Abstract
CP190 protein is one of the key components of Drosophila insulator complexes, and its study is important for understanding the mechanisms of gene regulation during cell differentiation. However, Cp190 mutants die before reaching adulthood, which significantly complicates the study of its functions in imago. To overcome this problem and to investigate the regulatory effects of CP190 in adult tissues development, we have designed a conditional rescue system for Cp190 mutants. Using Cre/loxP-mediated recombination, the rescue construct containing Cp190 coding sequence is effectively eliminated specifically in spermatocytes, allowing us to study the effect of the mutation in male germ cells. Using high-throughput transcriptome analysis we determined the function of CP190 on gene expression in germline cells. Cp190 mutation was found to have opposite effects on tissue-specific genes, which expression is repressed by CP190, and housekeeping genes, that require CP190 for activation. Mutation of Cp190 also promoted expression of a set of spermatocyte differentiation genes that are regulated by tMAC transcriptional complex. Our results indicate that the main function of CP190 in the process of spermatogenesis is the coordination of interactions between differentiation genes and their specific transcriptional activators.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Specialization of cells during multicellular organism development is accompanied by an activation of developmental genes by enhancers [1] and insulator proteins play a key role in the regulation of promoter-enhancer interactions [2]. They form physical contacts between special DNA regions—insulators, thereby providing spatial interactions between promoters and cis-regulatory elements. In a similar way, insulator proteins block nonspecific promoter-enhancer interactions and establish barriers between adjacent domains on chromosomes.
In Drosophila melanogaster, nearly a dozen regulatory proteins that bind to insulators in various combinations have been found [3–6]. CP190, which interacts with most of the known insulators, is one of the key proteins among them. CP190 is involved in DNA contact formation, regulation of three-dimensional nuclear architecture, and blocking of promoter-enhancer interactions [7–9]. In some cases, CP190 acts as a transcription factor and attracts regulatory proteins and epigenetic modifiers to target genes [6, 10–12].
Loss of CP190 function is accompanied by transcriptional alterations that affect, among others, homeotic genes encoding key developmental regulators [8, 13, 14]. Null mutations of Cp190 are recessive lethal, leading to the death of D. melanogaster at the pupal stage [15, 16], making it challenging to investigate the regulatory effects of CP190 in adult cells. Therefore, its effects on gene expression and nuclear morphology have been studied either in cultured cells or at early developmental stages [8, 13], giving rise to numerous limitations when interpreting results.
Here we used spermatogenesis of D. melanogaster as a model of cellular differentiation. During the differentiation of male germline cells, more than 1000 specific genes are activated under the effect of specialized transcription factors tMAC and tTAF [17, 18]. However, only a small portion of genomic tMAC and tTAF binding sites are in close proximity to target genes [18]. This may indicate the regulation of genes through a network of spatial interactions between the regulatory elements of genome involving insulator proteins. To study the effect of CP190 on gene regulation during cell differentiation, we developed a genetic system that allowed us to selectively examine the phenotype of lethal Cp190 mutations in adult male germline cells. Using this approach, we evaluated the effect of CP190 protein on gene expression in testes and analyzed its effect on the regulation of spermatocyte differentiation genes.
EXPERIMENTAL
Molecular cloning and Drosophila transformation. A rescue construct (GenBank ON783212) containing the coding sequence of the Cp190 gene under the control of Ubi63E gene promoter was created based on the pUAST-attB vector, as described in RESULTS. PhiC31-mediated transgenesis of the y–w–;P{y[+t7.7]=CaryP}attP40;M{vas-int.B}ZH-102D line harboring the attP40 integration site in the second chromosome was performed using this construct, producing y1w67;CP190-mCD8-GFP line [19].
Fly stocks. All D. melanogaster lines were maintained at 25°C on standard feed. The following fly stocks were used for the experiments: y1w67;CP190-mCD8-GFP carried a rescue construct; y1w67;nanos-Cre harbored nanos-Cre transgene integrated into attP40 site [18, 20], y1w67;+/+;Cp1902,e1/TM6B,Tb1 [15] and y1w67;+/+;Cp1903,e1/TM6,Tb1 [16], carried mutant alleles of Cp190. By standard genetic crosses, flies y1w67;CP190-mCD8-GFP;Cp1902,e1/TM6,Tb1 and y1w67;nanos-Cre;Cp1903,e1/TM6,Tb1 were obtained from these stocks. Males with Cre/loxP-mediated recombination were obtained by crossing nanos-Cre carried males with rescue construct carried females. When performing control experiments in RNA-seq assay, we used y1w67;{Ubi>stop>mCD8-GFP} line with {Ubi>stop>mCD8-GFP} transgene integrated into attP40 site [20].
RNA extraction and reverse transcription. For the expression analysis of whole testes, 25 pairs of testes were dissected and RNA was isolated using TRIZOL reagent (Invitrogen, USA, 15596018). 1 μg of total RNA was used for cDNA production; reverse transcription was performed using Superscript II Reverse Transcriptase (Invitrogen, 18080093) with dT20 oligomers as primers.
Quantitative PCR. Quantitative PCR was performed using BioMaster HS-qPCR SYBR Blue reaction mixture (Biolabmix, Russia, MHC030). The sequences of primers used for assay are listed in Table S1 (Supplementary Information on the web-site http://www.molecbio.ru/ downloads/2023/1/supp_Romanov_engl). The efficacy of rescue cassette elimination was evaluated using ΔΔCt method, and Cp190 expression was measured using absolute quantification. Actin42A gene primers were used as a reference in both cases. Serial dilutions of gDNA from larval imaginal discs of у1w67;CP190-mCD8-GFP females were used for standard curve producing.
Microscopy. For the analysis of live GFP fluorescence, testes were transferred to slides into a drop of PBS and covered with a cover glass, then analyzed immediately using Olympus BX-51 fluorescence microscope. For immunostaining, testes were fixed using standard method with 4% formaldehyde solution in PBST [21]. Primary rat anti-CP190 antibodies (kindly provided by A.K. Golovnin [22]) and secondary antibodies (anti-Rat Cross-Adsorbed A568, Thermo Fisher Scientific, A-11077) were used in the experiment. Immunostaining samples were analyzed using a Сarl Zeiss LSM 710 (Germany) confocal microscope.
Transcriptome analysis using RNA-seq. Testes of three-week-old imago y1w67;CP190-mCD8-GFP/nanos-Cre and y1w67;CP190-mCD8-GFP/nanos-Cre and y1w67;CP190-mCD8-GFP/nanos-Cre;Cp1902/Cp1903 were used for experiment. Testes of y1w67;{Ubi>stop>mCD8-GFP}/nanos-Cre with {Ubi>stop>mCD8-GFP} were used as controls. Total RNA was isolated from 25 paired testes per sample using TRIZOL. Sequencing libraries were prepared using TruSeq RNA Sample Preparation v2 Kit (Illumina, USA, RS-121-2002) and sequenced on the Illumina MiSeq platform using 2×75 bp paired-end mode. In total, from 3.7 to 6.5 millions of paired-end reads were obtained per sample.
Genomic alignment was performed using HISAT2 tool [23] (genome assembly dm6, Release 6 plus ISO1 MT, Aug. 2014) under standard parameters. Read counts for each gene were summarized using FeatureCounts [24] based on UCSC refGene genomic annotation (https://hgdownload.soe.ucsc.edu/goldenPath/ dm6/bigZips/genes/dm6.refGene.gtf.gz). From two to three biological replicates for each genotype were prepared; gene expression profiles in samples of different genotypes showed a high correlation (Fig. S2, see Supplementary Information on the web-site http://www. molecbio.ru/downloads/2023/1/supp_Romanov_engl). Analysis of differential gene expression was performed using DESeq2 algorithm [25] with “ashr” [26] and “ihv” [27] options. Data on gene expression in whole testes of can, comr, mip40, and bam mutants have been obtained previously and are available in the Gene Expression Omnibus database (GSE97182) [18]. Data on gene expression in Drosophila tissues were obtained from the modENCODE database [28] and clustered using the Cluster 3.0 program [29]. The enrichment of tissue-specific genes was analyzed using the TissueEnrich package [30]. Sequencing data of testis transcriptome with single-cell resolution were obtained from Fly Cell Atlas database and analyzed using Seurat package [31, 32]. Sequencing results obtained in this work are available in the Gene Expression Omnibus database (PRJNA847720).
Binding of Can, Comr and Mip40 to CP190-dependent genes. Data on the binding of Can, Comr and Mip40 proteins with chromosomes of male germline were previously obtained using DamID-seq method (GSE97182) [18]. Protein binding sites were mapped using DamID-Seq software [33]. Every gene region in the genome was divided into a specified number of regions. According to DamID-seq data, protein enrichment in each region of certain gene set was calculated by comparing the frequency of protein-bound GATC-fragment centers in each fragment of gene set with the same value for all D. melanogaster genes. Statistical significance of enrichment was assessed using Fisher’s exact test.
RESULTS
Conditional Rescue System Ensures Viability of Cp190 Mutants
Mutants of insulator protein CP190 coding gene do not survive to the adult stage [15, 16], so the study of its function in tissues of adult D. melanogaster tissues is fraught with experimental difficulties. To overcome this problem and to establish the regulatory function of CP190 in adult male germ cells, we developed a genetic system for tissue-specific conditional rescue of the Cp190 mutation (Fig. 1a).
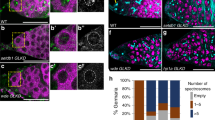
Genetic system for conditional rescue of Cp190 loss of function mutants. (a) Schematic representation of Cp190-mCD8-GFP rescue construct and nanos-Cre construct for Cre recombinase expression in Drosophila germline. (b) Conceptual scheme of Cp1902/Cp1903 mutants conditional rescue (see explanations in the text). (c) Imaging of testes not carrying the rescue construct, with a single dose of one and with conditional rescue of Cp190 mutation. Phase contrast (top row) and GFP fluorescence detection (bottom row). Yellow dotted lines indicate the boundaries of testes. Legend: St.t—spermatid tails; St—spermatid heads; Tr—tracheal tubes; Sg—area of testis containing spermatogonia (continuous line); Sc—area of testis containing spermatocytes (dashed line).
The system is comprised of two genetic constructs. The first one is CP190-mCD8-GFP rescue construct that contains Cp190 coding sequence (CDS) under the control of constitutive promoter Ubi63E, which has been successfully used previously to rescue Cp190 mutants [31, 32]. CDS of Cp190 is followed by hybrid terminator HIS3-SV40 and coding sequence of mCD8-GFP chimeric reporter [33, 34]. Region spanning CDS of CP190 and terminator is floxed and comprise a rescue gene cassette. The second construct contains Cre recombinase gene under the control of nanos gene promoter, which ensures recombinase production specifically in germline cells (Fig. 1а) [18, 19]. If both constructs are combined in Cp190-mutant flies, rescue cassette would be eliminated in male germline, resulting in CP190-deficient cells marked with GFP, while mutant phenotype would be rescued in somatic cells. When the rescue cassette is removed in germline cells, coding sequence of mCD8-GFP chimeric reporter gene becomes close to Ubi63E promoter, resulting in GFP-labeled Cp190-deficient cells (Fig. 1b).
Using phiC31-mediated transgenesis, CP190-mCD8-GFP genetic construct was integrated into attP40 site within second chromosome [19]. Transgenic flies were viable and fertile with both one and two copies of the construct (data not shown). At the same time, one copy was sufficient to rescue lethal phenotype of Cp1902/Cp1903 compound, which do not produce normal CP190 protein [15, 16]. Expression of one or two copies of rescue construct didn’t cause developmental defects under Cp1902/Cp1903 or wild-type background. This indicates a moderate level of ectopic CP190 expression, since an overexpression of CP190-encoding transgenes was shown to cause severe developmental defects [34].
A series of genetic crosses produced males carrying both CP190-mCD8-GFP and nanos-Cre constructs against mutant Cp190 (y1w67; CP190-mCD8-GFP/nanos-Сre; Cp1902/Cp1903). Activation of the mCD8-GFP reporter protein starting from the early spermatocyte stage was detected in the germline cells, indicating the removal of rescue cassette (Fig. 1c). Obtained flies possessed normal viability and exhibited no visible spermatogenesis abnormalities. GFP-negative spermatocytes and GFP-positive somatic envelope cells were not visually detected, indicating a high efficiency and specificity of rescue construct elimination.
In y1w67 control testes, as well as testes carrying rescue construct in the absence of Cre recombinase, only background fluorescence of tracheal tubules and spermatid fragments was present (Fig. 1c). This confirms robust repression of reporter protein production in all cell types of testes.
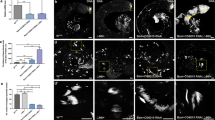
An additional validation of the efficiency of rescue cassette elimination from the germline cell genome with quantitative PCR showed that rescue construct is remaining in 49.6% of whole testis cells (Fig. 2a). This value corresponds to the proportion of somatic cells in testis (50.4%), which were estimated using single cell transcriptome sequencing data [31]. This result indicates that the rescue cassette is eliminated in the vast majority of germ cells.
Cre-mediated recombination results in elimination of rescue cassette and loss of CP190 in cells of the male germline. (a) The number of copies of the rescue cassette in testes with conditional rescue was compared with the imaginal discs of larvae carrying a double dose of the CP190-mCD8-GFP construct. The number of copies of the rescue cassette was normalized to the dose of the construct. The dotted line indicates the proportion of somatic cells in Drosophila testes according to single cell transcriptome sequencing [31]. (b) Location of targets for RT-qPCR inside Cp190 gene. Corresponding primers were used to measure the expression of endogenous and transgenic Cp190 mRNA. (c) Normalized expression of Cp190 in of three-day-old adult’s testes with CP190-mCD8-GFP and nanos-Cre constructs. Expression was measured using primers to the loci indicated in the diagram (b). Expression was normalized to the activity of the Actin42A housekeeping gene. P-value was obtained using Student’s T-test with a two-sided distribution. (d) Confocal images of testes immunostaining using anti-CP190. In the upper row is the testis of the wild type (yw). The bottom row is the testis of males under conditional rescue. DNA was stained with Hoechst dye. The boundaries of testes are underlined with a continuous yellow line. An asterisk marks the apical end of the testes. A long arrow shows somatic cells of the sheath. A dotted line shows a cluster of CP190-negative spermatocytes. The short arrow shows a CP190-positive spermatocyte. The scale is 10 µm.
Conditional Rescue System Significantly Reduces Ectopic Cp190 Expression in Adult Male Germline
We have analyzed expression levels of endogenous and transgenic Cp190 mRNA by RT-qPCR. For this purpose, primers on Cp190 coding and 3'-untranslated regions of endogenous Cp190 were selected (Fig. 2b). The former pair of primers enabled us to measure the expression level of both endogenous and transgenic Cp190, while the latter pair captured only endogenous Cp190 mRNA. It was found that rescue cassette elimination under nanos-Cre resulted in a 3.8-fold decrease in Cp190 CDS expression in whole testes (Fig. 2c). However the level of Cp190 CDS was still 2.4-fold higher compared to wild-type, which is likely due to increased expression of transgenic Cp190 over endogenous in somatic cells as well as remaining rescue cassette in single spermatocytes (Fig. 2d).
Production of CP190-depleted germline cells was confirmed using immunostaining (Fig. 2d). In y1w67;CP190-mCD8-GFP/nanos-cre;Cp1902/Cp1903 testes, CP190 was detected in nuclei of cells from apical end, testis sheath and in single spermatocytes. At the same time, CP190 expression was observed only in single nuclei of spermatocytes. In wild-type testes, a stable level of CP190 expression was observed in both somatic and germ cells. Thus, our system enables rescue of Cp190 mutant phenotype in somatic cell lineage with simultaneous elimination of rescue cassette in male germline, which allows us to reproduce mutant phenotype specifically in adult sex cells.
Mutation of Cp190 Gene Affects the Expression of Differentiation Genes in Drosophila Male Germline
To determine the effect of CP190 on gene expression in adult testes, we performed high-throughput transcriptome sequencing (RNA-seq) followed by differential gene expression analysis (table S2, see Supplementary Information on the web-site http://www. molecbio.ru/downloads/2023/1/supp_Romanov_engl). In the experiment we analyzed testes of three-week-old imago as y1w67;CP190-mCD8-GFP/nanos-Cre and y1w67;CP190-mCD8-GFP/nanos-Cre;Cp1902/Cp1903 testes showed equivalent level of Cp190 expression at that age (Fig. S2, see Supplementary Information on the web-site http://www.molecbio.ru/downloads/ 2023/1/supp_Romanov_engl). It is important to note that the activity of rescue cassette is controlled by the Ubi63E gene promoter, which makes level of Cp190 transcription higher than the endogenous one (Fig. 2c), thus can affect overall gene expression. To accurately account for this effect in our experiment, we evaluated the effect of increased Cp190 expression in somatic cells on the transcriptome of whole testes.
For this purpose, we measured gene expression in testes of y1w67;CP190-mCD8-GFP/nanos-Cre;+/+ males, in which endogenous Cp190 gene were functional while the rescue cassette were active only in somatic cells. We used gene expression in testes of control line (see Experimental) for comparison. The presence of rescue construct resulted in moderate changes in transcription: expression of only 12 genes changed more than 8-fold (Fig. 3a). In contrast, analysis of testes with conditional rescue of Cp190 mutation in somatic cells (y1w67;CP190-mCD8-GFP/nanos-cre;Cp1902/Cp1903) revealed 89 genes whose expression changed more than 8-fold compared to the control (Fig. 3a).
Depletion of CP190 in male germline is accompanied by activation of testis-specific genes and inactivation of housekeeping genes. (a) Differential gene expression in testes with increased CP190 expression in somatic cells (left diagram) or in testes with conditional rescue (right diagram) in RNA-seq. Log2 FoldChanges (LFC) of transgenic testes compared to the control is plotted on the vertical axis. On the horizontal axis, the normalized mean expression in control testes is plotted. Genes with a statistically significant differential expression (|LFC| > 1, P < 0.05) are marked in red. LFC values of genes with insignificant expression change (gray dots, N.S.) were truncated using an empirical Bayesian approach for clarity. Dotted lines indicate LFC values +3 and 0. Red indicates the number of red dots with LFC values between the closest dotted lines. (b) Intersection between sets of genes whose expression significantly increased and decreased in testes with conditional rescue or in testes with increased expression of CP190 in somatic cells (red dots in diagram a). Genes whose expression changed only in the background of conditional rescue are designated as Set 1 and Set 2. (c) Normalized expression of genes from Sets 1 and 2 in different Drosophila tissues according to modENCODE [28]. (d) Analysis of tissue-specific expression of CP190-dependent genes with expression in the male germline cells.
Moreover, among 226 genes with differential expression under conditional rescue (154 increased expression, 72 decreased), only 43 were also altered under Cp190 overexpression in somatic cells (Fig. 3b). These 43 genes were excluded from further analysis, which was focused on isolating the specific effect of a Cp190 mutation on gene expression in germline.
After this correction, we found 120 genes which had increased expression under mutant Cp190 background (Set 1) as well as 63 genes which were downregulated (Set 2) (Fig. 3b). To characterize these sets, we analyzed tissue-specific expression patterns using modENCODE data on fly tissue transcriptomes [26]. It turned out that among the genes in Set 1, three major clusters can be distinguished, represented by those active in male reproductive, nervous, and digestive systems (Fig. 3c). The genes in Set 2 are expressed ubiquitously, and only 14 of them can be classified as testis-specific. Analysis of gene expression patterns in various Drosophila tissues using the TissueEnrich tool [30] confirmed the prevalence of testis-specific genes in Set 1 but not in Set 2 (Fig. 3d). Thus, CP190 protein can be involved both in maintaining the expression of ubiquitously active genes in testes and in repression or modulation of the activity of tissue-specific genes, including ones involved in spermatogenesis.
At the same time, it should be taken into account that the cellular composition of the testes is quite diverse; it is represented by several types of somatic cells, as well as germline cells at different stages of differentiation. Various cell types have distinguished features in the expression profile, which is confirmed by the data of transcriptome sequencing of single testes cells. The characterized expression profiles of individual cell populations make it possible to identify specific marker genes and to determine in which cells a particular gene is normally active.
To determine in which particular testis cells Cp190-dependent genes are normally active, we used marker genes of different Drosophila testis cell populations from the DRscDB database [31, 38]. In Set 1, only single somatic cell marker genes were identified, whereas spermatocyte markers accounted for 10% in total (Fig. 4a). In Set 2, fat and pigment cell markers comprised 22% of genes, while germline cell markers were sporadic (Fig. 4a). Clustering of Set 1 genes by the expression level in different testis cell subpopulations using Fly Cell Atlas single cell transcriptome dataset [31] revealed that 66% of genes were spermatocyte-specific (Fig. 4b). At least 60% of expression of such genes is provided by germline cells, which allows us to refer the majority of Set 1 genes to spermatocyte differentiation genes (Fig. 4b). In contrast, only one-fifth of Set 2 genes can be identified as spermatocyte differentiation genes, and their total number is 5.7 times lower than in Set 1. Taken together, this suggests that CP190 has a predominantly repressive effect with respect to the spermatocyte differentiation genes.
Loss of CP190 in male germline cells leads to impaired activity of spermatocyte differentiation genes. (a) Enrichment of cell type marker genes in Sets 1 and 2 according to DRscDB data [38]. Cell types were annotated in the single cell transcriptome sequencing experiment [31], in some cases including several subpopulations with their own subset of marker genes. The subpopulations cell types are indicated by colored rectangles. The size of the circles characterizes the representation of markers in the whole set. Statistical significance of enrichment was calculated by Fisher’s exact test. Markers with significant enrichment are encircled with red. (b) Gene expression from Sets 1 and 2 in testis cell subpopulations according to Fly Cell Atlas [31]. The identity of populations according cell type and anatomical structures is encoded by colored rectangles. The color of the dots characterizes the scaled level of gene expression compared to the average expression in the testis cells. The size of the dots describes the proportion of cells in the population in which transcripts of a given gene are detected. The bar chart shows the contribution of germline and somatic cells to the total transcription of each gene in the whole testis. Using hierarchical clustering by expression levels in cell subpopulations, genes in each set were divided into two clusters. In Set 1, genes with high activity in spermatocytes (spermatocyte differentiation genes) are clearly distinguished. In Set 2, the clusters can be characterized as somatic specialization genes and spermatocyte differentiation genes. (c) Comparison of expression change estimates in two clusters from Set 2. Log2 FoldChange estimates were obtained separately for spermatocyte differentiation genes (red) and somatic specialization genes (blue) from the assumption that, in testes with conditional rescue, inactivation occurred only in germline cells (dark gray rectangles) or only in somatic cells (light gray rectangles). The observed Log2 FoldChange values are shown in white rectangles. ***P < 5 × 10–8, Wilcoxon test, N.S.—P > 0.05.
The main part of Set 2 genes is normally active predominantly in somatic cells of testes (Fig. 4b). At the same time, it was still unclear whether the expression of these genes changed in germline cells under Cp190 depletion. Using single cell transcriptome data [31] to estimate the contribution of individual testis cell populations to gene expression, we modeled two expression profiles: in the first profile, Set 2 genes were inactivated exclusively in germline, while in the second profile—exclusively in somatic cells. In the next step, we compared the observed changes in gene expression of Set 2 with two profiles. Inactivation of genes with somatic specialization in somatic cells alone or in germline alone does not allow us to simulate the same severe drop in transcription that we observed in testes with conditional rescue (P < 5 × 10–8, Wilcoxon’s test) (Fig. 4c). In contrast, the estimate made for spermatocyte differentiation genes under hypothesis of germline-specific inactivation did not differ from the observed values (P = 0.23, Wilcoxon’s test) (Fig. 4c). This may indicate that the drop in the transcription level of genes from somatic specialization group of Set 2 occurs in both germline and somatic cells.
It is interesting to note that similar regulatory effects of CP190 were found in Drosophila somatic cell culture. Using previously obtained data from transcriptome analysis of Kc167 cells under Cp190 knockdown by RNA interference, we found an upregulation of 820 genes and a downregulation of only 260 (Fig. 5a) [13]. Similarly, upregulated genes were represented predominantly by genes specific for testes, brains, and digestive system (Figs. 5b, 5c). On the other hand, genes which were downregulated under Cp190 depletion had nonspecific expression profiles (Figs. 5b, 5c).
CP190 downregulates tissue-specific genes and promotes the activation of housekeeping genes in Kc167 cells. (a) The effect of Cp190 inactivation in Kc167 cells according to published data [13]. Differential expression was analyzed in the same manner as in Fig. 3a. (b) Expression of CP190-dependent genes (red dots in diagram (a) in Drosophila tissues according to modENCODE [28]. (c) Analysis of tissue-specific expression for CP190-dependent genes from Kc167 cells.
Cp190 Depletion Disrupts Expression of tMAC Targets
In testes activation of differentiation genes occurs in a coordinated manner during the growth phase of spermatocytes I. Several mutations that disrupt this process have been described [17]. For example, mutation in bam gene aborts spermatogenesis progression at spermatogonia stage, resulting in the enrichment of the testes with undifferentiated germline cells [39]. Meiosis arrest genes mip40, comr and can encode transcription factors, and their mutations lead to spermatogenesis arrest at spermatocyte I stage [17].
Defects in Mip40 and Comr, the components of testis meiosis arrest complex tMAC, cause dramatic alterations in testis gene expression [17, 18]. On the other hand, impaired function of testis TBP-associated factors tTAFs, including Can, causes less severe disruption of transcriptome. Together, tTAF and tMAC promote the activation of spermatocyte differentiation program. However, only a fraction of spermatocyte differentiation genes are directly regulated by tTAFs and tMAC [18].
Given the association of CP190 with differentiation genes, we proposed a functional interaction between CP190, tTAF, and tMAC. We previously obtained gene expression profiles in testes of bam, mip40, comr, and can mutants, and mapped binding sites of Mip40, Comr, and Can in chromosomes of male germline cells [18]. Thus, we compared expression of Set 1 genes, which were upregulated under conditional rescue, in testes of wild-type and bam, mip40, comr, and can mutants (Fig. 6a).
Depletion of CP190 in male germline cells leads to impaired activity of tMAC-dependent spermatocyte differentiation genes. (a) Expression of genes from two clusters of Set 1 in testes of wild type and mutants with impaired differentiation germline cells. Colored rectangles show the stage of spermatogenesis arrest in testes of each genotype. Genes mip40 and comr encode components of testis-specific transcriptional activator tMAC. Gene can encodes one of the tTAF proteins. Statistical significance of differences in gene expression in testes between mutants and wild type was established by Wilcoxon’s test. (b) Enrichment of Set 1 wuth genes bound by Can, Comr and Mip40 in specific region in chromosomes of male germline [18]. Statistical significance of enrichment was measured using Fisher’s exact test. TSS—transcription start site; TES—transcription termination site. (c) Same as in (a), for genes from the two clusters of Set 2. (d) Same as (b), for genes from Set 2.
The activity of spermatocyte differentiation genes from Set 1 (Fig. 4b) is significantly reduced in bam, mip40, and comr mutants, but not in can mutants (Fig. 6a). The rest genes of Set 1 is almost unaffected by mip40, comr and can mutations, except bam mutation, which leads to a slight decrease in their expression, which can be explained by the presence of genes with high activity in spermatocytes in that subset (Fig. 4b). Thus, CP190 limits the activity of tMAC-dependent spermatocyte differentiation genes.
The distribution of Comr, Mip40, and Can on chromosomes show that these proteins bind to genes from Set 1 more frequently than expected, while Comr binding was most significant (Fig. 6b). Thus, Cp190 mutation affects the activity of genes directly regulated by the tMAC complex.
The expression of Set 2 genes is characterized by less pronounced differences between bam, mip40, comr, and can mutants and wild-type (Fig. 6c), although both mutants with tMAC disruption and bam mutants have reduced expression of spermatocyte differentiation genes and genes specific to somatic cells. Set 2 genes had some association with transcription factor Comr (Fig. 6d), indicating that their expression can be directly activated by tMAC complex, and CP190 protein is somehow involved in this process.
DISCUSSION
In this work, we studied in detail the effect of the CP190 protein on gene activity in D. melanogaster. The main effect of the CP190 mutation was observed on the subpopulation of spermatocyte differentiation genes, which are regulated by tMAC transcription complex. Interestingly, in the absence of CP190 protein, the activity of those genes increases 2-fold or more, which allows us to conclude that their activity is modulated by this protein in normal conditions, limiting the level of their expression.
In general, Drosophila cell cultures have shown moderate changes in the transcriptome under depletion of insulator protein CP190 [8, 13, 40]. We also identified only 183 genes which expression depends on CP190 in spermatocytes. The expression of 120 CP190-dependant genes was upregulated upon Cp190 mutation and 63 were downregulated, suggesting a prevailing repressor function of CP190. It was also shown on cell cultures that CP190, like other insulator proteins, tends to suppress transcription, despite its simultaneous activating functions [13, 41].
We found that CP190 affects the activity of tissue-specific genes and housekeeping genes in opposite ways, indicating the possibility that there are several mechanisms by which CP190 controls the activity of these genes. It can be assumed that CP190 is capable of acting as a transcription factor for genes whose activation depends on CP190. This is indirectly confirmed by the fact that the cooperative action of CP190 and the factor M1BP, a housekeeping gene regulator, is required to activate CP190-dependent genes [13]. Moreover, promoters of genes whose expression is downregulated upon knockdown in cells are enriched with CP190, while the repressor effect of CP190 is most likely not to be directly implemented [13].
It is important to note that the activation of spermatocyte differentiation program is accompanied by a significant rearrangement of chromatin architecture [42]. Thus, the number of distal contacts decreases at the spermatocyte stage, and promoters of active differentiation genes appear in an isolated environment [42]. For CP190, along with the Chro protein, involvement in the formation and maintenance of topologically associated domains (TADs) has been shown [41]. Moreover, it was found that disruption of gene expression resulting from CP190 knockdown correlates with alteration of boundaries of surrounding TADs [41]. Moreover, it was found that TADs containing housekeeping genes tended to be more stable [41]. This suggests that observed differential effect of CP190 on the expression of tissue-specific and housekeeping genes may be caused by divergent regulation at the level of chromatin structure, making housekeeping genes to be activated by CP190, while tissue-specific genes to be repressed.
Given the absence of an effect on male germline cell differentiation, the normal fertility of mutant males, and the moderate influence on gene expression pattern changes, the CP190 insulator protein can hardly be considered as a key regulatory factor of spermatogenesis. In comparison, the comr (tMAC) mutation affects the expression of over 2500 genes and causes a delay in meiosis [43]. However, given the key role of CP190 in the regulation of nuclear architecture, it can be assumed that CP190 deficiency leads to failures in the communication of specific transcription factors and their regulatory elements and general alterations in chromatin structure, which causes disruptions in the fine-tuning mechanisms of expression regulation.
In our study, we found that CP190 depletion leads to inactivation of tMAC-dependent differentiation genes. Notably, in Drosophila somatic cells, repressor complex dREAM, homologous to tMAC, whose main function is to regulate differentiation and cell cycle genes, functions as partner of CP190 [44–47]. The interaction of CP190 and dREAM in cell cultures plays an important role in the control of divergent gene pairs [46]. CP190-dependent recruitment of dREAM to divergent genes in Drosophila enables the reduction of downregulation of single gene in pair. The presence of common subunits (e.g. Mip40 and Caf1) in dREAM and tMAC, as well as the relationship between CP190 and tMAC-dependent genes, allows us to speculate that CP190 plays a role in tMAC recruitment to chromosomes. Future studies in this direction could explain why the main effect of CP190 depletion in male germline is to inhibit the activity of spermatocyte differentiation genes.
REFERENCES
Long H.K., Prescott S.L., Wysocka J. 2016. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 167, 1170–1187.
Kyrchanova O., Georgiev P. 2014. Chromatin insulators and long-distance interactions in Drosophila. FEBS Lett. 588, 8–14.
Yang J., Corces V.G. 2011. Chromatin insulators: a role in nuclear organization and gene expression, in Advances in Cancer Res. Woude G.W., Ed. 110, London: Academic, 43–76.
Nègre N., Brown C.D., Shah P.K., Kheradpour P., Morrison C.A., Henikoff J.G., Feng X., Ahmad K., Russell S., White R.A.H., Stein L., Henikoff S., Kellis M., White K.P. 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6, e1000814.
Maksimenko O., Bartkuhn M., Stakhov V., Herold M., Zolotarev N., Jox T., Buxa M.K., Kirsch R., Bonchuk A., Fedotova A., Kyrchanova O., Renkawitz R., Georgiev P. 2015. Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 25, 89–99.
Zolotarev N., Maksimenko O., Kyrchanova O., Sokolinskaya E., Osadchiy I., Girardot C., Bonchuk A., Ciglar L., Furlong E.E.M., Georgiev P. 2017. Opbp is a new architectural/insulator protein required for ribosomal gene expression. Nucleic Acids Res. 45, 12285–12300.
Cuartero S., Fresán U., Reina O., Planet E., Espinàs M.L. 2014. Ibf1 and Ibf2 are novel CP190-interacting proteins required for insulator function. EMBO J. 33, 637–647.
Schwartz Y.B., Linder-Basso D., Kharchenko P.V., Tolstorukov M.Y., Kim M., Li H.-B., Gorchakov A.A., Minoda A., Shanower G., Alekseyenko A.A., Riddle N.C., Jung Y.L., Gu T., Plachetka A., Elgin S.C.R., Kuroda M.I., Park P.J., Savitsky M., Karpen G.H., Pirrotta V. 2012. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 22, 2188–2198.
Kaushal A., Dorier J., Wang B., Mohana G., Taschner M., Cousin P., Waridel P., Iseli C., Semenova A., Restrepo S., Guex N., Aiden E. L., Gambetta M.C. 2022. Essential role of Cp190 in physical and regulatory boundary formation. Sci. Adv. 8, eabl8834.
Sabirov M., Kyrchanova O., Pokholkova G.V., Bonchuk A., Klimenko N., Belova E., Zhimulev I.F., Maksimenko O., Georgiev P. 2021. Mechanism and functional role of the interaction between CP190 and the architectural protein Pita in Drosophila melanogaster. Epigenet. Chromatin. 14, 16.
Bohla D., Herold M., Panzer I., Buxa M.K., Ali T., Demmers J., Krüger M., Scharfe M., Jarek M., Bartkuhn M., Renkawitz R. 2014. A functional insulator screen identifies NURF and dREAM components to be required for enhancer-blocking. PLoS One. 9, e107765.
Ali T., Krüger M., Bhuju S., Jarek M., Bartkuhn M., Renkawitz R. 2017. Chromatin binding of Gcn5 in Drosophila is largely mediated by CP190. Nucleic Acids Res. 45, 2384–2395.
Bag I., Chen S., Rosin L.F., Chen Y., Liu C.-Y., Yu G.-Y., Lei E.P. 2021. M1BP cooperates with CP190 to activate transcription at TAD borders and promote chromatin insulator activity. Nat. Commun. 12, 4170.
Savitsky M., Kim M., Kravchuk O., Schwartz Y.B. 2016. Distinct roles of chromatin insulator proteins in control of the Drosophila bithorax complex. Genetics. 202, 601–617.
Butcher R.D.J., Chodagam S., Basto R., Wakefield J.G., Henderson D.S., Raff J.W., Whitfield W.G.F. 2004. The Drosophila centrosome-associated protein CP190 is essential for viability but not for cell division. J. Cell Sci. 117, 1191–1199.
Oliver D., Sheehan B., South H., Akbari O., Pai C.Y. 2010. The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions. BMC Cell Biol. 11, 101.
White-Cooper H. 2010. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction. 139, 11–21.
Laktionov P.P., Maksimov D.A., Romanov S.E., Antoshina P.A., Posukh O.V., White-Cooper H., Koryakov D.E., Belyakin S.N. 2018. Genome-wide analysis of gene regulation mechanisms during Drosophila spermatogenesis. Epigenetics Chromatin. 11, 14.
Markstein M., Pitsouli C., Villalta C., Celniker S.E., Perrimon N. 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40, 476–483.
Laktionov P.P., Maksimov D.A., Andreyeva E.N., Shloma V.V., Belyakin S.N. 2013. A genetic system for somatic and germinal lineage tracing in the Drosophila melanogaster gonads. Tsitologiia. 55, 185–189.
Solovei I., Cremer M. 2010. 3D-FISH on cultured cells combined with immunostaining, in Methods Mol. Biol. 659, Bridger J.M., Volpi E.M., Eds. New York: Humana Press, 117–126.
Golovnin A., Volkov I., Georgiev P. 2012. SUMO conjugation is required for the assembly of Drosophila Su(Hw) and Mod(mdg4) into insulator bodies that facilitate insulator complex formation. J. Cell Sci. 125, 2064–2074.
Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915.
Liao Y., Smyth G.K., Shi W. 2014. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30, 923–930.
Love M.I., Huber W., Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550.
Stephens M. 2016. False discovery rates: a new deal. Biostatistics. 18, 275–294.
Ignatiadis N., Klaus B., Zaugg J.B., Huber W. 2016. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods. 13, 577–580.
Brown J.B., Boley N., Eisman R., May G.E., Stoiber M.H., Duff M.O., Booth B.W., Wen J., Park S., Suzuki A.M., Wan K.H., Yu C., Zhang D., Carlson J.W., Cherbas L., Eads B.D., Miller D., Mockaitis K., Roberts J., Davis C.A., Frise E., Hammonds A.S., Olson S., Shenker S., Sturgill D., Samsonova A.A., Weiszmann R., Robinson G., Hernandez J., Andrews J., Bickel P.J., Carninci P., Cherbas P., Gingeras T.R., Hoskins R.A., Kaufman T.C., Lai E.C., Oliver B., Perrimon N., Graveley B.R., Celniker S.E. 2014. Diversity and dynamics of the Drosophila transcriptome. Nature. 512, 393–399.
de Hoon M.J.L., Imoto S., Nolan J., Miyano S. 2004. Open source clustering software. Bioinformatics. 20, 1453–1454.
Jain A., Tuteja G. 2019. TissueEnrich: tissue-specific gene enrichment analysis. Bioinformatics. 35, 1966–1967.
Li H., Janssens J., De Waegeneer M., Kolluru S.S., Davie K., Gardeux V., Saelens W., David F.P.A., Brbić M., Spanier K., Leskovec J., McLaughlin C.N., Xie Q., Jones R.C., Brueckner K., Shim J., Tattikota S.G., Schnorrer F., Rust K., Nystul T.G., Carvalho-Santos Z., Ribeiro C., Pal S., Mahadevaraju S., Przytycka T.M., Allen A.M., Goodwin S.F., Berry C.W., Fuller M.T., White-Cooper H., Matunis E.L., DiNardo S., Galenza A., O’Brien L.E., Dow J.A.T.; FCA Consortium, Jasper H., Oliver B., Perrimon N., Deplancke B., Quake S.R., Luo L., Aerts S., Agarwal D., Ahmed-Braimah Y., Arbeitman M., Ariss M.M., Augsburger J., Ayush K., Baker C.C., Banisch T., Birker K., Bodmer R., Bolival B., Brantley S.E., Brill J.A., Brown N.C., Buehner N.A., Cai X.T., Cardoso-Figueiredo R., Casares F., Chang A., Clandinin T.R., Crasta S., Desplan C., Detweiler A.M., Dhakan D.B., Donà E., Engert S., Floc’hlay S., George N., González-Segarra A.J., Groves A.K., Gumbin S., Guo Y., Harris D.E., Heifetz Y., Holtz S.L., Horns F., Hudry B., Hung R.J., Jan Y.N., Jaszczak J.S., Jefferis G.S.X.E., Karkanias J., Karr T.L., Katheder N.S., Kezos J., Kim A.A., Kim S.K., Kockel L., Konstantinides N., Kornberg T.B., Krause H.M., Labott A.T., Laturney M., Lehmann R., Leinwand S., Li J., Li J.S.S., Li K., Li K., Li L., Li T., Litovchenko M., Liu H.H., Liu Y., Lu T.C., Manning J., Mase A., Matera-Vatnick M., Matias N.R., McDonough-Goldstein C.E., McGeever A., McLachlan A.D., Moreno-Roman P., Neff N., Neville M., Ngo S., Nielsen T., O’Brien C.E., Osumi-Sutherland D., Özel M.N., Papatheodorou I., Petkovic M., Pilgrim C., Pisco A.O., Reisenman C., Sanders E.N., Dos Santos G., Scott K., Sherlekar A., Shiu P., Sims D., Sit R.V., Slaidina M., Smith H.E., Sterne G., Su Y.H., Sutton D., Tamayo M., Tan M., Tastekin I., Treiber C., Vacek D., Vogler G., Waddell S., Wang W., Wilson R.I., Wolfner M.F., Wong Y.E., Xie A., Xu J., Yamamoto S., Yan J., Yao Z., Yoda K., Zhu R., Zinzen R.P. 2022. Fly Cell Atlas: a single-nucleus transcriptomic atlas of the adult fruit fly. Science. 375, eabk2432.
Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E.P., Jain J., Srivastava A., Stuart T., Fle-ming L.M., Yeung B., Rogers A.J., McElrath J.M., Blish C.A., Gottardo R., Smibert P., Satija R. 2021. Integrated analysis of multimodal single-cell data. Cell. 184, 3573–3587. e29.
Maksimov D.A., Laktionov P.P., Belyakin S.N. 2016. Data analysis algorithm for DamID-seq profiling of chromatin proteins in Drosophila melanogaster. Chromosome Res. 24 (4), 481‒494.
Akbari O.S., Oliver D., Eyer K., Pai C.-Y.Y. 2009. An entry/gateway cloning system for general expression of genes with molecular tags in Drosophila melanogaster. BMC Cell Biol. 10, 8.
Evans C.J., Olson J.M., Ngo K.T., Kim E., Lee N.E., Kuoy E., Patananan A.N., Sitz D., Tran P., Do M.-T., Yackle K., Cespedes A., Hartenstein V., Call G.B., Banerjee U. 2009. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat. Methods. 6, 603–605.
Pindyurin A.V., Pagie L., Kozhevnikova E.N., van Arensbergen J., van Steensel B. 2016. Inducible DamID systems for genomic mapping of chromatin proteins in Drosophila. Nucleic Acids Res. 44, 5646–5657.
Lee T., Luo L. 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251–254.
Hu Y., Tattikota S.G., Liu Y., Comjean A., Gao Y., Forman C., Kim G., Rodiger J., Papatheodorou I., Santos G.D., Mohr S.E., Perrimon N. 2021. DRscDB: A single-cell RNA-seq resource for data mining and data comparison across species. Comput. Struct. Biotechnol. J. 19, 2018–2026.
Gönczy P., Matunis E., DiNardo S. 1997. bag-of-marbles and benign gonial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Development (Cambridge). 124, 4361–4371.
Bartkuhn M., Straub T., Herold M., Herrmann M., Rathke C., Saumweber H., Gilfillan G.D., Becker P.B., Renkawitz R. 2009. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 28, 877–888.
Chathoth K.T., Mikheeva L.A., Crevel G., Wolfe J.C., Hunter I., Beckett-Doyle S., Cotterill S., Dai H., Harrison A., Zabet N.R. 2022. The role of insulators and transcription in 3D chromatin organization of flies. Genome Res. 32, 682–698.
Ilyin A.A., Kononkova A.D., Golova A.V., Shloma V.V., Olenkina O.M., Nenasheva V.V., Abramov Y.A., Kotov A.A., Maksimov D.A., Laktionov P.P., Pindyurin A.V., Galitsyna A.A., Ulianov S.V., Khrameeva E.E., Gelfand M.S., Belyakin S.N., Razin S.V., Shevelyov Y.Y. 2022. Comparison of genome architecture at two stages of male germline cell differentiation in Drosophila. Nucleic Acids Res. 50, 3203–3225.
Laktionov P.P., White-Cooper H., Maksimov D.A., Belyakin S.N. 2014. Transcription factor Comr acts as a direct activator in the genetic program controlling spermatogenesis in D. melanogaster. Mol. Biol. (Moscow). 48, 130–140. https://doi.org/10.1134/S0026893314010087
Lee H., Ohno K., Voskoboynik Y., Ragusano L., Martinez A., Dimova D.K. 2010. Drosophila RB proteins repress differentiation-specific genes via two different mechanisms. Mol. Cell. Biol. 30, 2563–2577.
DeBruhl H., Wen H., Lipsick J.S. 2013. The complex containing Drosophila Myb and RB/E2F2 regulates cytokinesis in a histone H2Av-dependent manner. Mol. Cell. Biol. 33, 1809–1818.
Korenjak M., Kwon E., Morris R.T., Anderssen E., Amzallag A., Ramaswamy S., Dyson N.J. 2014. dREAM co-operates with insulator-binding proteins and regulates expression at divergently paired genes. Nucleic Acids Res. 42, 8939–8953.
Beall E.L., Lewis P.W., Bell M., Rocha M., Jones D.L., Botchan M.R. 2007. Discovery of tMAC: a Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B. Genes Dev. 21, 904–919.
ACKNOWLEDGMENTS
Authors would like to acknowledge A.K. Golovnin for the provided antibodies (IGB RAS), A.V. Pindurin (IMCB SB RAS) for assistance in cloning, and the “Molecular and Cellular Biology” core facility for the provided equipment.
Funding
The reported study was funded by Russian Foundation for Basic Research, project nos. 19-34-90108, 17-00-00181, and 19-04-00872, and supported by Basic Science Research Program FWGZ-2021-0017 (122011900429-5).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
Conflict of interest. The authors declare that they have no conflicts of interest.
Statement on the welfare of animals. The conditions in which Drosophila melanogaster was kept complied with the standards specified in Order of the Ministry of Health of Russia no. 267 of June 19, 2003 “On Approval of the Rules of Laboratory Practice in the Russian Federation.”
ADDITIONAL INFORMATION
The text was submitted by the author(s) in English.
Supplementary Information
Rights and permissions
About this article
Cite this article
Romanov, S.E., Shloma, V.V., Koryakov, D.E. et al. Insulator Protein CP190 Regulates Expression оf Spermatocyte Differentiation Genes in Drosophila melanogaster Male Germline. Mol Biol 57, 113–126 (2023). https://doi.org/10.1134/S0026893323010120
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893323010120