the same time, in phage λ1l, the frequency of “clear” mutations increases because of the activity of the SOS repair system in bacteria pre-irradiated with different UV doses (254 nm). In the case of UV-irradiated (254 nm) phage (Fig. 3a) and plasmid (Fig. 4) containing cyclobutane pyrimidine dimers in DNA, the maximum values of α and mutation frequency m are achieved at a UV dose of about 30 J/m2 to bacteria for strain AB1157 and about 6 J/m2 for strain AB1886 uvrA6. A similar result was obtained for 8-MOP + UV (λ ≥ 320 nm) inactivated phage λ1l and plasmid, but only using AB1157 bacteria as a host, since in the case where AB1886 uvrA6 bacteria were used, SOS repair and induction of “clear” mutations in phage λ1l and SOS repair in the plasmid were not observed (Figs. 3b, 4).
In the case of inoculation of pre-UV-irradiated (254 nm) bacteria with phages and plasmids treated with different doses of 8-MOP + UV (λ ≥ 320 nm), SOS repair of monoadducts and diadducts in DNA should be taken into consideration. In this series of experiments, we used only strain AB1157 and did not use the mutant strain AB1886 uvrA6, since 8-MOP diadducts are not repaired because of the absence of the UVR enzymes that excise linker arm, while 8-MOP monoadducts (but not angelicin monoadducts) are not repaired by the bacterial SOS system, because the PolV enzyme is unable to bypass the replication block caused by 8-MOP molecule intercalated into DNA [22].
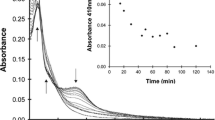
Figure 5a shows the curves of 8-MOP + UV (λ ≥ 320 nm) inactivation kinetics of phage λ upon inoculation onto AB1157 bacteria not irradiated and UV-irradiated (254 nm) with the optimum dose (30 J/m2). The dependences of N/N0 on the amount of diadducts D formed in DNA are as follows:
The kinetic curves of inactivaition for bacteriophage λ treated with 8-MOP + UV(λ ≥ 320 nm) upon inoculation onto (a) unirradiated and UV-irradiated (254 nm) (with optimum dose of 30 J/m2) AB1157 uvr+recA+ bacteria and (b) unirradiated AB1157 uvr+recA+ bacteria (1) and unirradiated DM1187 recA441 lexA51 sfiA11 arg+ bacteria. Sowing of the bacteriophage-infected DM1187 bacteria on Petri dishes at 30°С (without adenine) (3) and at 42°С (+adenine) (2). On the y axis, the survival rate N/N0; on the x axis, phage exposure time (min).
N/N0 = e–D[1+K(1 – P)], bacteria are not irradiated (each diadduct is lethal and monoadducts not repaired by the UVR enzymes are lethal (see formulas above)).
N/N0 = e–D[(1 – S)+K(1 – P)], bacteria are UV-irradiated (254 nm) with the optimum dose of 30 J/m2. This variant takes into account that diadducts are repaired by the SOS enzymes with probability S, and monoadducts are repaired by the UVR excision repair enzymes with probability P, but are not repaired by the PolV enzyme of the bacterial SOS system (according to [22]).
The slope ratio of σ–/σ+inactivation curves for unirradiated and irradiated bacteria is 1.42 (Fig. 5a). Then,
D[(1 – S) + K(1 – P)]/D[1 + K(1 – P)] = [(1 – S) + K(1 – P)]/[1 + K(1 – P)] = 1/1.42, or
S = [1 + K(1 – P)] (1 – 1/1.42) = (1 + 0.9 × 0.14) × (1 – 1/1.42) = 1.026 × 0.42/1.42 = 0.29.
Therefore, the probability of 8-MOP diadduct repair by the SOS enzymes and the simultaneous formation of a mutation in the DNA of bacteriophage λ is 0.29, and, hence, the probability of the lethal effect of 8-MOP diadduct on bacteriophage λ is 1 – 0.29 = 0.71.
In the next series of experiments, the mutant E. coli strain DM1187 recA441 sfiA11 lexA51arg+ was used for the phage λ inoculation; the rest of the markers were as in AB1157. This strain is characterized by constitutive synthesis of all SOS proteins (the lexA51 mutation) [23], and the RecA protein (RecA441) is activated at elevated temperatures (42°С) in the presence of adenine (100 μg/mL) in the medium [24]. The sfiA11 mutation provides the strain viability under the constitutive synthesis of the SOS regulon. First, the induction level of the SOS system was tested. For this purpose, the DM1187 and AB1157 cells (control strain with closed SOS system) were transformed with the pColD plasmid and comparative intensities of cell luminescence were determined. Strain DM1187 is characterized by the luminescence intensity approximately 100 times higher than that of strain AB1157, which points to the openness of the SOS system in strain DM1187.
Figure 5b shows the curves of 8-MOP + UV (λ ≥ 320 nm) inactivation kinetics of phage λ upon inoculation onto strains AB1157 and DM1187 without preliminary UV (254 nm) irradiation. Petri dishes with DM1187 cells containing adenine (100 μg/mL) in the top layer were incubated at 42°С. The dependences of N/N0 on the amount of diadducts D formed in DNA are as follows:
N/N0 = e–D[1 + K(1 – P)], AB1157 bacteria (each diadduct is lethal, and monoadducts not repaired by the UVR enzymes are lethal);
N/N0 = e–D[(1 – S) + K(1 – P)], DM1187 bacteria (monoadducts not repaired by UVR enzymes are lethal, as well as diadducts not repaired by the SOS system).
The kinetic curve slope ratio is σAB1157/σDM1187 = 1.38. Then, S = 1.026 × 0.38/1.38 = 0.28. Consequently, S = 0.28, which almost coincides with the result for pre-UV-irradiated (254 nm) AB1157 bacteria (Fig. 5a).
Comparison of the kinetic curve slopes of 8-MOP + UV (λ ≥ 320 nm) inactivation of phage λ obtained by sowing of phage-infected DM1187 bacteria on Petri dishes at 30°С (without adenine) and at 42°С (+adenine) makes it possible to assess the contribution of activated RecA* to the process of SOS repair of 8-MOP diadducts. As can be seen from Fig. 5b, the ratio of these slopes, equal to σ30/σ42, is almost the same as the slope ratio of σΑΒ1157/σ42 = 1.38. Therefore, it can be suggested that the activated RecA* protein is critical for the SOS repair of 8-MOP diadducts.
W-Reactivation of 8-MOP Monoadducts with the Help of MucA’2B Polymerase
In the next series of experiments, the E. coli TK603 uvrA6 and GW514 uvrA6 strains containing the pKM101mucAB+ plasmid were used for phage λ inoculation [25, 26].
The bacteriophage preparation was either irradiated with UV light (254 nm) or treated with 8-MOP + UV (λ ≥ 380 nm), and the phage DNA thus contained either cyclobutane pyrimidine dimers or mainly 8-MOP monoadducts [5]. Figure 6 shows the kinetic curves of 8-MOP + UV (λ ≥ 380 nm) inactivation (a) and UV inactivation (254 nm) (b) of phage λ upon inoculation onto GW514 bacteria not irradiated and pre-UV-irradiated (254 nm) with the optimum dose (6 J/m2). The dependences of N/N0 on the amount of monoadducts M formed in the phage DNA (Fig. 6a) are as follows:
The kinetic curves of inactivaition for bacteriophage λ treated with 8-MOP + UV(λ ≥ 380 nm) (a) and irradiated with UV (254 nm) (b) upon inoculation onto unirradiated and UV-irradiated (254 nm) (with optimum dose of 6 J/m2) GW514 and TK603 uvrA6 bacteria. (а): (1) inoculation onto unirradiated GW514 bacteria; (2) inoculation onto UV-irradiated (254 nm) GW514 bacteria with the dose of 6 J/m2. (b): (1) inoculation onto unirradiated TK603 uvrA6 bacteria; (2) inoculation onto UV-irradiated (254 nm) TK603 uvrA6 bacteria with the dose of 6 J/m2; (3) inoculation onto unirradiated GW514 bacteria; (4) inoculation onto GW514 bacteria irradiated with the dose of 6 J/m2. On the y axis, the survival rate, N/N0; on the x axis, the exposure time of the phage preparation (min) (a) and the UV dose (254 nm), J/m2 (b).
N/N0 = e–М, bacteria are not irradiated (each monoadduct is lethal);
N/N0 = \({{{\text{e}}}^{{-{\text{M}}{{{({\text{1}}-S)}}_{{\text{m}}}}}}}\), bacteria are UV-irradiated (254 nm) at the optimum dose of 6 J/m2. In this variant, it is taken into account that monoadducts are repaired by the MucA’2B enzyme with the probability of Sm, but are not repaired by the PolV (UmuD’2С) enzyme of the bacterial SOS system (upon inoculation of 8‑MOP + UV (λ ≥ 380 nm) treated phage onto E. coli strain TK603 uvrA6, the effects of W-reactivation and W-mutagenesis are absent). In addition, in the case where unirradiated GW514 bacteria are inoculated with bacteriophage containing 8-MOP monoadducts in the DNA, the so-called P-reactivation phenomenon is not observed. In other words, phage survival rates on GW514 and TK603 are equal. The slope ratio of the inactivation curves σ−/ σ+ for unirradiated and UV-irradiated (254 nm) GW514 bacteria is 1.7 (Fig. 6a). Then, (1 – Sm) = 1/1.7, and, therefore, the probability of SOS repair of 8-MOP monoadduct with the participation of the MucA’2B polymerase Sm = 1 – 1/1.7 = 0.41, and, hence, the probability of the lethal effect of monoadduct is 1 – 0.41 =0.59.
The kinetic curves of UV inactivation (254 nm) of a phage whose DNA contains lethal cyclobutane pyrimidine dimers are somewhat different (Fig. 6b). In this case, the phenomenon of “P-reactivation” takes place. Namely, the survival rate of UV-irradiated (254 nm) phage on non-UV-irradiated (254 nm) GW514 bacteria containing the pKM101 mucAB+ plasmid exceeds the survival rate of this phage on unirradiated TK603 uvrA6 bacteria. Therefore, MucA’2B polymerase is able to bypass the replication block caused by cyclobutane pyrimidine dimers (as well as angelicin monoadducts (data not shown)), even if there is no SOS induction in the cell, but it is not able to bypass the replication block caused by 8-MOP monoadducts without additional participation of the enzymes of the bacterial SOS system. Pre-UV-irradiation (254 nm) of GW514 bacteria only considerably enhances W-reactivation of UV-irradiated (254 nm) phage, in particular, owing to the increase in expression of the mucA and mucB genes [27]. Because of this, Fig. 6b shows four kinetic curves of UV-irradiated (254 nm) phage inactivation: on strain GW514 not irradiated and pre-irradiated with UV light (254 nm) at a dose of 6 J/m2 and also on strain TK603 uvrA6 not irradiated and pre-irradiated with the optimumUV (254 nm) dose of 6 J/m2.
DISCUSSION
The data of the presented study indicate that, to assess the relative contributions of 8-MOP monoadducts and diadducts as lethal and mutagenic defects in the DNA of bacteriophages and plasmids, it is necessary to take into account, first, the inability of the bacterial SOS system, which includes PolV polymerase, to bypass the replication block caused by 8-MOP monoadducts and, second, the inability of the plasmid MucA’2B polymerase to bypass the replication block caused by 8-MOP monoadducts (the lack of “P-reactivation”) without additional assistance from the cellular SOS system.
It should be noted that, in the study [28], in which it was first demonstrated that 8-MOP diadducts (“crosslinks”) determined the formation of mutations in bacteriophage λ during SOS repair, there was some misperception of the putative role of 8-MOP monoadducts in SOS mutagenesis. In [28], it was demonstrated that angelicin, which can form exclusively monoadducts, highly efficiently induced SOS mutagenesis in the case where the phage was inoculated onto pre-UV-irradiated bacteria. On the basis of these findings, it was suggested that 8-MOP monoadducts were also able to induce SOS mutagenesis in bacteriophages. However, as was demonstrated in the present study, 8-MOP monoadducts induced SOS mutagenesis only upon the use of plasmid (pKM101) MucA’2B polymerase, but not bacterial PolV polymerase (UmuD’2C).
The data presented in this study on different abilities of MucA’2B and UmuD’2C polymerases to bypass the replication blocks formed by “weak intercalator” angelicin or “strong intercalator” 8-MOP monoadduct represent an interesting, but not the only, example of the influence of the degree of overlap between the aromatic rings of flat intercalators and nitrogenous bases in DNA on the SOS pathway of the mutation formation in bacteria. In particular, it was demonstrated that N-2-acetylaminofluorenecarcinogen, the flat molecule of which is intercalated into the DNA double helix, upon SOS repair induced the formation of frame shift mutations in the 5'-GGCGCC sequence of bacterial chromosome, the frequency of which did not depend on UmuD’2C polymerase, but increasedconsiderably in the presence of MucA’2B polymerase [29].
The question of the relative contribution of monoadducts and diadducts to the lethal and mutagenic effects of 8-MOP + UV (λ ≥ 320 nm) on bacterial cells remains unresolved. In this case, the task is complicated because of the presence of a second genome in the bacterial cell and the efficient process of post-replication recombination repair, which depends on a considerable group of genes (RecA, RecBC, RecE, etc.). In particular, it can be suggested that PolV polymerase, with the participation of the proteins of the recombination repair system, is to some extent able to bypass the replication block caused by 8-MOP monoadducts.
REFERENCES
Dall’Acoua, F., Marciani, S., and Rodighiero, G., Interstrand cross-linkages occurring in the photoreactions between psoralen and DNA, FEBS Lett., 1970, vol. 9, pp. 121—123. https://doi.org/10.1016/0014-5793(70)80330-1
Cole, R.S., Psoralen monoadducts and interstrand cross-links in DNA, Biochim. Biophys. Acta, 1971, vol. 254, pp. 30—39. https://doi.org/10.1016/0005-2787(71)90111-0
Dall’Acqua, F., Marciani, S., Ciavatta, G., and Rodighiero, G., Formation of inter-strand cross-linkings in the photoreactions between furocoumarins and DNA, Z. Naturforsch., 1971, vol. 26b, pp. 561—565. https://doi.org/10.1515/znb-1971-0613
Rodighiero, G. and Dall’-Acoua, F., Biochemical and medical aspect of psoralens, Photochem. Photobiol., 1976, vol. 24, pp. 647—653. https://doi.org/10.1111/j.1751-1097.1976.tb06887.x
Chatterjee, P.K. and Cantor, C.R., Photochemical production of psoralen DNA monoadducts capable of subsequent photocross-linking, Nucleic Acids Res., 1978, vol. 5, pp. 3619—3633.https://doi.org/10.1093/nar/5.10.3619
Hradechna, Z. and Kittler, L., Photobiology of furocoumarins: various types of cross-linking with DNA and their interference with the development of lambda phage, Acta Virol.,1982, vol. 26, pp. 305—311.
Belogurov, A.A., Zuev, A.V., and Zavil’gel’skii, G.B., Repair of 8-methoxypsoralen monoadducts and diadducts in bacteriophages and bacteria, Mol. Biol. (Moscow), 1976, vol. 10, no. 4, pp. 857—867.
Cassuto, E., Gross, N., Bardwell, E., and Howard-Flanders, P., Genetic effects of photoadducts and photocross-links in the DNA of phage λ exposed to 360 nm light and trimethyl-psoralen or khellin, Biochim. Biophys. Acta, 1977, vol. 475, pp. 589—600. https://doi.org/10.1016/0005-2787(77)90319-7
Weigle, J.J., Induction of mutation in a bacterial virus, Proc. Natl. Acad. Sci. U.S.A., 1953, vol. 39, pp. 628—636. https://doi.org/10.1073/pnas.39.7.628
Radman, M., SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis, Molecular Mechanisms for Repair of DNA, Hanawalt, P. and Setlow, R.B., Eds., New York: Plenum, 1975, part A, pp. 355—367.https://doi.org/10.1007/978-1-4684-2895-7_48.
Kato, T. and Shinoura, Y., Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light, Mol. Gen. Genet., 1977, vol. 156, pp. 121—131.https://doi.org/10.1007/BF00283484
Tang, M., Bruck, L., Eritja, R., Tarner, J., et al., Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD’2C mutagenic complex and RecA protein, Proc. Natl. Acad. Sci. U.S.A., 1998, vol. 95, pp. 9755—9760.https://doi.org/10.1073/pnas.95.17.9755
Tang, M., Shen, X., Frank, E.G., Woodgate, R., et al., UmuD’2C is an error-prone DNA polymerase, Proc. Natl. Acad. Sci. U.S.A., 1999, vol. 96, pp. 8919—8924.https://doi.org/10.1073/pnas.96.16.8919
Reuven, N.B., Arad, G., Maor-Shoshani, A., and Livneh, Z., The mutagenesis protein UmuC is a DNA polymerase activated by UmuD’, RecA, and SSB, and is specialized for translesion replication, J. Biol. Chem., 1999, vol. 274, pp. 31763—31766. https://doi.org/10.1074/jbc.274.45.31763
Defais, M., Lesca, C., Monsarrat, B., and Hanavalt, P., Translesion synthesis is the main component of SOS repair in bacteriophage lambda DNA, J. Bacteriol., 1989, vol. 171, pp. 4938—4944. https://doi.org/10.1128/jb.171.9.4938-4944.1989
Jiang, Q., Karata, K., Woodgate, R., Cox, M.M., et al., The active form of DNA polymerase V is UmuD’2C-RecA*-ATP, Nature, 2009, vol. 460, pp. 359—363. https://doi.org/10.1038/nature08178
Patel, M., Jiang, Q., Woodgate, R., Cox, M.M., et al., A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V, Crit. Rev. Biochem. Mol. Biol., 2010, vol. 45, pp. 171—184. https://doi.org/10.3109/10409238.2010.480968
Zavil’gel’skii, G.B., Belogurov, A.A., and Kryuger, D.N., W-reactivation and W-mutagenesis in bacteriophages λ and T7: a comparative study of the action of ultraviolet radiation (254 nm) and of the photosensitizing agents, 8-methoxypsoralen and angelicin, Genetika (Moscow), 1982, vol. 18, no. 1, pp. 24—34.
Krivisky, A.S., Esipova, V.V., and Zuev, A.V., Induction of mutations and repair in bacteriophages after photosensitizing action of 8-methoxypsoralen, Biol. Zentralbl., 1979, vol. 98, pp. 175—183.
Ashwood, M.J. and Grant, F., Conversion of psoralen DNA monoadducts in Escherichia coli to interstrand DNA cross-links by near UV light (320—360 nm): inability of angelicin to form crosslinks in vivo, Experientia, 1977, vol. 33, pp. 384—386. https://doi.org/10.1007/BF02002841
Sambrook, J., Fritsch, E.F., and Maniatis, T.,Molecular Cloning: A Laboratory Manual, New York: Cold Spring Harbor Lab., 1989, 2nd ed.
Zavil’gel’skii, G.B. and Kotova, V.Y., SOS repair of 8‑methoxypsoralene monoadducts in DNA of lambda bacteriophage and plasmids is mediated by MucA’2B, but not UmuD’2C (PolV) polymerase, Russ. J. Genet., 2013, vol. 49, no. 12, pp. 1195—1199. https://doi.org/10.1134/S1022795413120144
Pacelli, L., Edmiston, S., and Mount, D., Isolation and characterization of amber mutation in the lexA gene of Escherichia coli K12, J. Bacteriol., 1979, vol. 137, pp. 568—573.https://doi.org/10.1128/JB.137.1.568-573.1979.573
George, J. and Buttin, G., Prophage induction and cell division in Escherichia coli: I. Further characterization of the thermosensitive mutation tif-1 whose expression mimics the effectof the UV-irradiation, Mol. Gen. Genet., 1972, vol. 119, pp. 139—152. https://doi.org/10.1007/BF00269133
Dobson, P.P. and Walker, G.C., Plasmid pKM101 mediated W-reactivation in Escherichia coli K12 and Salmonella typhimurium LT: genetic dependence, kinetics of induction and effect of chloramphenicol, Mutat. Res., 1980, vol. 71, pp. 25—41. https://doi.org/10.1016/0027-5107(80)90004-4
Hauser, J., Levine, A.S., Ennis, D.G., Chumakov, K.M., et al., The enhanced mutagenic potential of the MucAB proteins correlates with the highly efficient processing of the MucA protein, J. Bacteriol., 1992, vol. 174, pp. 6844—6851.https://doi.org/10.1128/jb.174.21.6844-6851.1992
Elledge, S.J.. and Walker, G.C., The muc genes of pKM101 are induced by DNA damage, J. Bacteriol., 1983, vol. 155, pp. 1306—1315.https://doi.org/10.1128/JB.155.3.1306-1315.1983
Belogurov, A.A. and Zavilgelsky, G.B., Mutagenic effect of furocoumarin monoadducts and cross-links on bacteriophage lambda, Mutat. Res., 1981, vol.84, pp. 11—15. https://doi.org/10.1016/0027-5107(81)90045-2
Janel-Bintz, R., Maenhaut-Michel, G., and Fuchs, R.P., MucAB but not UmuDC proteins enhance (–2) frame shift mutagenesis induced by N-2-acetylaminofluorene at alternating GC sequences, Mol. Gen. Genet., 1994, vol. 245, pp. 279—285. https://doi.org/10.1007/BF00290107.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by N. Maleeva
Rights and permissions
About this article
Cite this article
Kotova, V.Y., Abilev, S.K. & Zavilgelsky, G.B. The Ratio between Lethal and Mutagenic Damages in the DNA of Plasmids and Bacteriophages Induced by 8-Methoxypsoralen Plus UV (λ ≥ 320 nm) Treatment. Russ J Genet 57, 795–803 (2021). https://doi.org/10.1134/S1022795421070097
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795421070097