Abstract
In silico analysis of the promoter region of the At4g01870 gene of Arabidopsis thaliana (L.) Heynh. showed the presence of ABRE, W-box, RAV1-A, MYB, and LFY cis-elements in the sequence. These regulatory motifs bind the transcription factors involved in responses to abscisic acid (ABA) and stresses. Stable transgenic plants carrying the β-glucuronidase gene under the control of the 5'-deletion fragments of the At4g01870 promoter were obtained. According to the results of histochemical staining of transformants, gene expression was induced by abiotic stress and was most significant in the conductive tissues of the root, leaves, and sepals as well as in flowers. The study of At4g01870 gene expression by RT-PCR confirmed that the gene transcript content increased after the exposure of plants to a solution of NaCl or at 37°C and after ABA treatment; however, hypothermia almost unchanged the level of accumulation of the transcripts. Along with ABA, expression of the At4g01870 gene was induced by indolylacetic and salicylic acids and ethylene precursor 1-aminocyclopropane-1-carboxylic acid; it was hardly regulated by methyl jasmonate and inhibited by cytokinin. The TolB-like protein, encoded by the At4g01870 gene, functions as a type of platform, based on which protein complexes are assembled. Given the previously identified ABA-binding properties of the protein At4g01870 and the presence of the ABA-dependent cis-elements in the promoter of its coding gene, it can be assumed that the protein encoded by the At4g01870 gene allows to control the hormonal signals in the cell, providing a structural platform for the interaction of specific effector proteins, trans-factors and ion channels.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Abscisic acid (ABA) accumulates in plant cells under the influence of stress conditions, such as drought, salinity, abnormal temperatures, or the action of pathogens [1]. In such a case, the hormone induces the expression of many ABA-dependent genes. The canonical ABA signal pathway contains several mandatory elements. It includes the PYR/PYL/RCAR receptors, which after binding ABA acquire the ability to interact with PP2C protein phosphatases and inhibit their activity [2]. Suppression of PP2C unblocks the process of autoactivation of the SnRK2 kinase, which, in the active state, phosphorylates the trans-factors regulating the transcription of ABA-dependent genes [3]. The result of the analysis of promoters of such ABA-induced genes showed that the regulation of ABA gene expression is determined by the presence of cis-elements responsible for the binding of ABA-dependent transcription factors in the promoter zones of the genes. Among the cis-elements involved in ABA- and stress-regulated gene activation, sequences, such as ABRE (ABA response element), MYB, and W-box, were identified [4–6]. ABA-regulated genes, which have similar motifs in the promoter regions, are targets for signaling networks of ABA signal pathways and they are of particular interest for understanding the molecular mechanisms of the action of the hormone.
Earlier, among ABA-binding proteins, we identified a family of ABA-activated proteins containing a TolB-like beta-propeller domain (based on PD40 repeats) and a C-terminal domain that carries homology with dipeptidyl peptidase IV (DPP IV) [7]. Proteins of the family were similar to the TolB protein of bacteria Escherichia coli by its amino acid sequence. The TolB protein is an important structural and functional domain of the bacterial Tol-Pal system, protecting the cell from the penetration of peptide toxins–colicins and maintaining the water-salt metabolism of the bacterial cell [8]. Genes, homologues of this family, were found in 46 plant genomes, and the proteins encoded by them revealed a high degree of interspecific similarity, indicating the conservative function of this group of proteins [7]. Three homologous genes in Arabidopsis thaliana encode proteins of this family. Two of these genes, At1g21670 and At1g21680, are expressed in the leaves, and the third gene, At4g01870, is mainly expressed in roots and flowers. Despite the similarity of nucleotide sequences, homologous genes are characterized by specific hormone-induced regulation and are located in the genome under different promoters, which probably determines their strictly specific time and place of expression.
According to the results obtained using affinity chromatography methods, as well as direct and competitive enzyme immunoassay systems, At4g01870 expressed in bacterial cells reversibly and specifically interacts with ABA, and its N-terminal region binds to the hormone [7, 9]. Along with ABA, ethylene, methyl jasmonate, indolylacetic acid, and salicylic acid may be involved in regulation of At4g01870 expression [10]. This distinctive feature of the studied gene opens up additional possibilities for identifying the points of intersection of the ABA regulatory network with signaling pathways of other hormones and is indicative of the complex interaction of various hormonal systems controlling gene expression.
The purpose of the study was the analysis of the promoter of the At4g01870 gene for the subsequent identification of its possible functions.
MATERIALS AND METHODS
Plant material, cultivation, and treatment conditions.Arabidopsis thaliana (L.) Heynh. ecotype Columbia 0 plants were cultivated in soil or Petri dishes on Murashige and Skoog medium (MS) with half the concentration of mineral salts, containing agar (0.5% agar) at an illumination of 100 μE/(m2 s), 23°С, and light duration of 16 h. At the age of 1–2 weeks, depending on the purpose of the experiment, the plants were transferred onto filter paper moistened with liquid MS and incubated under increased (37°C) or decreased temperature (4°C), and also on hormone or NaCl (200 mM) solutions during the time specified in the description of the results. In the study, trans-zeatin and indolylacetic acid (IAA) at a concentration of 5 × 10–6 M, ABA and methyl jasmonate at a concentration of 5 × 10–5 M, salicylic acid and ethylene precursor 1-aminoacylcyclopropane-1-carboxylic acid (ACC) at a concentration of 10–5 M were used. Optimal concentrations of phytohormones were determined at preliminary experiments. For analysis of hormone-dependent gene expression by RT-PCR after reverse transcription, filters with seedlings on hormone solutions in MS medium were exposed to light for 3 h in accordance with the protocol of Kilian et al. [10]. Control plants were placed on MS medium containing ethyl alcohol at a concentration used to prepare phytohormone solutions (0.1%). For fluorometric analysis of β-glucuronidase activity, the leaves of 7-week-old plants (natural aging period) grown in soil were sprayed with hormone solutions and fixed in liquid nitrogen after 24 h.
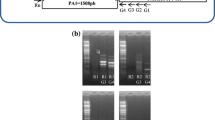
Production and histochemical analysis of Arabidopsis transgenic plantscontaining PAt4g01870-GUS chimeric construct. The 5'-deletion fragments of promoter region of the At4g01870 gene –384 + 15 (P384), –298 + 15 (P298), and –197 + 15 (P197) bp amplified by PCR were cloned into the pCXGUS-P plasmid (Arabidopsis Biological Resource Center). The required primers (Table 1) were selected using the VectorNTI Suite 9 program (Invitrogen, United States). The correctness of obtained amplicons and their cloning into a plasmid was proved by sequencing at ZAO Evrogen (Moscow, Russia). The resulting vector constructs containing 5'‑deletion fragments of the regulatory zone of the At4g01870 gene were used for the production of Ara-bidopsis transgenic plants by agrobacterial transformation of peduncles [11]. Transgenic plants with inserted genetic constructs were selected on a selective medium in the presence of antibiotic hygromycin at a concentration of 25 μg/mL. The presence of inserted genetic constructs was proved by PCR using a set of special primers.
Fluorometric analysis of β-glucuronidase enzyme activity was performed according to the method proposed by Jefferson et al. [12].
For histochemical analysis of GUS-activity in the tissues of transgenic Arabidopsis plants, the method of Vitha et al. [13] was used.
Analysis ofAt4g01870gene expression by RT-PCR. Total RNA was isolated from Arabidopsis seedlings using TRIzol® (Invitrogen) according to the manufacturer’s recommendations (http://www.invitrogen.com/). Synthesis of cDNA and subsequent RT-PCR analysis was performed using a LightCycler 96 amplifier (Roche, Switzerland) according to the procedure described previously [14]. The effectiveness of hormonal treatment and the effects of stressors were assessed by measuring the expression of stress-inducible genes and marker genes for hormones. The following primers were used for RT-PCR analysis: for At4g01870 gene—F: CGGACTAAAGTCGGCGGTGTCT; R: CACGACCGTCCTATCATCACTCA; for marker gene of ABA action RAB18—F: TCGCATTCGGTCGTTGTATTGT; R: AGTAAACAACACACATCGCAGGA. The expression level of the target genes was normalized to the expression level of the polyubiquitin gene (UBQ10). Primers used for the UBQ10 gene–F: GCGTCTTCGTGGTGGTTTCTAA; R: GAAAGAGATAACAGGAACGGAAACA.
Experiments were performed in three biological replicates. The significance of differences between the experimental and control samples was assessed using Student’s t-test. The figures show the mean and standard errors of the mean.
RESULTS
In Silico Analysis of the Promoter Region of the At4g01870 Gene
Using the AGRIS online software resource (http:// agris-knowledgebase.org/AtcisDB/atcisview.html?id= At4g01870) [15], a bioinformatical search for the proposed regulatory sequences for At4g01870 gene was conducted and the scheme for their location in the promoter was made (Fig. 1). In the sequence of the promoter region of the At4g01870 gene (384 bp), which is almost equal to the full intergenic spacer between the 3'-end of At4g1880 gene and the 5'-end of the ArabidopsisAt4g01870 gene, eight potential regulatory motifs were identified. It should be noted that regulatory elements of the gene, which can affect its expression, might also exist more remotely from the structural zone.
Scheme of the arrangement of the proposed cis-elements in the promoter of the At4g01870 gene. The AGRIS program (http://agris-knowledgebase.org/AtcisDB/atcisview.html?id=At4g01870) was used for the identification of regulatory elements. “A” in the ATG translation initiation site is designated as +1. The first base in the 5'-direction before ATG is designated as –1.
Among the detected cis-elements, there is an ABA‑dependent ABRE (A-like binding site element) element, which has a conservative TACGTGGA sequence, with which transcription factors of the AREB/ABF family (ABRE-binding factors) interact [4]. These trans-factors belong to the bZIP subfamily of proteins containing domains with a “basic leucine zipper” (bZIPs) structure. Involvement in the response to dehydration, salinity, and treatment with ABA are known among their possible functions. Plants over-expressing these factors demonstrated increased resistance to drought [16]. In addition, Ca2+, one of the most important secondary messengers in transmitting various endogenous signals, for example, ABA signal in stomatal plant cells, is involved in ABRE-mediated transcription [17].
The promotor of At4g01870 includes two sequences (CACCAACC and ACCAACC) interacting with MYB transcription factors [5, 18]. It is known that the A-rabidopsis genome contains 126 R2R3-MYB genes, some of which are mediators of ABA-dependent initiation of gene expression. One of the trans-factors of the MYB family, MYB96, is also induced by auxin and represents a common link or convergence point of the signaling pathways of auxin and abscisic acid. It should be noted that the response system via MYB/MYC factors functions more slowly than the bZIP-ABRE system, which is associated with the need for de novo protein synthesis [19].
In the At4g01870 promoter, there are three W-box elements, including the conservative sequence TTGACC (TTGAC(C/T)). W-box sequences are bound by transcription factors of the WRKY family, which includes 72 representatives in Arabidopsis [6]. It is known that WRKY trans-factors act in descending regions of at least two ABA receptor circuits: cytoplasmic PYR/PYL/RCAR and ABAR/GUN5/CHLH complex localized in chloroplasts. WRKY trans-factors are important components of plants' signaling networks coordinating the expression of a wide variety of genes. However, they can act as both positive and negative regulators [6].
In the promoter region of the At4g01870 gene, CCATTG regulatory sequence was also detected, with which LFY (or LEAFY, floral meristem identity) trans-factor, which regulates the transcription of a number of genes responsible for the organogenesis of Arabidopsis flowers, interacts [20, 21]. Possibly, LFY-dependent induction of the At4g01870 gene is associated with its direct participation in the development of flower structures during the transition to the reproductive stage: according to a study of the expression of this gene by DNA microarray, gene transcripts accumulated mainly in flowers and roots.
Another regulatory cis-element identified in the At4g01870 promoter, RAV1-A (CAACA), binds with the trans-factors of the RAV (Related to ABI3/VP1) subfamily, which belongs to the AP2/ERF family (APETALA2/Ethylene Responsive Factor). AtRAV1 is expressed predominantly during germination and in the early stages of growth and development of Arabidopsis and is suppressed by exogenous ABA and brassinosteroids. In addition, it can be induced by wounding and low temperature [22] and inhibited by salinity and lack of moisture [23]. The RAV1 protein contains a conservative repressor RLFGV domain and is able to act as a negative regulator of plant development [24] and a positive regulator of the aging process [25].
Thus, the bioinformatics search data confirmed the hormone- and stress-regulated function of the cis-elements of the At4g01870 promoter and suggested the involvement of the gene in signaling processes and responses to environmental factors or certain groups of phytohormones.
Histochemical Analysis of the Expression of the At4g01870 Gene Promoter under Stress Conditions and Treatment with Hormones
For studying the regulation of the expression of the At4g01870 gene, we produced Arabidopsis transgenic lines carrying the GUS reporter gene under the control of 5'-deletion fragments of the At4g01870 promoter –384 + 15 (P384), –298 + 15 (P298), and ‒197 + 15 (P197) bp (Fig. 2). The size of the maximal promoter fragment practically coincides with the size of the intergenic spacer of At4g1880 and At4g01870 genes. Fragments of shorter length allowed us to consistently exclude part of the cis-elements recognized by the trans-factors of MYB and WRKY families.
According to the obtained results, transformants with shortened variants of promoters (P298 and P197) resistant to the selective antibiotic hygromycin did not show GUS activity either by the histochemical staining of tissues or by measuring the enzyme activity using the fluorescent method. This suggests that the deletion of a –384–298-bp promoter fragment, including two potential MYB cis-elements and possibly other unidentified regulatory sequences, led to a loss of promoter activity (Figs. 1 and 2), which is extremely interesting and requires further detailed study of this fragment.
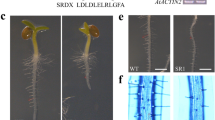
In plants with the largest promoter fragment (P384), β-glucuronidase activity was most significant in the conductive tissues of the root, leaves, and sepals as well as in the flower, root tip, and trichomes (Fig. 3A). The preferential expression of the GUS gene under the control of the promoter of the At4g01870 gene in conductive tissues indicated a tissue-specific expression pattern of the gene. The treatment of 2-week-old plants with cytokinin (trans-zeatin 5 × 10–6 M) for 5 h did not have a visible effect on the intensity of histochemical staining of leaf tissues (Fig. 3B-b), whereas incubation on ABA solution (5 × 10–5 M) for 5 h increased the intensity of staining compared with the control variant (Fig. 3B-c). Under salt stress (200 mM NaCl, 5 h), the intensity of staining of the stomata and bases of trichomes did not change compared to the control variant. The highest content of the At4g01870 product was in the root and leaf conducting systems. It consistently decreased in the midvein, veins of the second and third order, and mesophyll (data not shown). At increased and decreased temperatures, changes in the expression of the GUS-construct were multidirectional. Under heat stress (37°C, 5 h), the intensity of staining increased, especially in the stomata and bases of trichomes (Fig. 3B-d), while cold stress (4°C, 5 h) decreased the staining intensity and increased its unevenness (Fig. 3B-e). The obtained pattern of the distribution of GUS activity indicated tissue-specific regulation of the expression of the At4g01870 gene by stressors of a different nature.
Histochemical analysis of GUS activity. GUS activity expression was detected in (Aa–Ae) 2-week-old and (Af) 5-week-old transgenic A. thaliana plants incubated after cultivation and appropriate treatment with phytohormones and stressors in X‑Gluc solution overnight. For the transformation of plants, a construct where the GUS gene was under the control of the maximal promoter fragment (P384) of the At4g01870 gene was used. (A) Localization of GUS activity in various parts and organs of transgenic plants grown under normal conditions without effect of any exogenous factors: (a) the root tip, (b) root, (c) cross section of the leaf, (d) leaf, (e) area of trichomes and (f) flower; (B) leaf of 2-week-old transgenic A. thaliana plants (a) grown under control conditions, (b) treated with BAP, and (c) ABA for 5 h, exposed to temperature of (d) 37°C and (e) 4°C.
Measurements of β-glucuronidase activity by the fluorometric method in transgenic A. thaliana plants carrying the largest deletion fragment of the promoter (P384 bp) corresponded, in general, to the data of calorimetric staining. ABA, NaCl, and hyperthermia increased the expression of the GUS gene under the control of the At4g01870 promoter (151.5 ± 14.1, 147.2 ± 0.7, and 168.5 ± 3.8%, respectively, relative to the control), cytokinin and hypothermia did not have a significant effect (97 ± 1.2 and 112.2 ± 1.8%) at least within the investigated time interval.
Thus, the results of the analysis of the expression of the marker GUS gene under the control of a promoter fragment of the At4g01870 gene suggested that the At4g01870 gene and its product may be involved in the formation of resistance to adverse environmental effects with the involvement of ABA.
Quantitative Analysis of At4g01870 Gene Expression
The results of histochemical studies of the activity of the of promoter At4g01870 gene were compared with the data of the analysis of the expression of this gene by RT-PCR in wild-type A. thaliana plants. At all studied stages of ontogenesis, exogenous ABA activated the accumulation of transcripts of the RAB18 gene (Responsive to ABA) used as a marker for the action of this phytohormone (Fig. 4a). A completely different pattern of regulation by abscisic acid was observed for the At4g01870 gene. In 7-day-old Col 0 seedlings, ABA activated the accumulation of mRNA of this gene by approximately four times; however, ABA did not cause significant changes in its expression in 2-week-old plants, and, on the contrary, suppressed its activity in the leaves of 7-week-old plants (Fig. 4b). Thus, the features of regulation of the expression of the At4g01870 gene by exogenous ABA during ontogenesis, positive or negative, are probably mediated by the action of ontogenetic regulatory factors.
Effect of ABA on the relative content of transcripts of (a) RAB18 and (b) At4g01870 genes in 1-, 2-, and 7-week-old wild-type A. thaliana plants. The plants were treated for 3 h (1) control) with water or (2) with ABA solution (5 × 10–5 M). Total isolated RNA was analyzed by RT-PCR. UBQ10 was used as a reference gene. Mean values of three independent determinations ± SE are shown.
The involvement of ABA in the formation of stress response suggested the stress-dependent pattern of expression of the At4g01870 gene. Indeed, the content of gene transcripts after 3 h of exposure of 2‑week transgenic plants at 37°C increased in the aerial part of plants (Fig. 5a), and the content of transcripts of the studied gene increased only in the roots when plants were incubated with NaCl solution (200 mM) (Fig. 5b). However, hypothermia hardly changed the level of accumulation of transcripts during this time, which corresponded to the results of the analysis of the expression of the РAt4g01870-GUS construct. However, it should be noted that the response to salinity had a pronounced organ-specific character and was limited to roots.
Effect of (a) hyperthermia in the aerial part and (b) salinity in the root on the expression of the At4g01870 gene in 2‑week-old wild-type A. thaliana plants. The plants were kept for 3 h (control, 1) under normal conditions, (a, 2) under increased temperature 37°С, (b, 2) in NaCl solution (200 mM). The isolated RNA was analyzed by RT-PCR. UBQ10 was used as a reference gene. Mean values of three independent determinations ± SE are shown.
Reprogramming of the genome expression and the initiation of the cascade of defense mechanisms under stress are usually associated with the participation of the complex of hormonal systems. Since some cis-elements of the At4g01870 promoter (MYB, WRKY), along with ABA, are involved in the regulation by IAA, jasmonic and salicylic acids, and ethylene, we estimated how treatment with these hormones alters the expression of the At4g01870 gene in wild-type plants (Fig. 6). After 3 h of exposure of 7-day-old seedlings on IAA (10–6 M), salicylic acid (10–5 M), and ethylene precursor ACC (10–5 M) solutions, the accumulation of At4g01870 transcripts increased at least twice. The exception was methyl jasmonate (5 × 10–5 M), the reaction to which was less significant, and trans-zeatin (5 × 10–6 M), which suppressed the expression of this gene. Similar results obtained using DNA microarray technology are presented in the eFP database [10]. They were also partially confirmed by the fluorometric method used for determination of β-glucuronidase activity in A. thaliana transgenic plants, carrying РAt4g01870-GUS construct (P384). After 24 h of incubation of transformants on IAA, salicylic acid, and ethylene precursor ACC solutions, the enzyme activity significantly increased and amounted to 145.7 ± 1.5, 155.5 ± 9.2, and 152 ± 2.0% of the control, respectively. At the same time, methyl jasmonate had no significant effect (107.8 ± 9.2%). All these results allowed us to attribute the studied gene to genes involved in the wide range of protective reactions involved to some extent in hormonal signaling.
Hormonal regulation of At4g01870 gene expression in 7-day-old wild-type A. thaliana seedlings. The plants were treated for 3 h in (1) water, (2) methyl jasmonate solution (5 × 10–5 M), (3) ABA solution (5 × 10–5 M), (4) ACC solution (10–5 M), (5) trans-zeatin solution (5 × 10–5 M), (6) IAA solution (5 × 10–6 M), (7) brassinolide (5 × 10–5 M), and (8) salicylic acid (10–5 M) solutions. The relative content of transcripts of the At4g01870 gene was analyzed by RT-PCR. UBQ10 was used as a reference gene. Mean values of three independent determinations ± SE are shown.
DISCUSSION
As a result of our in silico analysis of the promoter region of the At4g01870 gene, the presence of cis-elements in its sequence responsible for the binding of ABA-dependent transcription factors was shown. Among them, such ABA-dependent motifs as ABRE, W-box, RAV1-A, MYB, and LFY were identified.
The genes with ABRE motifs in their promoter regions are the main targets of ABF/AREB transcription factors [4] activated by SnRK2-type protein kinases and, therefore, can be the target genes of the ABA signal transduction chain, including PYR/PYL/RCAR receptors [2]. The AREB/ABF factors belong to the bZIP trans-factor subfamily, group A, which has nine homologs in the Arabidopsis genome. It was shown that some of them are induced in response to dehydration, salinity, and treatment with ABA. Therefore, the presence of ABRE motifs in the At4g01870 promoter suggests that the products of this gene are involved in the physiological reactions associated with dehydration and salinity. Indeed, salt stress contributed to an increase in gene expression and resulted in the increased accumulation of the protein encoded by it according to the results of Western blot analysis [9]. The stress-dependent activation of the accumulation of the At4g01870 protein corresponded to the function of the TolB protein of Escherichia coli, necessary for maintaining the water–salt balance and integrity of the outer membrane. It is known that, in addition to ABA, Ca2+ is involved in ABRE-mediated transcription. This means that various ion channels or proteins regulating their activity can be among the targets of At4g01870.
Similarly to the TolB E. coli protein, At4g01870 may also be associated with responses to the action of pathogens. The involvement of Arabidopsis proteins, TolB homologues, in detoxification reactions in response to the treatment with the biopathogen Pseudomonas was identified in proteomic studies of Mueller et al. [26]. The regulation of such a response is probably the result of the specific action of not a single but a group of phytohormones, which are in close interaction with each other. Salicylic and jasmonic acids, and ethylene can be associated with protective mechanisms providing resistance to pathogens along with ABA. The nature of the interaction of ABA with these phytohormones can be both synergistic and antagonistic. All these phytohormones, as was shown in our study, regulated the expression of At4g01870, acting, probably, via MYB or WRKY trans-factors.
The functions of At4g01870 mediated by trans-factors of the WRKY family are not limited to participation in responses to biotic stresses. It is known that one of the members of the WRKY family, WRKY39, is a positive regulator of the signaling pathways of salicylic acid and methyl jasmonate, which is activated in response to heat stress [6]. It is possible that the induction of At4g01870 by increased temperatures, demonstrated in our studies, is also hormone-dependent, but this assumption requires additional experimental verification.
In addition to response to external signals associated with hormone-dependent reactions, At4g01870 is probably directly involved in the implementation of ontogenetic programs. According to bioinformatics data, the accumulation of At4g01870 transcripts reaches its maximum at the stage of early green cotyledons during seed embryogenesis and then during their germination. At the vegetative stage, preferential expression of the gene is concentrated in the conductive tissues of the root, leaves, and sepals, which is consistent with the results of histochemical analysis of transformants (Fig. 3). During the transition to the reproductive stage, mRNA and At4g01870 protein accumulated in inflorescences [9]. The molecular mechanism of this process was probably determined by the LFY-dependent induction of At4g01870 gene integrated into the morphogenesis process of flower structures.
Functions of the At4g01870 gene and the protein encoded by it are probably associated with complex interactions of various stimuli and growth inhibitors and the switching of the hormone regulation vector from positive to negative and vice versa. Thus, at the juvenile stage, expression of the At4g01870 gene in response to ABA increased, while it decreased in aging leaves (Fig. 4b). Along with feedback suppression of the synthesis of endogenous ABA or hormone balance shift, the specificity of the response could be provided by binding to various transcription factors acting as activators or repressors of gene expression. Such a mechanism, in particular, is characteristic for the WRKY family, representatives of which can act as both positive and negative regulators, and the same WRKY protein can sometimes have both roles depending on the specific reaction [6].
The specificity of the response to ABA suggests the existence of not the only linear transmission pathway of the signal of this hormone in the plant cell but the presence of a complete regulatory network capable of forming various physiological reactions [2]. In the At4g01870 promoter, we identified three W-boxes bound by transcription factors of the WRKY family. These trans-factors are capable of acting not only in the descending parts of the PYR/PYL/RCAR receptor chain, but they are also important components of the ABA signal transduction chain GUN5-WRKY40-ABI5 ABA [27]. When an ABA signal is transmitted through this chain, the H-subunit of the Mg-chelatase acts as an antagonist of the transcriptional repressors of the WRKY family and removes the inhibition of the ABI5 gene. We have previously shown that abi5-1 and gun5-1 mutants lack significant ABA-dependent regulation of expression of the At4g01870 gene and its homologues At1g21670 and At1g21680 [28]. All these results suggest that these genes are target genes for the ABA GUN5-WRKY40-ABI5 signal transduction chain. It should also be noted that, in addition to ABA, the ABI5 transcription factor could function as an integrator of signaling of phytohormones, such as auxins, cytokinins, gibberellic acid, and brassinosteroids, providing a balanced response of plants to abiotic stress [29].
Thus, the At4g01870 gene is integrated into a complex network system of plant responses to ABA and other phytohormones. It is known that the product of this gene contains conservative WD40 repeats—sequences of 40 amino acids with a high content of tryptophan, proline, and aspartate responsible for a certain structural organization of the protein. If at least three such repeats are present, the protein molecule may have a conformation of the so-called beta-propeller domain. Such a structural unit is able to perform the function of a platform, based on which the protein, participating in a wide variety of processes are assembled: signal transfer, assembly of a transcriptional apparatus, synthesis and degradation of proteins, and many other complexes. Considering ABA-binding properties of the protein provided by N-terminal fragment and the presence of cis-elements binding ABA-dependent trans-factors in the promoter of the gene encoding it, we can assume that the protein encoded by the At4g01870 gene allows to controll the transduction of the hormonal signal in cell, providing a structural platform for the interaction of specific effector proteins, trans-factors, and ion channels.
REFERENCES
Vishwakarma, K., Upadhyay, N., Kumar, N., Yadav, G., Singh, J., Mishra, R.K., Kumar, V., Verma, R., Upadhyay, R.G., Pandey, M., and Sharma, S., Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects, Front. Plant Sci., 2017, vol. 8: 161.
Hubbard, K.E., Nishimura, N., Hitomi, K., Getzoff, E.D., and Schroeder, J.I., Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions, Genes Dev., 2010, vol. 24, pp. 1695–1708.
Fujii, H., Chinnusamy, V., Rodrigues, A., Rubio, S., Antoni, R., Park, S.Y., Cutler, S.R., Sheen, J., Rodriguez, P.L., and Zhu, J.K., In vitro reconstitution of an abscisic signalling pathway, Nature, 2009, vol. 462, pp. 660–664.
Fujita, Y., Yoshida, T., and Yamaquchi-Shinozaki, K., Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants, Physiol. Plant., 2013, vol. 147, pp. 15–27.
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L., MYB transcription factors in Arabidopsis, Trends Plant Sci., 2010, vol. 15, pp. 573–581.
Bakshi, M. and Oelmüller, R., WRKY transcription factors: Jack of many trades in plants, Plant Signal. Behav., 2014, vol. 9: e27700.
Demidenko, A.V., Kudryakova, N.V., Karavaiko, N.N., Kazakov, A.S., Cherepneva, G.N., Shevchenko, G.V., Permyakov, S.E., Kulaeva, O.N., Oelmüller, R., and Kusnetsov, V.V., The ABA-binding protein AA1 of Lu-pinus luteus is involved in ABA-mediated responses, Russ. J. Plant Physiol., 2015, vol. 62, pp. 161–170.
Bonsor, D.A., Hecht, O., Vankemmelbeke, M., Sharma, A., Krachler, A.M., Housden, N.G., Lilly, K.J., Moore, G.R., and Kleanthous, C., Allosteric β-propeller signaling in TolB and its manipulation by translocating colicins, EMBO J., 2009, vol. 28, pp. 2846–2857.
Bartashevich, D.A., Karavaiko, N.N., and Kusnetsov, V.V., The novel ABA-binding protein encoded by At4g01870 gene in A. thaliana is able to interact with RNA in vitro, Dokl. Biochem. Biophys., 2014, vol. 457, pp. 128–131.
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., D’Angelo, C., Bornberg-Bauer, E., Kudla, J., and Harter, K., The AtGenE-xpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses, Plant J., 2007, vol. 50, pp. 347–363.
Clough, S.J. and Bent, A.F., Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana, Plant J., 1988, vol. 16, pp. 735–743.
Jefferson, R.A., Kavanagh, T.A., and Bevan, M.V., GUS-fusions: beta-glucuronidase as a sensitive and versalite gene fusion marker in higher plants, EMBO J., 1987, vol. 6, pp. 3901–3907.
Vitha, S., Benes, K., Phillips, J.P., and Gartland, K.M.A., Histochemical GUS analysis, Methods Mol. Biol., 1995, vol. 44, pp. 185–193.
Danilova, M.N., Kudryakova, N.V., Voronin, P.Yu., Oelmüller, R., Kusnetsov, V.V., and Kulaeva, O.N., Membrane receptors of cytokinin and their regulatory role in Arabidopsis thaliana plant response to photooxidative stress under conditions of water deficit, Russ. J. Plant Physiol., 2014, vol. 61, pp. 434–442.
Yilmaz, A., Mejia-Guerra, M.K., Kurz, K., Liang, X., Welch, L., and Grotewold, E., AGRIS: Arabidopsis Gene Regulatory Information Server, an update, Nucleic Acids Res., 2011, vol. 39, database issue, pp. 1118–1122.
Kim, J.B., Kang, J.Y., and Kim, S.Y., Over-expression of a transcription factor regulating ABA responsive gene expression confers multiple stress tolerance, Plant Biotech. J., 2004, vol. 2, pp. 459–466.
Tuteja, N., Abscisic acid and abiotic stress, Plant Signal. Behav., 2007, vol. 2, pp. 135–138.
Roy, S., Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome, Plant Signal. Behav., 2016, vol. 11: e1117723.
Shinozaki, K. and Yamaguchi-Shinozaki, K., Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways, Curr. Opin. Plant Biol., 2000, vol. 3, pp. 217–223.
Lamb, R.S., Hill, T.A., Tan, Q.K.G., and Irish, V.F., Regulation of APETALA3 floral homeotic gene expression by meristem identity genes, Development, 2002, vol. 129, pp. 2079–2086.
Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M., LEAFY controls floral meristem identity in Arabidopsis, Cell, 1992, vol. 69, pp. 643–659.
Feng, C.Z., Chen, Y., Wang, C., Kong, Y.H., Wu, W.H., and Chen, Y.F., Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development, Plant J., 2014, vol. 80, pp. 654–668.
Fu, M., Kang, H.K., Son, S.H., Kim, S.K., and Nam, K.H., A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA, Plant Cell Physiol., 2014, vol. 55, pp. 1892–1904.
Hu, Y.X., Wang, Y.H., Liu, X.F., and Li, J.Y., Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development, Cell Res., 2004, vol. 14, pp. 8–15.
Woo, H.R., Kim, J.H., Kim, J.Y., Kim, J.G., Lee, U., Song, I.J., Kim, J.H., Lee, H.Y., Nam, H.G., and Lim, P.O., The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis, J. Exp. Bot., 2010, vol. 61, pp. 3947–3957.
Mueller, S., Hilbert, B., Dueckershoff, K., Roitsch, T., Krischke, M., Mueller, M.J., and Berger, S., General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in A-rabidopsis, Plant Cell, 2008, vol. 20, pp. 68–85.
Shang, Y., Yan, L., Liu, Z.Q., Cao, Z., Mei, C., Xin, Q., Wu, F.Q., Wang, X.F., Du, S.Y., Jiang, T., Zhang, X.F., Zhao, R., Sun, H.L., Liu, R., Yu, Y.T., et al., The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition, Plant Cell, 2010, vol. 22, pp. 1909–1935.
Vinogradov, N.V., Danilova, M.V., Kudryakova, N.V., Kusnetsov, V.V., and Kulaeva, O.N., ABA-dependent regulation of the expression of AA1 homologs (abscisic acid activated 1) in A. thaliana mutants with impaired synthesis or transduction of ABA signal, Mater. Mezhd. nauch. konf. “Fiziologiya rastenii—teoreticheskaya osnova innovatsionnykh agro- i fitobiotekhnologii” (Proc. Int. Sci. Conf. “Plant Physiology—Theoretical Basis for Innovative Agro- and Phytobiotechnologies”), Kaliningrad, 2014, pp. 33–35.
Skubacz, A., Daszkowska-Golec, A., and Szarejko, I., The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk, Front. Plant Sci., 2016, vol. 7: 1884.
Funding
This study was supported by the Russian Science Foundation (project no. 14-14-00584).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
This article is devoted to professor Olga Nikolaevna Kulayeva to the outstanding plant physiologist and biochemist in connection with 90-year anniversary.
Translated by V. Mittova
Abbreviations: ABA—abscisic acid; ACC—1-aminocyclopropane-1-carboxylic acid; MS—Murashige and Skoog nutrient medium; RT-PCR—real-time polymerase chain reaction after reverse transcription.
Rights and permissions
About this article
Cite this article
Vinogradov, N.V., Andreeva, A.A., Danilova, M.N. et al. The ABA- and Stress-Induced Expression of the ArabidopsisthalianaAt4g0180 Gene Is Determined by the Cis-Elements Responsible for Binding the ABA-Dependent Trans-Factors . Russ J Plant Physiol 66, 521–529 (2019). https://doi.org/10.1134/S102144371902016X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S102144371902016X