Abstract
RNA is a crucial component of every living organism and is necessary for gene expression and its regulation in the cell. Mechanisms of RNA synthesis (especially mRNA synthesis) were a subject of extensive study for a long time. More recently, RNA degradation pathways began to be considered as equally important part of eukaryotic cell metabolism. These pathways have been studied intensely, and ample information accumulated about RNA degradation systems and their role in cell life. It is currently obvious that RNA decay is of no less importance as RNA synthesis and contributes to regulating the RNA level in the cell. The review considers the main RNA degradation enzymes, the decay pathways of various coding and non-coding RNAs, the mechanisms providing RNA quality control in the nucleus and cytoplasm, and certain structural elements responsible for RNA stability or short life in the cell.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Degradation of RNA in the cell is performed by enzymatic systems that cleave phosphodiester bonds and eventually yield mononucleotides. Many mechanisms work in the nucleus and cytoplasm to rapidly detect and eliminate nonfunctional (defective) RNAs. These molecules are so short-lived in the cell that they remain undetectable unless components of cell degradation systems are inactivated. Functional RNAs greatly vary in stability. Their stability in the cell is associated with their functions and is regulated by various structural elements and specific chemical modifications. The function of the structural elements and modifications consists, to a substantial extent, in protecting the RNA molecules from their rapid decay. According to our estimates, the half-life of RNAs lacking protective structures is approximately 20–40 min in mammalian cells [1, 2]. It was proposed that RNAs are classified as unstable with a half-life of less than 2 h, stable with a half-life of 2–16 h, and highly stable with a half-life of more than 16 h [3].
The mRNA stability in the cytoplasm is determined by a cap, which is at the 5′ end, and a poly(A) tail, which interacts with poly(A)-binding proteins (PABPs) and occurs at the 3′ end. These structures act to physically bring the 5′ and 3′ ends close together and to circularize translated mRNAs, thus preventing the access of deadenylases, decapping factors, and exonucleases to mRNAs [4]. Histone mRNAs lack poly(A) tails and are an exception; their stability is regulated by a stem-loop structure located at the 3′ end. Certain structures that improve the stability and are likely to similarly fold into stem-loop structures are found in the 3′ region in many “typical” mRNAs [5]. The poly(A) tail increases the life not only for mRNAs and long non-coding RNAs (ncRNAs), but also for certain RNA polymerase III transcripts, such as those of short interspersed elements (SINEs), which are short mobile genetic elements and do not code for any protein [6].
The codon composition was shown to affect the mRNA life. Different tRNAs that bind synonymous codons substantially vary in abundance. The mRNA molecules that have many codons decoded using rare tRNAs are translated slower and are more prone to decay. In contrast, mRNAs with an optimal codon usage are translated faster and live longer, although their poly(A) tails are shorter than those of mRNAs with less optimal codon compositions [7, 8].
The stability of an RNA molecule often depends on the stability of its secondary or tertiary structure. For example, hypomodified RNA molecules were found to have less stable secondary structures and to decay faster [9]. It was shown with an example of the rodent 4.5SI RNA, which is a small ncRNA, that a relatively simple structure of a double-stranded stem formed via complementary interactions of the 5′- and 3′-terminal regions of a molecule ensures its stability in the cell [1]. The long ncRNAs MALAT1 and NEAT1 escape degradation in the nucleus owing to a stable triple-helical structure that forms at their 3′ ends [10].
Specific modifications can stabilize small RNAs. For example, 2′-O-methylation of the 3′-terminal ribose moiety is a common modification and substantially improves the stability of microRNAs, small interfering RNAs (siRNAs), and piRNAs. The modification prevents the synthesis of a 3′-terminal oligo(U) tail, which is thought to be a widespread RNA degradation signal [11].
Permanent association with proteins provides an important means to protect cell RNAs from cell nucleases. For example, rRNAs are included in the ribosome; small nuclear RNAs (snRNAs), in the spliceosome [12]; the 7SL RNA, in the signal recognition particle (SRP) [13]; and the 7SK RNA, in small nuclear RNPs [14].
ENZYMES AND COMPLEXES RESPONSIBLE FOR RNA DECAY
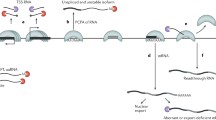
Based on their enzymatic activity, RNA-degrading enzymes (ribonucleases, or RNAses) are classified into three main types: endonucleases cleave internal sites in an RNA strand, 5'–3' exonucleases hydrolyze an RNA strand from its 5' end, and 3'–5' exonucleases degrade RNA molecules from their 3' ends. Enzymes of all three types are intensely utilized to degrade RNA in the cell. The end points of many RNA degradation metabolic pathways involve exosomes, which employ 3'–5' exonuclease and endonuclease activities, or Xrn exonucleases, which possess 5'–3' exonuclease activity. Recent studies identified the Dis3L2 3'–5' exonuclease as a final participant of several important pathways of RNA degradation in the cytoplasm [15, 16]. The main pathways that are responsible for degrading various cell RNAs and the proteins that ensure their degradation are considered below and schematically shown in Fig. 1.
Main pathways of cell RNA degradation. Genes for various RNAs are transcribed in the nucleus by one of the three RNA polymerases. Mature and immature transcripts pass through quality control in both the nucleus and cytoplasm, and aberrant RNA molecules are eliminated (PPD, RTD, NRD, NSD, NGD, and NMD). Mature functional mRNA decay mechanisms (SMD, NMD, and RNA interference) are used to regulate gene expression in the cell. Virtually all RNA decay pathways converge to RNA degradation by the exosome (which utilizes TRAMP, NEXT, or SKI as a cofactor) or exonuclease Xrn1/2 or Dis3L2.
Exosome is a main ribonuclease complex in eukaryotic cells and is involved in both performing RNA quality control and degradation and processing certain RNA types. Exosomes are conserved and are found in all eukaryotes examined to date; structurally and functionally similar protein complexes were detected in eubacteria and archaea. A core part of the exosome consists of nine subunits arranged in two layers of rings: one is a protein hexamer (the polypeptides Rrp41, Rrp42, Rrp43, Rrp45, Rrp46, and Mtr3) and the other is a cap of three proteins (Rrp4, Rrp40, and Csl4). The nuclease function of the exosome is performed by the subunits Rrp44 (Dis3 in humans), which possesses 3'–5' exonuclease and endonuclease activities, and Rrp6 (PM-Scl 100 or EXOSC10 in humans), which acts as a 3'–5' exonuclease [17].
The Rrp44/Dis3 exonuclease is highly processive. Once bound to a substrate, the enzyme completely degrades it to release nucleoside 5′-monophosphates. The 3′ end of a substrate RNA should pass through the core ring to reach the exonuclease active site of Rrp44/Dis3, and the unstructured single-stranded 3′‑end should therefore be at least 35 nt in length [18]. The endonuclease catalytic center of Rrp44/Dis3 is closer to the surface and is therefore accessible to RNA both from within (RNA passes through the core channel in this case) and outside the complex (in this case, endonuclease may cleave the loops and other structured regions to prepare the substrate RNA for exonuclease cleavage) [19]. The Rrp6 exonuclease also hydrolyzes RNA to release nucleoside 5′-monophosphates, but is distributive in contrast to Rrp44/Dis3; i.e., the enzyme detaches from the substrate after catalyzing one reaction [20]. Rrp6 binds with the cap of the exosome core. The 3′ end of RNA presumably passes through the hole in the cap and reaches the Rrp6 catalytic center through the cleft between the cap and the hexamer. It is unclear what factor determines if a particular RNA is degraded by Rrp44/Dis3 or by Rrp6. A nuclear exosome consists of 11 subunits and includes both Rrp44/Dis3 and Rrp6; a cytoplasmic exosome consists of 10 subunits and lacks Rrp6 [21].
Helicase Mtr4 acts as a key accessory factor for nuclear exosomes. In yeasts, Mtr4 is a component of the Trf4/5–Air1/2–Mtr4 polyadenylation complex (TRAMP). The complex includes the noncanonical poly(A) polymerase Trf4 or Trf5, the RNA-binding protein Air1 or Air2, and the helicase Mtr4 [22–24]. The variant TRAMP4 (Trf4–Air2–Mtr4) is involved mostly in RNA quality control in the nucleus, while the variant TRAMP5 (Trf5–Air1–Mtr4) mostly contributes to RNA processing [25]. TRAMP facilitates and enhances activity of the nuclear exosome. On the one hand, Mtr4 helicase unfolds the secondary structures present in a substrate RNA. On the other hand, Trf4/5 synthesizes an oligo(A) sequence at the 3′ end of the target RNA to provide a convenient substrate for exosome landing. Trf4/5 and Air1/2 activate Mtr4, which requires a terminal single-stranded fragment of 5–6 nt for its binding. In turn, Mtr4 regulates the length of the oligo(A) sequence by preventing Trf4/5 from producing too long poly(A) tails [26]. In yeasts, the TRAMP complex ensures rapid degradation of virtually all RNAs that are substrates of nuclear exosomes: aberrant mRNAs, rRNAs, tRNAs, sn/snoRNAs, and cryptic unstable transcripts (CUTs) resulting from pervasive transcription by RNA polymerase II. TRAMP is additionally involved in processing pre-rRNAs and pre-sn/snoRNAs [27].
In mammals, helicase Mtr4 forms two different complexes in the nucleoli and the nucleoplasm. In the nucleoli, Mtr4 binds with Trf4-2 (a homolog of yeast Trf4) and ZCCHC7 (the closest homolog of Air1/2) to produce human TRAMP (hTRAMP), which is analogous to yeast TRAMP. In the nucleoplasm, Mtr4 interacts with the RNA-binding proteins RBM7 and ZCCHC8 to produce the nuclear exosome targeting complex (NEXT). The difference between hTRAMP and NEXT concerns not only their localization in the nucleus, but their function as well. While hTRAMP is involved in oligoadenylating rRNAs and tRNAs to ensure their exosomal degradation, NEXT plays a role in processing and degrading RNA polymerase II transcripts, including mRNAs, sn/snoRNAs, and promoter upstream transcripts (PROMPTs) [28].
In the cytoplasm, the exosome functions in complex with the Superkiller complex (SKI), which consists of the RNA helicase Ski2 and RNA-binding proteins Ski3 and Ski8 [29]. The Ski7 protein is responsible for the interaction between the SKI complex and the exosome in yeasts. However, Ski7 is absent in mammals, and Hbs1 seems to play its function [30].
Exonuclease Dis3L2. (Dis3-like 3′–5′ exoribonuclease 2) is a free cytoplasmic enzyme and is responsible for degrading many RNAs in the 3′–5′ direction. Dis3L2 is similar to the exosomal Dis3 nuclease, but is devoid of the N-terminal PIN domain, which is responsible for endonuclease activity and binding to the rest of the exosome. A catalytic domain is in the central part of the Dis3L2 molecule, while two cold-shock domains occur in its N-terminal part. The enzyme was shown to degrade both single- and double-stranded substrate RNAs in vitro. Oligo(U) present at the 3′ end of an RNA provides a signal for its degradation by Dis3L2 [16].
Xrn exonucleases are main 5′–3′ exonucleases in eukaryotic cells. The Xrn group includes cytoplasmic Xrn1 (Pacman in Drosophila melanogaster) and nuclear Xrn2 (Rat1 in yeasts). The enzymes processively degrade 5′-monophosphorylated substrate RNAs. The mechanism of action of Xrn1 was studied comprehensively. In particular, it is known that only uncapped RNAs (a cap is too large for the active center of the enzyme) with a single-stranded region of at least 4 nt at the 5′ end are utilized as substrates by the enzyme. An intricate secondary structure of an RNA bound with the enzyme is degraded as the RNA passes into the active center. Xrn1 and Xrn2 are structurally similar in the N-terminal part and especially in the active center region and differ in their C-terminal regions [31]. Rai1 pyrophosphate hydrolase is necessary as a cofactor for the function of Rat1 nuclear exonuclease in yeasts, converting 5′-triphosphate to monophosphate; a similar mammalian cofactor of Xrn2 has not been identified as of yet [32].
PROCESSING, QUALITY CONTROL, AND DEGRADATION OF mRNA IN THE NUCLEUS
When mRNA is processed in the nucleus, a cap is added to its 5′ end, its non-coding regions (introns) are removed (splicing), the 3′ end is polyadenylated, and mRNA is bound with packaging and export factors. Systems of mRNA quality control work at all of these steps and ensure rapid degradation of defective molecules.
The cap protects mRNA from 5′–3′ exonuclease degradation and provides for pre-mRNA processing, export into the cytoplasm, and translation initiation [33]. Capping takes place soon after transcription initiation, when nascent mRNA reaches 25–50 nt in length, and is directly associated with a transition of RNA polymerase II into the elongation state [34]. The RNA polymerase II transition to elongation is thus restricted to successfully capped transcripts, while uncapped transcripts are immediately degraded by Xrn2/Rat1 5′–3′ exonuclease.
Like capping, mRNA splicing occurs cotranscriptionally and is performed by the spliceosome, which is a special ribonucleoprotein complex. Introns that are removed via splicing have a lasso shape and are therefore first cleaved by Dbr1 endonuclease [35] and then degraded from both ends of the molecule by the nuclear exosome and Xrn2/Rat1 exonuclease. The pre-mRNAs that somehow escaped splicing are degraded by the same Xrn2/Rat1 (after being decapped by the Dcp1/Dcp2 complex with the Lsm2-8 nuclear decapping cofactor) and the nuclear exosome.
Apart from histone mRNAs, mRNAs undergo 3′‑terminal polyadenylation in eukaryotic cells. The poly(A) tail performs several functions, protecting the 3′ end of the molecule from exosomal degradation, playing a role in mRNA transport from the nucleus into the cytoplasm, and facilitating translation. An AAUAAA hexanucleotide in the 3′ part of the transcribed mRNA region provides a polyadenylation signal. The cleavage and polyadenylation specificity factor (CPSF) binds to the hexanucleotide to stimulate the mRNA cleavage and the recruitment of poly(A) polymerase (PAP) [36]. The poly(A) tail may reach 70–80 nt in yeast cells and is approximately 250 nt in mammals. The poly(A) tail interacts with poly(A)-binding proteins (PABPs), which bind to RNAs during their polyadenylation. A minimal site of PABP association with the poly(A) tail is 12 consecutive adenosine residues; mRNAs lacking a poly(A) tail sufficient for the binding of at least one PABP molecule are rapidly degraded [37, 38]. Deadenylation is therefore the first step in virtually all mechanisms of mature mRNA degradation.
After the AAUAAA-dependent mRNA cleavage, RNA polymerase II continues synthesizing the rest of the transcript. The 5′ end of the nascent transcript remains unprotected and recruits Xrn2/Rat1 5′–3′ exonuclease, which degrades RNA. The RNA nuclease catches up and collides with RNA polymerase II during degradation, thus stimulating transcription termination according to a torpedo mechanism [39]. The transcripts that have not undergone AAUAAA-dependent cleavage in a timely manner are degraded by the nuclear exosome [40].
During its synthesis, the nascent mRNA binds with proteins, which provide for its proper packaging and export. The THO complex, which consists of the Mft1, Tho2, Hpr1, Thp2, and Tex1 proteins, acts together with Sub2 RNA helicase to ensure mRNA export from the nucleus in yeasts. Mutations of THO/Sub2 lead to transcript degradation, which seems to be due to polyadenylation defects. AAUAAA-dependent cleavage and polyadenylation are most likely possible only when a proper mRNP complex is assembled [41]. When mRNP assembly is distorted, the respective mRNA is retained in the vicinity of its transcription site and subsequently degraded from the 5′ or 3′ end [42].
All of the above systems responsible for quality control of nascent mRNAs function cotranscriptionally in the immediate vicinity of the transcription elongation complex in both yeasts and higher eukaryotes. After transcription termination, mRNA quality control is far less efficient [43].
A kinetic hypothesis is the most popular hypothesis that explains the principles of RNA quality control [44]. Degradation factors compete with processing, packaging, and export factors for mRNA binding, and the binding with proper proteins is advantageous in terms of energy and stimulates the transition to the next maturation step. Defective molecules are less likely to rapidly and correctly associate with necessary proteins and are therefore more likely to undergo degradation. Thus, degradation factors do not necessarily have to possess the capability of detecting various defects in mRNA because, by default, they degrade any molecules that have failed to bind with proper proteins and to proceed to the next processing step or nuclear export in a timely manner [45]. A substantial fraction of mRNAs degraded this way are normal molecules that somehow failed to rapidly associate with proper factors [40].
PABPN1 and PAPα/γ-mediated RNA decay (PPD). Relatively recent studies identified an mRNA degradation pathway that occurs in the mammalian nucleus, is associated with excessive mRNA polyadenylation by canonical PAP (one of its two forms, PAPα or PAPγ), is mediated by nuclear PABP (PABPN1), and leads to RNA degradation by the nuclear exosome [46]. A poly(A) tail of 300–800 nt provides a degradation signal in this case. The pathway was termed PABPN1 and PAPα/γ-mediated decay (PPD). PPD is involved in degrading the majority of intron-containing mis-spliced mRNAs and mRNAs that escaped mRNP formation and nuclear export. Certain ncRNAs (see below) are also degraded via PPD [47].
CYTOPLASMIC mRNA DEGRADATION PATHWAYS
Deadenylation and decapping of mRNA. Deadenylation is the first step of a degradation pathway for the majority of mRNAs. A long poly(A) tail is truncated to a short oligo(A) region in the process. Poly(A)-specific 3′–5′ exonucleases involved in the Pan2–Pan3 and Ccr4–Not complexes are responsible for deadenylation [38, 48]. In yeasts, the Pan2–Pan3 complex seem to play only a minor role and functions mostly in the nucleus, while the Ccr4–Not complex drives deadenylation prior to mRNA decay in the cytoplasm. Mammalian mRNA deadenylation occurs in the cytoplasm and proceeds via two steps with the involvement of two respective protein complexes.
At the first step, the poly(A) tail is shortened by the Pan2–Pan3 complex, which binds with PABP through Pan3. Pan2 is a catalytic subunit. Pan2–Pan3 is distributive and functions rather slowly, gradually truncating the poly(A) tail to approximately 110 nt in length. Then PABP and certain translation initiation factors go away to be replaced by translational repressors. The mRNA becomes untranslated and may be targeted to P-bodies (see below) or the second deadenylation step. The second step employs the Ccr4–Not complex, which is processive and fast. Not is a structural protein, while Ccr4 and Caf/Pop2 deadenylases perform the catalytic function. Pan2–Pan3 and Ccr4–Not may form a supercomplex, thus providing for coordination of the two deadenylation steps [49]. It should be noted that the first deadenylation step starts, in fact, as soon as mRNA appears in the cytoplasm. By default, the first step determines the life for mRNAs that do not have any determinant of short-lived molecules [50].
Once deadenylated, mRNA decays via one of the two degradation pathways, from the 5′ or 3′ end. In the first case, the Lsm1-7 complex binds with oligo(A) to recruit the Dcp1/Dcp2 decapping proteins. Xrn1 exonuclease degrades decapped mRNA in the 5′–3′ direction. The pathway seems to be the most common mRNA decay pathway in the cytoplasm [31]. The other variant is degradation from the 3′ end by the cytoplasmic exosome with the involvement of the SKI complex. In this case, the 5′-terminal region (no more than 10 nt) with the cap is degraded by the DcpS decapping protein [51]. The Lsm1-7 complex is known to bind to oligo(A) to inhibit the exosomal degradation pathway, thus regulating the choice between the pathways of mRNA decay. Factors that may affect the choice are still unknown [52].
Specific cytoplasmic granules were found to contain the decapping factors (Lsm1 and Dcp1/Dcp2), Xrn1 5′–3′ exonuclease, and mRNA decay intermediates in yeast cells. The granules were termed the processing bodies, or P-bodies. The P-bodies provide room for mRNA decapping and 5′–3′ degradation. More recent studies detected deadenylation enzymes (Pan2, Pan3, Ccr4, and Caf1) and deadenylated mRNAs in the P-bodies. Mammalian P-bodies are often termed the GW-bodies. The P-bodies are specific sites that provide not only for mRNA decay, but also for the accumulation and temporal storage of nontranslated mRNAs [53]. Isolation in the P-bodies protects the cell from occasional translation of the molecules that have still not been degraded [49]. Thus, mRNAs occur in two sites that are discrete both spatially and functionally in the cytoplasm: polyribosomes (a translated state) and P-bodies (nontranslated state) [54].
Uridylation of mRNA is the second most common modification of the 3′ end in cell RNAs after polyadenylation. Like polyadenylation, uridylation proceeds via non-template-driven synthesis of an oligo(U) or, rare, poly(U) sequence at the 3′ end of RNA by terminal uridylyl transferases (TUTases). TUTases are structurally similar to poly(A) polymerases and are usually devoid of a functional classical RNA recognition motif (RRM). TUTases are either capable of interacting with any RNA or require accessory factors, varying in substrate specificity. Uridylation is undergone by transcripts of all three RNA polymerases in eukaryotes and serves to regulate the mRNA stability as one of its most important functions. TUTases and, primarily, TUT4 (ZCCHC6) and TUT7 (ZCCHC11) uridylate mRNA molecules. Almost all TUTases perform their functions in the cytoplasm. Uridylated RNAs are recognized and utilized by free nucleases Dis3L2 and 3′hExo (Eri1) and the exosome [55–57].
Fully functional typical mRNAs are usually not uridylated. Their long poly(A) tails probably prevent their uridylation, and at least partial deadenylation is therefore necessary for this modification. However, the poly(A) tail is relatively short in yeasts, and both deadenylated and adenylated mRNAs can undergo uridylation in yeast cells. A shortened oligo(A) tail with at least two uridine residues at the end is well recognized by the Lsm1-7 complex, which drives decapping; the efficiency of their binding increases with the oligo(U) sequence increasing to 5 nt and then reaches a plateau. In human cells, uridylation is observed for approximately 20% of mRNAs with a poly(A) tail of less than 25 nt, and these mRNAs undergo degradation [58–60]. Uridylation suppresses translation of particular mRNAs without their degradation in certain cases; i.e., the oligo(U) sequence binds with an upstream part of the molecule and translation stops [61, 62]. An interesting mechanism was described in Arabidopsis thaliana: uridylyltransferase Urt1 protects deadenylated (that is, containing oligo(A) tails) mRNAs from being degraded from the 3′ end. However, uridylation does most likely not increase the mRNA stability in the cell in this case, but is necessary to ensure mRNA degradation in the direction from the 5′ to the 3′ end only [63].
Uridylation with subsequent degradation by Dis3L2 is an important pathway that was relatively recently described to degrade mRNAs in the 3′–5′ direction and does not involve the exosome. Acting together, TUT4/7 and Dis3L2 are probably responsible for degradation of a substantial mRNA portion in human cells and cells of other organisms [15, 64]. The two proteins are actively involved in degrading the majority of mRNAs during apoptosis. Schizosaccharomyces pombe Dis3L2 seems to function with Xrn1 in a concerted manner. The two enzymes often affect the same mRNA targets and are both found in P-bodies [65].
Uridylation is additionally involved in degrading the mRNA fragments formed at previous degradation steps, such as NMD (see below), RNA interference, and ribosome stalling. In particular, fragments resulting from microRNA-mediated cleavage are uridylated by TUT2 and TUT3 (up to 15 residues) and then degraded in human cells; their exact decay mechanism remains unknown [57]. TUT4 and TUT7 are responsible for degrading not only cell, but also virus RNAs and are thus involved in the immune response. Activity of the enzymes is observed upon human infection with influenza virus [66].
Degradation of histone mRNAs. Histone mRNAs are unique in being nonpolyadenylated. The 3′ ends of mammalian histone mRNAs contain a stem-loop structure, which protects the mRNAs from cell exonucleases in the absence of a poly(A) tail and determines their life [67]. The element binds with the stem loop-binding protein (SLBP) and Eri1 3′–5′ exonuclease [68]. The stem-loop structure should be no more than 45–80 nt away from the stop codon, indicating that the ribosome should be in the immediate vicinity of SLBP during termination. It is important that histone mRNAs are translated only during DNA replication and are to be degraded by the end of the S phase. Oligouridylation of histone mRNAs was observed in the late S phase and upon inhibition of DNA replication and is possibly driven by TUT7, which is recruited by SLBP [69]. The oligo(U) tail provides a platform for the binding of the Lsm1-7 complex with subsequent histone mRNA decapping and degradation from the 5′ end by Xrn1 or serves to prime exosomal degradation from the 3′ end [70]. TUTases and Eri1 are additionally involved in mRNA decay in the latter case, performing several consecutive rounds of oligouridylation and degradation of the stem-loop structure; the remaining mRNA is then degraded by the exosome. SLBP can interact with helicase Upf1, which unwinds the stem-loop structure to facilitate its degradation [71].
AU-rich element (ARE)-mediated decay (AMD). AREs are sequences of 50–150 nt and often harbor several AUUUA motifs. AREs occur in the 3′-untranslated region (3′-UTR) in many short-lived eukaryotic mRNAs. The majority of these mRNAs code for proteins that regulate cell growth or the cell response to external factors. Expression of the genes for such proteins needs the strongest control, in particular, via a fast mRNA degradation mechanism. Many of the relevant genes act as proto-oncogenes [72]. The stability of certain long ncRNAs also correlates with the presence of AREs [3]. An ARE was found in the 5′-terminal region of a short-lived virus RNA (HSUR1, Herpesvirus saimiri U РНК1) [73].
Several cell factors bind with ARE-binding proteins (ARE-BPs) and regulate the mRNA stability, stimulating or suppressing mRNA decay. TTP, BRF1, and KSRP are the best studied ARE-BPs that stimulate mRNA degradation [74, 75]. ARE-BPs seem to stimulate rapid deadenylation of ARE-containing mRNAs. Then the mRNAs are decapped and undergo exonuclease degradation from the 3′- or the 5′ end [76, 77]. In contrast, the ARE-BP HuR stabilizes mRNAs. ARE-containing mRNAs do not decay within a short period of time prior to translation. Their protection from destabilizing proteins is due to HuR. The ARE-BP AUF1 is capable of causing both rapid degradation and stabilization of ARE-containing mRNAs, depending on the particular substrate [78].
mCRD-dependent degradation of the c-fos mRNA. It is of interest that many short-lived mRNAs carrying the above AREs additionally harbor other destabilizing elements in their protein-coding region or 3′‑UTR. Thus, a double mechanism regulating the mRNA life arose in mammalian cells. The presence of two determinants of rapid mRNA degradation confers more flexibility on the cell response to external and internal signals.
Two independent pathways are known for rapid degradation of the c-fos mRNA in the cell; one is ARE dependent and the other, ARE independent. A second determinant providing for rapid decay of the c-fos mRNA is known as the major protein-coding-region determinant of instability (mCRD) and occurs in the protein-coding region. It is of interest that actinomycin D and other transcriptional inhibitors do not affect the mCRD function, while suppressing the destabilizing effect of the ARE. The finding supports the idea that each of the determinants utilizes its own independent degradation pathway [79].
The c-fos mCRD is a 320-nt region. An 87-nt purine-rich sequence is a functional element of the mCRD and occurs in its 5′ part; the minimal distance between the mCRD and the poly(A) tail should be approximately 450 nt [80]. The mechanism of mCRD-dependent degradation is associated with mRNA translation. The mCRD binds with Unr and other accessory factors, which additionally interact with PABP on the poly(A) tail of the mRNA to allow its cyclization and with Ccr4 deadenylase to block its activity. A ribosome should pass through the mCRD to activate degradation. The ribosome displaces Unr and the other proteins to release the end of the poly(A) tail and to activate deadenylase [80, 81].
A similar mechanism of ARE-independent degradation was observed for the c-myc mRNA, which also harbors a rapid degradation determinant in its protein-coding region in addition to an ARE in its 3′-UTR. Like in the case of the c-fos mRNA, the ARE-independent pathway of c-myc mRNA degradation depends on translation [82, 83].
GU-rich element (GRE)-mediated decay (GMD). GREs are usually GU repeats or UGUUUGUUUGU-containing sequences and are found in Drosophila and mammals. Like AREs, GREs occur in the 3′-UTR in many short-lived mRNAs. The set includes the mRNAs for proteins involved in cell differentiation, proliferation, signal transduction, and other processes. GRE-containing mRNAs are generally more stable than ARE-containing mRNAs. Rare mRNAs harbor elements of both types. Main GRE-binding proteins belong to the CELF family (CELF1, CELF2, etc.). It is of interest that CELF1, CELF2, and HuR can bind with both GREs and AREs. CELF1 is thought to stimulate rapid deadenylation and degradation of mammalian mRNAs by recruiting PARN deadenylase. CELF2 was shown to bind to GRE-containing mRNAs in stress, thus providing for their stabilization and transport into stress granules. Drosophila cells were not found to contain PARN, but have the Cup protein, which interacts with the cap-binding protein eIF4E and is capable of recruiting Ccr4 deadenylase. The mechanism of GRE-dependent mRNA decay is still unknown in detail [84].
Nonsense-mediated decay (NMD) is an mRNA control mechanism that rapidly degrades the aberrant (defective) molecules that contain a premature termination codon (PTC). The process prevents an accumulation of the proteins that have only partly synthesized C ends and are potentially dangerous for the cell. PTCs may arise as a result of genomic mutations, transcription errors, or incorrect mRNA splicing. It is possible that NMD evolved in parallel with splicing in eukaryotes as a mechanism that detects and degrades the mRNA molecules that find their way into the cytoplasm and still have unremoved introns (the majority of these mRNAs are degraded in the nucleus via PPD in mammals) [85]. It is known now that NMD is often used not only to destroy aberrant molecules in the cell, but also to regulate expression of several genes. NMD substrates may include the mRNAs that have an additional short open reading frame upstream of the main open reading frame, an intron in the 3′-UTR, or an extremely long 3′-UTR [86, 87]. The set of mRNAs subject to NMD includes the mRNAs for factors involved in the cell response to various types of stress [88–90] and the NMD factors [91]. In addition, alternative splicing associated with the NMD mechanism [92] serves to mediate the autoregulation of expression of several cell proteins, including hnRNP L [93], PTB [94], chromatin remodeling factors [95, 96], etc.
Degradation of mRNA via NMD was observed in yeasts [97], plants [98], and animals [86]. Proteins involved in the process greatly differ between different groups of organisms. The main factors necessary for NMD in animals include Upf1, Upf2, Upf3, Smg1, Smg5, Smg6, Smg7, and PNRC2. Upf1 is an ATP-binding protein and possess RNA-helicase activity. Upf1 is responsible for recognizing the degradation substrate according to the majority of NMD models [99]. Upf1 interacts with eRF1 and eRF3, suggesting its involvement in translation termination [100]. In complex with Upf2 and Upf3, Upf1 is phosphorylated by Smg1 kinase [101]. The Smg5, Smg6, Smg7, and PNRC2 factors interact with phosphorylated Upf1 to directly trigger mRNA degradation. Two mRNA degradation variants are possible to occur in animal cells after NMD starts. Most commonly, Smg6 cleaves the mRNA in the vicinity of its PTC [102]. The resulting 5′ and 3′ mRNA fragments undergo exonuclease degradation (the 5′-terminal fragments are degraded in exosomes; and the 3′-terminal fragments, by Xrn1 exonuclease) without being deadenylated and decapped [103]. Alternatively, Smg5 and Smg7 recruit deadenylases and exonucleases; deadenylation (Ccr4–Caf1) starts the NMD pathway in this case and is followed by exosomal degradation in the 3′–5′ direction [104] or PNRC2 recruits the Dcp1/Dcp2 decapping factors and the mRNA is then degraded from the 5′ end by Xrn1 [105]. Recent findings indicate that during NMD mRNA fragments may be uridylated by TUT4 and TUT7 and then degraded by the exosome or Dis3L2 exonuclease [106].
Several models were assumed to explain how the NMD substrate is recognized, but none of the models explains the mechanism in full [107]. A downstream marker model ascribes the major role to marker proteins that occur on mRNA downstream of the PTC and upstream of the normal termination codon. A large exon junction protein complex (EJC) provides an example of marker proteins in mammalian cells, binding to mRNA approximately 20–25 nt upstream of an exon junction in the nucleus [108]. A false 3′‑UTR model is based on the assumption that translation termination at a premature stop codon differs from termination at a normal stop codon because it takes place at a greater distance from the mRNA 3′ end [109]. Another model [107] suggests that NMD does not employ a special mechanism to recognize the PTC-containing mRNA, but is a consequence of premature translation termination, as a result of which a major part of the mRNA is released from ribosomes and becomes accessible to cell nucleases.
Staufen1-mediated decay (SMD). Staufen1 (Stau1) is an RNA-binding protein that is involved in mRNA transport and translation control. Stau1 was found to determine the short life of the Arf1 (ADP ribosylation factor) mRNA in the cell [110]. Stau1 binds with a certain region known as the Stau1-binding site (SBS) in the 3′-UTR of mRNA. The Arf1 SBS is a 19-nt hairpin with a 100-nt loop and is 20–25 nt away from the termination codon [111]. SMD is a translation-dependent degradation mechanism, occurring during normal termination of the ribosome at a stop codon [110].
Other mRNAs also act as SMD targets in the cell. These mRNAs mostly code for proteins involved in the control of cell metabolism, the transport of organelles, and cell division [110]. However, SBSs were not found in SMD targets other than the Arf1 mRNA. It was demonstrated that a SBS may result from a complementary binding of the Alu SINE contained within the mRNA 3′-UTR with another Alu element contained in a cytoplasmic polyadenylated ncRNA. Such RNAs were termed the 1/2-sbsRNA because they contain only one of the complementary strands that form a SBS. One ncRNA of the kind can destabilize several mRNAs [112]. A similar mRNA degradation mechanism was recently observed in rodents. The 3′‑UTRs of respective mRNAs contain SINEs (B1, B2, B4, and ID), which interact with similar elements contained in long ncRNAs to form a double-stranded SBS and ensure rapid mRNA degradation [113]. The process is presumably involved in regulating myogenesis [114]. Apart from Stau1, its paralog Stau2 was found to potentially play a role in SMD [115].
The mechanism of RNA degradation via SMD is similar to that of NMD. Stau1/Stau2 binds with the Upf1 protein, which is phosphorylated by Smg1 and interacts with PNRC2. In turn, PNRC2 recruits the Dcp1/Dcp2 decapping proteins to trigger RNA degradation from the 5′ end by Xrn1. It is still unknown whether Smg5, Smg6, and Smg7 are involved in SMD [116].
Nonstop decay (NSD) is a cell RNA degradation pathway aimed at eliminating the mRNAs that lack a stop codon in the open reading frame [117, 118]. These mRNAs may arise as a result from transcription termination, premature polyadenylation, and, possibly, incomplete exosomal degradation. In addition, NSD helps the ribosome associated with an aberrant mRNA to dissociate and to become capable of landing on other templates.
The order of events in NSD depends on whether a poly(A) tail is present and where the ribosome stopped on the mRNA. When an mRNA lacking a stop codon is polyadenylated, the ribosome continues translation on the poly(A) tail and synthesizes a polylysine sequence at the end of the peptide. A positively charged polylysine peptide present in the ribosomal exit tunnel may cause the ribosome to stall. Degradation proceeds via the NGD mechanism in this case (see below). If the ribosome successfully reached the end of the poly(A) tail, the empty A site of the ribosome binds with the Ski7 protein, whose GTPase domain resembles the analogous domains of the EF1A and eRF3 translation factors. Ski7 binds with the SKI complex and recruits the exosome, which degrades the mRNA from the 3′ end without its preliminary deadenylation. The Dom34–Hbs1 protein complex ensures dissociation of the ribosome in this case [119]. If the mRNA is not polyadenylated, the exosome is recruited to its unprotected 3′ end immediately after dissociation of the ribosome (without the involvement of Ski7 and the SKI complex). The polypeptide is destroyed as well, undergoing ubiquitination and proteasomal degradation [117].
No-go decay (NGD) is an mRNA degradation mechanism that works when the ribosome stalls and further translation is impossible [120]. The causes of ribosome stalling include an mRNA secondary structure, a rare codon, translation of the poly(A) tail, or disorders of the ribosome. The NGD pathway includes endonuclease cleavage of the mRNA in the vicinity of the ribosome stalling site, which is recognized by the Dom34–Hbs1 complex. The complex recruits an unidentified endonuclease, which cleaves the mRNA. The Dom34–Hbs1 additionally stimulates ribosome dissociation from the mRNA. Then the 5′ fragment of the mRNA is degraded by the exosome (like in the NSD mechanism) and the 3′ fragment is degraded by Xrn1 exonuclease (often in P-bodies). The polypeptide is degraded as well [117].
Yeasts were the main model used to study NSD and NGD, but plant and animal cells were also found to possess these degradation pathways [121–124].
RNA interference is a mechanism that regulates (suppresses) gene expression and is based on the complementary binding of small RNAs (microRNAs or endogenous small interfering RNAs (siRNAs)) with their target mRNAs. When the sequences involved are fully complementary, the mRNA is endonucleolytically cleaved by the RNA-induced silencing complex (RISC). When the sequences are only partly complementary, translation is suppressed, but the mRNA is not cleaved. MicroRNAs are encoded by special genes. Their primary transcripts are known as the pri-microRNAs and are capped and polyadenylated like mRNAs. Pri-microRNAs are processed by Drosha endonuclease to yield pre-microRNAs, which are exported from the nucleus and are cleaved to produce mature microRNAs in the cytoplasm by Dicer endonuclease. RNA interference is well understood and is a process that basically differs from the majority of other RNA degradation pathways. The process was considered in detail in many reviews (e.g., see [125–128]).
MECHANISMS OF NON-CODING RNA DEGRADATION
rRNA. Almost all rRNAs are synthesized in the nucleus as longer precursors, which are then processed. RNA polymerase I synthesizes a polycistronic RNA, which includes three of the four eukaryotic rRNAs. Mature rRNAs result from a series of consecutive cleavages and shortenings of the primary transcript [129]. Two terminal fragments known as the external transcribed spacers (ETSs) and two internal transcribed spacers (ITSs) are excised and degraded. The 5′-terminal ETS fragment is degraded by the nuclear exosome with the involvement of TRAMP [130], while ITS1 is degraded by Xrn2 exonuclease [131].
The rRNA degradation process is still poorly understood because rRNAs are highly stable. Quality control of mature rRNAs in the nucleus is performed by the nuclear exosome in complex with TRAMP. It is still unclear what signatures are utilized by nuclear RNA degradation factors to recognize aberrant molecules. A signal to rRNA degradation is possibly provided by the incapability of an rRNA molecule to properly bind with ribosomal proteins. Distorted export of ribosomal subunits into the cytoplasm is also known to increase the level of rRNA degradation in the nucleus [132].
Nonfunctional rRNA decay (NRD) is a mechanism whereby defective mature rRNAs are recognized and degraded and is found in yeast cells [22, 133]. NRD takes place in the cytoplasm and involves two independent pathways, degradation of 25S rRNA molecules that have mutations of the peptidyltransferase center (25S NRD) and degradation of 18S rRNA molecules that have mutations of the decoding center (18S NRD). The substrates of 25S NRD accumulate in the cytoplasm close to the nuclear envelope, and their degradation is translation independent. Mutant 18S rRNAs are scattered through the cytoplasm, and their degradation stops in the absence of protein synthesis in the cell.
Defective 18S rRNAs are degraded by the same proteins as are involved in mRNA NGD: Xrn1 5′–3′ exonuclease, the cytoplasmic exosome, the Ski7 factor, and the Dom34–Hbs1 complex. The substrates of 18S NRD are often localized in P-bodies, as is the case with NGD. The similarity between 18S NRD and NGD probably stems from the fact that the ribosome stalled on mRNA provides a degradation signal in either process. Ribosome stalling is caused by an error occurring in the decoding center of the 18S rRNA in the case of 18S NRD and a barrier occurring on mRNA to prevent translocation in the case of NGD. The stalled ribosome contains a meaning codon in the A site in either case. It is possible that both mRNA and 18S rRNA are targeted to P-bodies and undergo degradation when the ribosome stalls [117, 134].
The 25S NRD process lacks similarity to any other degradation mechanism. Moreover, the process is independent not only of translation, but also of all proteins involved in various RNA decay mechanisms, Dis3 exosomal exonuclease being the only exception. The 25S NRD mechanism is thought to work when ribosomal proteins fail to associate with a defective 25S rRNA and is presumably associated with protein degradation [117, 134].
tRNA. In the nucleus, tRNAs undergo exosomal degradation after being preliminarily oligoadenylated by the TRAMP complex [23]. The majority of molecules degraded by this mechanism are aberrant precursors and hypomodified tRNAs. However, normal precursors were also found to be degraded in the nucleus. Inactivation of the exosome increase the mature tRNA level in the cell, indicating that functional molecules capable of normal processing account for an appreciable portion of normally degraded pre-tRNAs [135]. This promiscuity of degradation systems is possibly related to the fact that a kinetic factor, rather than errors, provides a degradation signal in RNA quality control, as is the case with mRNAs. In other words, a precursor that fails to quickly proceed to the next processing step (that is, to bind with necessary proteins) is to be degraded. However, there is evidence that Trf4 poly(A) polymerase, which is a component of TRAMP, is capable of distinguishing between hypomodified and fully modified tRNAs [136].
Functional tRNAs are extremely stable, living for several hours or even days. A tRNA molecule goes through approximately 40 translation cycles during this period. Each tRNA type has specific modifications, which ensure its correct recognition and aminoacylation. A mechanism known as rapid tRNA decay (RTD) is responsible for quality control of mature tRNAs in yeast cells. Hypomodified molecules are quickly deacylated and degraded [137]. The process involves 5′–3′ exonucleases Rat1 (in the nucleus) and Xrn1 (in the cytoplasm) and the Met22 protein [138]. Degradation enzymes presumably recognize hypomodified tRNAs by a lower stability of the tRNA secondary structure, especially in the acceptor and T stems [9]. In addition, a CCACCA sequence present in structurally unstable tRNAs at the 3′ end provides a signal for their fast degradation via RTD, while a CCA sequence is found in stable tRNAs. The addition of CCA is an important step of tRNA maturation in the nucleus and is performed by tRNA nucleotidyltransferase. The enzyme seems to try to add a second CCA triplet to all tRNAs, stimulating a temporary unfolding of the acceptor stem in the process. Stable molecules are sufficiently tough to avoid stem unfolding, while unstable molecules are thus marked by the CCACCA signal and degraded. It should be noted that this CCACCA labeling of unstable tRNAs is characteristic of all domains of life [139, 140].
Cleavage of the anticodon loop in tRNAs may occur in yeast, human, and plant cells exposed to oxidative stress. Rny1 endonuclease is responsible for the process in yeasts, leaving the vacuole for the cytosol in stress [141].
snRNAs and snoRNAs. snRNAs are transcripts synthesized by RNA polymerases II and III and are involved in the splicing of cell mRNAs. Mechanisms of their degradation are still poorly understood. The TRAMP complex and nuclear exosome are possibly responsible for degradation of snRNAs, as well as for their maturation [142, 143]. Degradation of the mature U6 snRNA similarly involves TRAMP and the nuclear exosome in Saccharomyces cerevisiae [144]. There are indications that the NEXT complex, which is a TRAMP analog, facilitates degradation of the U6 snRNA in mammals by binding to the U-rich sequences in its mature and immature variants [145]. In human cells, the pre-U6 snRNA is uridylated by nuclear TUTase TENT1 (TUT6) during processing so that 16 new uridine residues are added to the four uridine residues present at its 3′ end. Than 15 of them are hydrolyzed by Usb1 exonuclease, and a bond between the third and second phosphates arises in the remaining uridine residues to stabilize the U6 snRNA. Pre-U6 snRNA molecules lacking this modification are degraded by the nuclear exosome [146].
Defective yeast U1 snRNA molecules incapable of forming a proper complex with proteins are degraded in both the nucleus from the 3′ end by exosomal exonuclease Rrp6 and the cytoplasm by the Dcp2 decapping protein and Xrn1 5′–3′ exonuclease. Mammals also have a similar mechanism for cytoplasmic degradation of the U1 snRNA [147]. It was found additionally that misprocessed snRNA precursors are uridylated in the cytoplasm by TUT4/7 and then degraded by Dis3L2 exonuclease [148].
Little is known about degradation of small nucleolar RNAs (snoRNAs), which are involved in modifying cell rRNAs, tRNAs, and snRNAs. Yeast snoRNAs are encoded by autonomous genes, while higher eukaryotic snoRNAs are encoded in gene introns and are excised from respective transcripts. The nuclear exosome is involved in their maturation, as is the case with snRNAs [149]. In yeasts, snoRNA precursors are degraded in the exosome after being oligoadenylated by the TRAMP complex; the degradation process is in equilibrium with the process of snoRNA precursor maturation, which is mediated by the Pab2 and Rrp6 proteins [150, 151]. In mammals, the NEXT complex and nuclear exosome are most likely involved in degrading snoRNAs, like with snRNAs. In addition, PPD was implicated in degrading the non-coding transcripts of the genes that have snoRNA-coding introns in mammals [47].
Vault RNAs and Y RNAs are RNA polymerase III transcripts of approximately 100 nt in length and are involved in autophagy and the initiation of DNA replication, respectively [152, 153]. The human genome harbors four genes for each of these RNA groups. Vault RNAs are components of large cytoplasmic ribonucleoprotein complexes known as the Vault complexes. Y RNAs occur in both the cytoplasm (cYtoplasmic) and nucleus, and a substantial portion of their pool is associated with the La and Ro60 proteins in the cell. The proteins most likely protect Y RNAs from degradation [15]. Both Vault and Y RNAs are uridylated by TUT4/7 TUTases and then degraded by cytoplasmic Dis3L2 exonuclease [148]. However, a recent study showed that the Y RNA level decreases in response to a knockdown of PARN deadenylase, which is responsible for degrading the oligo(A) tail that is synthesized at the 3′ ends of these RNAs by noncanonical poly(A) polymerase PAPD5. This oligo(A) tail mediates Y RNA degradation by Dis3 exosomal nuclease [147]. It is known additionally that both Vault and Y RNAs are processed to yield shorter fragments, which are termed svRNA and YsRNA; degradation of these fragments has not been investigated as of yet [15].
MicroRNAs, endogenous siRNAs, and piRNAs. Degradation is still poorly understood for microRNA precursors, which are synthesized by RNA polymerase II. In mammals, pre-microRNAs may be uridylated by cytoplasmic TUTases TUT4/7. Once uridylated, the molecules are degraded by exosomal nucleases Rrp6 and Dis3 [15, 154]. Specifics of the function and “life cycle” were most comprehensively studied for microRNAs of the let-7 family. The Lin28 protein pluripotency factor was shown to recruit TUT4/7 to the let-7 RNA precursor (pre-let-7). As a result, the pre-let-7 molecule is no longer capable of interacting with Dicer and is degraded by cytoplasmic Dis3L2. Certain mutations of the Dis3L2 exonuclease gene cause Perlman syndrome, which includes fetal gigantism. Distorted let-7 metabolism is probably responsible for Perlman syndrome [155]. Other aberrant pre-microRNAs are also uridylated by TUT4/7 and subsequently degraded by Dis3L2 exonuclease, but oligo(U) synthesis is distributive in this case because of the absence of accessory factors, such as Lin28 [57]. Nuclear pri-microRNAs, which are pre-microRNA precursors, may undergo polyadenylation and degradation via the PPD pathway [47].
As for the majority of small RNAs regulating mRNA translation (that is, microRNAs, siRNAs, and piRNAs), different mechanisms of their degradation were described in different organisms. Uridylation of these RNAs is common and leads to their degradation in contrast to 2′-O-methylation of the 3′-terminal ribose moiety; the latter modification stabilizes the RNAs [56]. MicroRNAs consist of guide and passenger strands. The guide strand usually shows an extremely high stability (its average half-life is 119 h in mice), while the passenger strand is highly vulnerable to nucleases [156]. Many degradation pathways are known for mature microRNAs, which can be degraded both from the 5′ end and from the 3′ end. In the former case, degradation is driven by Xrn nucleases (cytoplasmic Xrn1 in human and Caenorhabditis elegans and nuclear Xrn2 in C. elegans only). MicroRNA degradation from the 3′ end can be performed by the exosome (human and Drosophila), polynucleotide phosphorylase (human), Eri1 exonuclease (mouse), PARN (human), Atrimmer2, and small RNA-degrading nucleases (SDNs) (Arabidopsis) [15, 157, 158]. Mature microRNAs that are bound with their target mRNAs via complementary interactions are degraded via a mechanism known as target RNA-directed miRNA degradation (TDMD). The mechanism is associated with uridylation, which is performed by TENT1 TUTase (although other enzymes cannot be excluded); Dis3L2 exonuclease is responsible for degradation [61].
Mature piRNAs are 2′-O-methylated at the 3′ end in the majority of organisms. The modification is introduced by Hen1 methyltransferase. In the absence of Hen1, an unidentified nuclease degrades these RNAs from the 3′ end [159]. Endogenous siRNAs decay similarly; the nuclease responsible for degradation of their uridylated forms is also unknown [15].
Short-lived RNA polymerase II transcripts. A great variety of types of short-lived RNA polymerase II transcripts were recently detected in eukaryotic cells [160, 161]. The transcripts are capped at the 5′ end, like mRNAs. There is evidence that some of the transcripts are involved in regulating expression of other genes. On the other hand, the majority of the transcripts may be nonfunctional, resulting from pervasive transcription by RNA polymerase II.
The degradation mechanism was studied in detail for an RNA type known as the cryptic unstable transcripts (CUTs) in yeasts. CUTs are transcripts of up to 400 nt in length that are synthesized from intergenic regions close to the promoters of mRNA genes and are degraded immediately. CUTs contain binding sites for the NNS complex, which consists of the Nrd1 and Nab3 RNA-binding proteins and Sen1 helicase. Nrd1 and Nab3 bind to specific motifs (GUAA/G and UCUUG, respectively) present in nascent RNAs. Nrd1 additionally interacts with the C-terminal domain of RNA polymerase II to stimulate transcription termination. The NNS complex recruits TRAMP and the nuclear exosome, which perform 3′–5′ exonuclease degradation of CUTs [162]. Certain CUTs (e.g., SRG1) are exported from the nucleus and are degraded with the involvement of the Dcp1/Dcp2 decapping complex and Xrn1 exonuclease [163].
Degradation of plant short-lived upstream non-coding transcripts (UNTs) and mammalian PROMPTs takes place in the nucleus and similarly depends on the nuclear exosome. The NEXT complex acts as a cofactor to recruit the exosome in mammals [164–167]. However, at least a portion of PROMPTs are polyadenylated and degraded via PPD. There is evidence that a cotranscriptional decapping and subsequent degradation by Xrn2 are possible for PROMPTs. It remains unclear whether different degradation pathways are used for different PROMPT subsets or any PROMPT may be degraded via any pathway [47].
Xrn1 unstable transcripts (XUTs) were found in yeast cells. XUTs are capped and polyadenylated in the nucleus, like mRNAs, and are then exported into the cytoplasm, where their degradation in the 5′–3′ direction is driven by Xrn1 exonuclease [168]. Recent studies showed that the NMD mechanism mediates cytoplasmic degradation of XUTs [169]. XUTs were found to contain short open reading frames, where ribosomes land to start translation. A stalling of these ribosomes occurs rather far away from the 3′ end of the molecule and triggers NMD with the help of Upf1, which stimulates the decapping of the transcript and its degradation by Xrn1 exonuclease [170]. Many XUTs form duplexes with mRNAs in the nucleus. The binding protects them from ribosome landing after their transfer into the cytoplasm and thus prevents their degradation via the NMD mechanism. However, Mtr4 and Dbp2 helicases can unwind such a duplex if a single-stranded 3′-terminal region is left after its formation. The region seems to provide a platform for helicase landing [169].
CONCLUSIONS
All RNAs are eventually degraded in the cell, while the lifetime of a particular molecule depends on its type. A substantial portion of the RNA pool synthesized in the cell is rapidly degraded already in the nucleus. The portion includes primarily the fragments that are excised from RNAs of various types in the course of their processing. Such fragments are degraded cotranscriptionally in the immediate vicinity of the RNA synthesis site by Xrn2/Rat1 exonuclease. Numerous and probably nonfunctional RNA molecules that are by-products of RNA polymerase II activity (CUTs, PROMPTs, and UNTs) are degraded in the nucleus with the involvement of the TRAMP complex (NEXT in mammals) and the exosome. Aberrant RNAs also undergo rapid degradation in the nucleus. These molecules contain errors, which result from distortions arising during RNA synthesis or processing, and are potentially dangerous for the cell. These RNAs are usually oligoadenylated by the TRAMP complex and then degraded by the nuclear exosome. Aberrant RNAs that escape degradation by nuclear quality control systems are degraded in the cytoplasm. It became clear in recent years that a variety of highly structured aberrant non-coding RNAs are uridylated in the cytoplasm by TUT4/7 and degraded by Dis3L2 exonuclease [61, 148, 171]. In yeast cells, the NRD mechanism works in the cytoplasm to eliminate defective rRNAs and the RTD mechanism degrades the tRNAs that have an insufficient number of nucleotide modifications. Defects in mRNAs are detected during translation. The mRNA molecules that contain premature stop codons and introns are degraded via the NMD mechanism. Lack of a stop codon results in mRNA degradation via NSD, and barriers to the progress of the ribosome lead to degradation via NGD. Different mechanisms trigger the above degradation pathways, but the final step is the same in all cases: defective RNAs are degraded from the 3′ or 5′ end by cell exonucleases Dis3 (in the exosome) and Xrn1, respectively.
The cell additionally possesses the mechanisms that are aimed at degrading fully functional RNAs. Stable long-lived RNAs, such as rRNAs and tRNAs, are not degraded until their functionality is lost as a result of accumulating defects. Defective molecules are eliminated via the NRD and RTD mechanisms in this case. According to data from recent studies, a number of small non-coding RNAs, such as Y RNAs, Vault RNAs, and BC1 and BC200 RNAs, are degraded with the help of TUT4/7 and Dis3L2 [148]. Degradation of microRNAs, siRNAs, and piRNAs similarly proceeds via uridylation and subsequent exonuclease hydrolysis from the 3′ end. The mRNA degradation process most commonly includes deadenylation, decapping, and subsequent exonuclease hydrolysis from the 5′ end by Xrn1 or from the 3′ end by the cytoplasmic exosome. Another common variant is degradation of oligoadenylated mRNAs via their uridylation by TUT4/7, subsequent decapping, and eventual hydrolysis from the 5′ end by Xrn1 nuclease or from the 3′ end by Dis3L2.
The process of mRNA degradation is one of the most important mechanisms that regulate gene expression. Primary or secondary structure elements are often present in mRNA molecules to ensure their rapid degradation or high stability. AU- and GU-rich elements are the most common signals for rapid degradation of mRNAs coding for many regulatory factors; the same elements may recruit stabilizing factors as well. Expression of certain proteins is regulated by changing the life of their mRNAs via degradation by the NMD mechanism. Signals for rapid degradation of such mRNAs may include a long 3′-UTR, an additional short open reading frame upstream of the main one, or an intron left by alternative splicing. Secondary structure elements less commonly act as determinants that regulate the mRNA life. Examples of such elements are provided by the stem-loop structure found in histone mRNAs and the double-stranded SBS, which determines degradation of certain mRNAs via the SMD mechanism.
Thus, a broad network of RNA degradation pathways exists in the eukaryotic cell. The network functions to protect the cell from potentially dangerous nonfunctional RNA molecules and, on the other hand, to regulate gene expression.
REFERENCES
Koval A.P., Gogolevskaya I.K., Tatosyan K.A., Kramerov D.A. 2012. Complementarity of end regions increases the lifetime of small RNAs in mammalian cells. PloS One.7, e44157.
Koval A.P., Gogolevskaya I.K., Tatosyan K.A., Kramerov D.A. 2015. A 5'-3'-terminal stem in small non-coding RNAs extends their lifetime. Gene.555, 464–468.
Clark M.B., Johnston R.L., Inostroza-Ponta M., Fox A.H., Fortini E., Moscato P., Dinger M.E., Mattick J.S. 2012. Genome-wide analysis of long noncoding RNA stability. Genome Res.22, 885–898.
Gorgoni B., Gray N.K. 2004. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Brief. Funct. Genomic Proteomic.3, 125–141.
Goodarzi H., Najafabadi H.S., Oikonomou P., Greco T.M., Fish L., Salavati R., Cristea I.M., Tavazoie S. 2012. Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature.485, 264–268.
Ustyantsev I.G., Tatosyan K.A., Stasenko D.V., Kochanova N.Yu., Krameriv D.A. 2020. Polyadenylation of Sine transcripts generated by RNA polymerase III dramatically prolongs their lifetime in cells. Mol. Biol. (Moscow). 54 (1), 67–74.
Mugridge J.S., Coller J., Gross J.D. 2018. Structural and molecular mechanisms for the control of eukaryotic 5'-3' mRNA decay. Nat. Struct.Mol. Biol.25, 1077–1085.
Nicholson A.L., Pasquinelli A.E. 2019. Tales of detailed poly(A) tails. Trends Cell Biol.29, 191–200.
Whipple J.M., Lane E.A., Chernyakov I., D’Silva S., Phizicky E.M. 2011. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev.25, 1173–1184.
Brown J.A., Valenstein M.L., Yario T.A., Tycowski K.T., Steitz J.A. 2012. Formation of triple-helical structures by the 3'-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc. Natl. Acad. Sci. U. S. A.109, 19202–19207.
Ji L., Chen X. 2012. Regulation of small RNA stability: methylation and beyond. Cell Res.22, 624–636.
Papasaikas P., Valcarcel J. 2016. The spliceosome: The ultimate RNA chaperone and sculptor. Trends Biochem. Sci.41, 33–45.
Wild K., Weichenrieder O., Strub K., Sinning I., Cusack S. 2002. Towards the structure of the mammalian signal recognition particle. Curr. Opin. Struct. Biol.12, 72–81.
Peterlin B.M., Brogie J.E., Price D.H. 2012. 7SK snRNA: A noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdisc. Rev. RNA.3, 92–103.
Labno A., Tomecki R., Dziembowski A. 2016. Cytoplasmic RNA decay pathways: Enzymes and mechanisms. Biochim. Biophys. Acta.1863, 3125–3147.
Luan S., Luo J., Liu H., Li Z. 2019. Regulation of RNA decay and cellular function by 3'-5' exoribonuclease DIS3L2. RNA Biol.16, 160–165.
Januszyk K., Lima C.D. 2014. The eukaryotic RNA exosome. Curr. Opin. Struct. Biol.24, 132–140.
Bonneau F., Basquin J., Ebert J., Lorentzen E., Conti E. 2009. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell.139, 547–559.
Wasmuth E.V., Lima C.D. 2012. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol. Cell.48, 133–144.
Januszyk K., Liu Q., Lima C.D. 2011. Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA.17, 1566–1577.
Chlebowski A., Lubas M., Jensen T.H., Dziembowski A. 2013. RNA decay machines: The exosome. Biochim. Biophys. Acta.1829, 552–560.
Doma M.K., Parker R. 2007. RNA quality control in eukaryotes. Cell.131, 660–668.
Kadaba S., Wang X., Anderson J.T. 2006. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA.12, 508–521.
Vanacova S., Wolf J., Martin G., Blank D., Dettwiler S., Friedlein A., Langen H., Keith G., Keller W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol.3, e189.
San Paolo S., Vanacova S., Schenk L., Scherrer T., Blank D., Keller W., Gerber A.P. 2009. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet.5, e1000555.
Jia H., Wang X., Anderson J.T., Jankowsky E. 2012. RNA unwinding by the Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex. Proc. Natl. Acad. Sci. U. S. A.109, 7292–7297.
Tudek A., Lloret-Llinares M., Jensen T.H. 2018. The multitasking polyA tail: Nuclear RNA maturation, degradation and export. Philos. Tran. R. Soc. Lond. B.373, 20180169.
Lubas M., Andersen P.R., Schein A., Dziembowski A., Kudla G., Jensen T.H. 2015. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rept.10, 178–192.
Halbach F., Reichelt P., Rode M., Conti E. 2013. The yeast ski complex: Crystal structure and RNA channeling to the exosome complex. Cell.154, 814–826.
Kalisiak K., Kulinski T.M., Tomecki R., Cysewski D., Pietras Z., Chlebowski A., Kowalska K., Dziembowski A. 2017. A short splicing isoform of HBS1L links the cytoplasmic exosome and SKI complexes in humans. Nucleic Acids Res.45, 2068–2080.
Nagarajan V.K., Jones C.I., Newbury S.F., Green P.J. 2013. XRN 5'→3' exoribonucleases: Structure, mechanisms and functions. Biochim. Biophys. Acta.1829, 590–603.
Xiang S., Cooper-Morgan A., Jiao X., Kiledjian M., Manley J.L., Tong L. 2009. Structure and function of the 5'→3' exoribonuclease Rat1 and its activating partner Rai1. Nature.458, 784–788.
Cougot N., van Dijk E., Babajko S., Seraphin B. 2004. “Cap-tabolism”. Trends Biochem. Sci.29, 436–444.
Perales R., Bentley D. 2009. “Cotranscriptionality”: The transcription elongation complex as a nexus for nuclear transactions. Mol. Cell.36, 178–191.
Chapman K.B., Boeke J.D. 1991. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell.65, 483–492.
Ustyantsev I.G., Golubchikova Yu.S., Borodulina O.R., Kramerov D.A. 2017. Canonical and noncanonical RNA polyadenylation. Mol. Biol. (Moscow). 51, (2), 2626–236.
Eckmann C.R., Rammelt C., Wahle E. 2011. Control of poly(A) tail length. Wiley Interdisc. Rev. RNA.2, 348–361.
Wilusz C.J., Wilusz J. 2008. New ways to meet your (3') end oligouridylation as a step on the path to destruction. Genes Dev.22, 1–7.
Tollervey D. 2004. Molecular biology: Termination by torpedo. Nature.432, 456–457.
Porrua O., Libri D. 2013. RNA quality control in the nucleus: The Angels’ share of RNA. Biochim. Biophys. Acta.1829, 604–611.
Saguez C., Schmid M., Olesen J.R., Ghazy M.A., Qu X., Poulsen M.B., Nasser T., Moore C., Jensen T.H. 2008. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol. Cell.31, 91–103.
Rougemaille M., Villa T., Gudipati R.K., Libri D. 2008. mRNA journey to the cytoplasm: Attire required. Biol. Cell.100, 327–342.
Schmid M., Jensen T.H. 2010. Nuclear quality control of RNA polymerase II transcripts. Wiley Interdisc. Rev. RNA.1, 474–485.
Hopfield J.J. 1974. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. U. S. A.71, 4135–4139.
Houseley J., Tollervey D. 2009. The many pathways of RNA degradation. Cell.136, 763–776.
Bresson S.M., Conrad N.K. 2013. The human nuclear poly(a)-binding protein promotes RNA hyperadenylation and decay. PLoS Genet.9, e1003893.
Bresson S.M., Hunter O.V., Hunter A.C., Conrad N.K. 2015. Canonical poly(A) polymerase activity promotes the decay of a wide variety of mammalian nuclear RNAs. PLoS Genet.11, e1005610.
Wahle E., Winkler G.S. 2013. RNA decay machines: Deadenylation by the Ccr4-not and Pan2-Pan3 complexes. Biochim. Biophys. Acta.1829, 561–570.
Zheng D., Ezzeddine N., Chen C.Y., Zhu W., He X., Shyu A.B. 2008. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol.182, 89–101.
Chen C.Y., Shyu A.B. 2011. Mechanisms of deadenylation-dependent decay. Wiley Interdisc. Rev. RNA.2, 167–183.
Milac A.L., Bojarska E., Wypijewska del Nogal A. 2014. Decapping scavenger (DcpS) enzyme: Advances in its structure, activity and roles in the cap-dependent mRNA metabolism. Biochim. Biophys. Acta.1839, 452–462.
Chowdhury A., Mukhopadhyay J., Tharun S. 2007. The decapping activator Lsm1p–7p–Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA.13, 998–1016.
Luo Y., Na Z., Slavoff S.A. 2018. P-bodies: composition, properties, and functions. Biochemistry.57, 2424–2431.
Sheth U., Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science.300, 805–808.
Lim J., Ha M., Chang H., Kwon S.C., Simanshu D.K., Patel D.J., Kim V.N. 2014. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell.159, 1365–1376.
Munoz-Tello P., Rajappa L., Coquille S., Thore S. 2015. Polyuridylation in eukaryotes: A 3'-end modification regulating RNA life. BioMed Res. Internat.2015, 968127.
Zigackova D., Vanacova S. 2018. The role of 3' end uridylation in RNA metabolism and cellular physiology. Philos. Trans. R. Soc. Lond. B.373, 20180171.
Chang H., Lim J., Ha M., Kim V.N. 2014. TAIL-seq: Genome-wide determination of poly(A) tail length and 3' end modifications. Mol. Cell.53, 1044–1052.
Rissland O.S., Norbury C.J. 2009. Decapping is preceded by 3' uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol.16, 616–623.
Song M.G., Kiledjian M. 2007. 3' Terminal oligo U‑tract-mediated stimulation of decapping. RNA.13, 2356–2365.
Haas G., Cetin S., Messmer M., Chane-Woon-Ming B., Terenzi O., Chicher J., Kuhn L., Hammann P., Pfeffer S. 2016. Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res.44, 2873–2887.
Lapointe C.P., Wickens M. 2013. The nucleic acid-binding domain and translational repression activity of a Xenopus terminal uridylyl transferase. J. Biol. Chem.288, 20723–20733.
Sement F.M., Ferrier E., Zuber H., Merret R., Alioua M., Deragon J.M., Bousquet-Antonelli C., Lange H., Gagliardi D. 2013. Uridylation prevents 3' trimming of oligoadenylated mRNAs. Nucleic Acids Res.41, 7115–7127.
Thomas M.P., Liu X., Whangbo J., McCrossan G., Sanborn K.B., Basar E., Walch M., Lieberman J. 2015. Apoptosis triggers specific, rapid, and global mRNA decay with 3' uridylated intermediates degraded by DIS3L2. Cell Repts.11, 1079–1089.
Malecki M., Viegas S.C., Carneiro T., Golik P., Dressaire C., Ferreira M.G., Arraiano C.M. 2013. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J.32, 1842–1854.
Le Pen J., Jiang H., Di Domenico T., Kneuss E., Kosalka J., Leung C., Morgan M., Much C., Rudolph K.L.M., Enright A.J., O’Carroll D., Wang D., Miska E.A. 2018. Terminal uridylyltransferases target RNA viruses as part of the innate immune system. Nat. Struct. Mol. Biol.25, 778–786.
Pandey N.B., Marzluff W.F. 1987. The stem-loop structure at the 3' end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol.7, 4557–4559.
Tan D., Marzluff W.F., Dominski Z., Tong L. 2013. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3'hExo ternary complex. Science.339, 318–321.
Lackey P.E., Welch J.D., Marzluff W.F. 2016. TUT7 catalyzes the uridylation of the 3' end for rapid degradation of histone mRNA. RNA.22, 1673–1688.
Mullen T.E., Marzluff W.F. 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5' to 3' and 3' to 5'. Genes Dev.22, 50–65.
Hoefig K.P., Rath N., Heinz G.A., Wolf C., Dameris J., Schepers A., Kremmer E., Ansel K.M., Heissmeyer V. 2013. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat. Struct. Mol. Biol.20, 73–81.
Barreau C., Paillard L., Osborne H.B. 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res.33, 7138–7150.
Fan X.C., Myer V.E., Steitz J.A. 1997. AU-rich elements target small nuclear RNAs as well as mRNAs for rapid degradation. Genes Dev.11, 2557–2568.
Gherzi R., Lee K.Y., Briata P., Wegmuller D., Moroni C., Karin M., Chen C.Y. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell.14, 571–583.
Sanduja S., Blanco F.F., Dixon D.A. 2011. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdisc. Rev. RNA.2, 42–57.
Mukherjee D., Gao M., O’Connor J.P., Raijmakers R., Pruijn G., Lutz C.S., Wilusz J. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J.21, 165–174.
Stoecklin G., Mayo T., Anderson P. 2006. ARE-mRNA degradation requires the 5'-3' decay pathway. EMBO Repts.7, 72–77.
Gratacos F.M., Brewer G. 2010. The role of AUF1 in regulated mRNA decay. Wiley Interdisc. Rev. RNA.1, 457–473.
Shyu A.B., Belasco J.G., Greenberg M.E. 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev.5, 221–231.
Grosset C., Chen C.Y., Xu N., Sonenberg N., Jacquemin-Sablon H., Shyu A.B. 2000. A mechanism for translationally coupled mRNA turnover: Interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell.103, 29–40.
Chang T.C., Yamashita A., Chen C.Y., Yamashita Y., Zhu W., Durdan S., Kahvejian A., Sonenberg N., Shyu A.B. 2004. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev.18, 2010–2023.
Lemm I., Ross J. 2002. Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol.Cell. Biol.22, 3959–3969.
Wisdom R., Lee W. 1991. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev.5, 232–243.
Vlasova-St Louis I., Dickson A.M., Bohjanen P.R., Wilusz C.J. 2013. CELFish ways to modulate mRNA decay. Biochim. Biophys. Acta.1829, 695–707.
Jaillon O., Bouhouche K., Gout J.F., Aury J.M., Noel B., Saudemont B., Nowacki M., Serrano V., Porcel B.M., Segurens B., Le Mouel A., Lepere G., Schachter V., Betermier M., Cohen J., et al. 2008. Translational control of intron splicing in eukaryotes. Nature.451, 359–362.
Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C. 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet.36, 1073–1078.
Schweingruber C., Rufener S.C., Zund D., Yamashita A., Muhlemann O. 2013. Nonsense-mediated mRNA decay: Mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta.1829, 612–623.
Gardner L.B. 2008. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol. Cell. Biol.28, 3729–3741.
Li Z., Vuong J.K., Zhang M., Stork C., Zheng S. 2017. Inhibition of nonsense-mediated RNA decay by ER stress. RNA.23, 378–394.
Martin L., Gardner L.B. 2015. Stress-induced inhibition of nonsense-mediated RNA decay regulates intracellular cystine transport and intracellular glutathione through regulation of the cystine/glutamate exchanger SLC7A11. Oncogene.34, 4211–4218.
Yepiskoposyan H., Aeschimann F., Nilsson D., Okoniewski M., Muhlemann O. 2011. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA.17, 2108-2118.
McGlincy N.J., Smith C.W. 2008. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci.33, 385–393.
Rossbach O., Hung L.H., Schreiner S., Grishina I., Heiner M., Hui J., Bindereif A. 2009. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol. Cell Biol.29, 1442–1451.
Wollerton M.C., Gooding C., Wagner E.J., Garcia-Blanco M.A., Smith C.W. 2004. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell.13, 91–100.
Izumikawa K., Yoshikawa H., Ishikawa H., Nobe Y., Yamauchi Y., Philipsen S., Simpson R.J., Isobe T., Takahashi N. 2016. Chtop (Chromatin target of Prmt1) auto-regulates its expression level via intron retention and nonsense-mediated decay of its own mRNA. Nucleic Acids Res.44, 9847–9859.
Yan Q., Weyn-Vanhentenryck S.M., Wu J., Sloan S.A., Zhang Y., Chen K., Wu J.Q., Barres B.A., Zhang C. 2015. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc. Natl. Acad. Sci. U. S. A.112, 3445–3450.
Guan Q., Zheng W., Tang S., Liu X., Zinkel R.A., Tsui K.W., Yandell B.S., Culbertson M.R. 2006. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet.2, e203.
Shvarts A.M., Komarova T.V., Skulachev M.V., Zvereva A.S., Dorokhov Yu.L., Atabekov I.G. 2007. mRNA stability in plants depends on the length of 3'-untranslated region. Biokhimiya. 72, 260–269.
Schell T., Kocher T., Wilm M., Seraphin B., Kulozik A.E., Hentze M.W. 2003. Complexes between the nonsense-mediated mRNA decay pathway factor human upf1 (up-frameshift protein 1) and essential nonsense-mediated mRNA decay factors in HeLa cells. Biochem. J.373, 775–783.
He F., Jacobson A. 2015. Nonsense-mediated mRNA decay: Degradation of defective transcripts is only part of the story. Annu. Rev. Genet.49, 339–366.
Grimson A., O’Connor S., Newman C.L., Anderson P. 2004. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans.Mol. Cell. Biol.24, 7483–7490.
Huntzinger E., Kashima I., Fauser M., Sauliere J., Izaurralde E. 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA.14, 2609–1267.
Lykke-Andersen S., Chen Y., Ardal B.R., Lilje B., Waage J., Sandelin A., Jensen T.H. 2014. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev.28, 2498–2517.
Loh B., Jonas S., Izaurralde E. 2013. The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev.27, 2125–2138.
Nicholson P., Gkratsou A., Josi C., Colombo M., Muhlemann O. 2018. Dissecting the functions of SMG5, SMG7, and PNRC2 in nonsense-mediated mRNA decay of human cells. RNA.24, 557–573.
Kurosaki T., Miyoshi K., Myers J.R., Maquat L.E. 2018. NMD-degradome sequencing reveals ribosome-bound intermediates with 3'-end non-templated nucleotides. Nat. Struct. Mol. Biol.25, 940–950.
Brogna S., McLeod T., Petric M. 2016. The meaning of NMD: Translate or perish. Trends Genet.: TIG.32, 395–407.
Chamieh H., Ballut L., Bonneau F., Le Hir H. 2008. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol.15, 85–93.
Amrani N., Ganesan R., Kervestin S., Mangus D.A., Ghosh S., Jacobson A. 2004. A faux 3'-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature.432, 112–118.
Kim Y.K., Furic L., Desgroseillers L., Maquat L.E. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell.120, 195–208.
Kim Y.K., Furic L., Parisien M., Major F., DesGroseillers L., Maquat L.E. 2007. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J.26, 2670–2681.
Gong C., Maquat L.E. 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature.470, 284–288.
Lucas B.A., Lavi E., Shiue L., Cho H., Katzman S., Miyoshi K., Siomi M.C., Carmel L., Ares M., Jr., Maquat L.E. 2018. Evidence for convergent evolution of SINE-directed Staufen-mediated mRNA decay. Proc. Natl. Acad. Sci. U. S. A.115, 968–973.
Wang J., Gong C., Maquat L.E. 2013. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev.27, 793–804.
Park E., Gleghorn M.L., Maquat L.E. 2013. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc. Natl. Acad. Sci. U. S. A.110, 405–412.
Kim Y.K., Maquat L.E. 2019. UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA.25, 407–422.
Inada T. 2013. Quality control systems for aberrant mRNAs induced by aberrant translation elongation and termination. Biochim. Biophys. Acta.1829, 634–642.
Klauer A.A., van Hoof A. 2012. Degradation of mRNAs that lack a stop codon: A decade of nonstop progress. Wiley Interdisc. Rev. RNA.3, 649–660.
Tsuboi T., Kuroha K., Kudo K., Makino S., Inoue E., Kashima I., Inada T. 2012. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3' end of aberrant mRNA. Mol. Cell.46, 518–529.
Harigaya Y., Parker R. 2010. No-go decay: A quality control mechanism for RNA in translation. Wiley Interdisc. Rev. RNA.1, 132–141.
Passos D.O., Doma M.K., Shoemaker C.J., Muhlrad D., Green R., Weissman J., Hollien J., Parker R. 2009. Analysis of Dom34 and its function in no-go decay. Mol. Biol. Cell.20, 3025–3032.
Saito S., Hosoda N., Hoshino S. 2013. The Hbs1–Dom34 protein complex functions in non-stop mRNA decay in mammalian cells. J. Biol. Chem.288, 17832–17843.
Szadeczky-Kardoss I., Csorba T., Auber A., Schamberger A., Nyiko T., Taller J., Orban T.I., Burgyan J., Silhavy D. 2018. The nonstop decay and the RNA silencing systems operate cooperatively in plants. Nucleic Acids Res.46, 4632–4648.
Szadeczky-Kardoss I., Gal L., Auber A., Taller J., Silhavy D. 2018. The no-go decay system degrades plant mRNAs that contain a long A-stretch in the coding region. Plant Sci.: Internat. J. Exp. Plant Biol.275, 19–27.
Agrawal N., Dasaradhi P.V., Mohmmed A., Malhotra P., Bhatnagar R.K., Mukherjee S.K. 2003. RNA interference: Biology, mechanism, and applications. Microbiol. Mol. Biol. Rev.: MMBR.67, 657–685.
Fang X., Qi Y. 2016. RNAi in plants: An argonaute-centered view. Plant Cell.28, 272–285.
Fischer S.E.J. 2015. RNA interference and microRNA-mediated silencing. Curr. Protocols Mol. Biol.112, 2611–2615.
Vilgelm A.E., Chumakov S.P., Prasolov V.S. 2006. RNA interference: Biology and prospects of application in biomedicine and biotechnology. Mol. Biol. (Moscow). 40 (3), 339–354.
Fatica A., Tollervey D. 2002. Making ribosomes. Curr. Opin. Cell Biol.14, 313–318.
Briggs M.W., Burkard K.T., Butler J.S. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8S rRNA 3' end formation. J. Biol. Chem.273, 13255–13263.
Wang M., Pestov D.G. 2011. 5'-end surveillance by Xrn2 acts as a shared mechanism for mammalian pre-rRNA maturation and decay. Nucleic Acids Res.39, 1811–1822.
Dez C., Houseley J., Tollervey D. 2006. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae.EMBO J.25, 1534–1546.
LaRiviere F.J., Cole S.E., Ferullo D.J., Moore M.J. 2006. A late-acting quality control process for mature eukaryotic rRNAs. Mol. Cell.24, 619–626.
Cole S.E., LaRiviere F.J., Merrikh C.N., Moore M.J. 2009. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol. Cell.34, 440–450.
Gudipati R.K., Xu Z., Lebreton A., Seraphin B., Steinmetz L.M., Jacquier A., Libri D. 2012. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol. Cell.48, 409–421.
Hamill S., Wolin S.L., Reinisch K.M. 2010. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc. Natl. Acad. Sci. U. S. A.107, 15045–15050.
Alexandrov A., Chernyakov I., Gu W., Hiley S.L., Hughes T.R., Grayhack E.J., Phizicky E.M. 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell.21, 87–96.
Chernyakov I., Whipple J.M., Kotelawala L., Grayhack E.J., Phizicky E.M. 2008. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5'-3' exonucleases Rat1 and Xrn1. Genes Dev.22, 1369–1680.
Betat H., Morl M. 2015. The CCA-adding enzyme: A central scrutinizer in tRNA quality control. BioEssays: News Rev. Mol.,Cell. Dev. Biol.37, 975–982.
Wilusz J.E., Whipple J.M., Phizicky E.M., Sharp P.A. 2011. tRNAs marked with CCACCA are targeted for degradation. Science.334, 817–821.
Thompson D.M., Parker R. 2009. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae.J. Cell Biol.185, 43–50.
Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., Tollervey D. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J.18, 5399–5410.
Zhang L., Wan Y., Huang G., Wang D., Yu X., Huang G., Guo J. 2015. The exosome controls alternative splicing by mediating the gene expression and assembly of the spliceosome complex. Sci. Repts.5, 13403.
Didychuk A.L., Butcher S.E., Brow D.A. 2018. The life of U6 small nuclear RNA, from cradle to grave. RNA.24, 437–460.
Hrossova D., Sikorsky T., Potesil D., Bartosovic M., Pasulka J., Zdrahal Z., Stefl R., Vanacova S. 2015. RBM7 subunit of the NEXT complex binds U-rich sequences and targets 3'-end extended forms of snRNAs. Nucleic Acids Res.43, 4236–4248.
Hilcenko C., Simpson P.J., Finch A.J., Bowler F.R., Churcher M.J., Jin L., Packman L.C., Shlien A., Campbell P., Kirwan M., Dokal I., Warren A.J. 2013. Aberrant 3' oligoadenylation of spliceosomal U6 small nuclear RNA in poikiloderma with neutropenia. Blood.121, 1028–1038.
Shukla S., Parker R. 2014. Quality control of assembly-defective U1 snRNAs by decapping and 5'-to-3' exonucleolytic digestion. Proc. Natl. Acad. Sci. U. S. A.111, E3277–E3786.
Labno A., Warkocki Z., Kulinski T., Krawczyk P.S., Bijata K., Tomecki R., Dziembowski A. 2016. Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acids Res.44, 10437–10453.
van Hoof A., Lennertz P., Parker R. 2000. Yeast exosome mutants accumulate 3'-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol.20, 441–452.
Larochelle M., Lemay J.F., Bachand F. 2012. The THO complex cooperates with the nuclear RNA surveillance machinery to control small nucleolar RNA expression. Nucleic Acids Res.40, 10240–10253.
Lubas M., Christensen M.S., Kristiansen M.S., Domanski M., Falkenby L.G., Lykke-Andersen S., Andersen J.S., Dziembowski A., Jensen T.H. 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell.43, 624–637.
Buscher M., Horos R., Hentze M.W. 2020. “High vault-age”: non-coding RNA control of autophagy. Open Biol.10, 190307.
Horos R., Buscher M., Sachse C., Hentze M.W. 2019. Vault RNA emerges as a regulator of selective autophagy. Autophagy.15, 1463–1464.
Liu X., Zheng Q., Vrettos N., Maragkakis M., Alexiou P., Gregory B.D., Mourelatos Z. 2014. A microRNA precursor surveillance system in quality control of microRNA synthesis. Mol. Cell.55, 868–879.
Chang H.M., Triboulet R., Thornton J.E., Gregory R.I. 2013. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature.497, 244–248.
Gantier M.P., McCoy C.E., Rusinova I., Saulep D., Wang D., Xu D., Irving A.T., Behlke M.A., Hertzog P.J., Mackay F., Williams B.R. 2011. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res.39, 5692–5703.
Sanei M., Chen X. 2015. Mechanisms of microRNA turnover. Curr. Opin. Plant Biol.27, 199–206.
Wang X., Wang Y., Dou Y., Chen L., Wang J., Jiang N., Guo C., Yao Q., Wang C., Liu L., Yu B., Zheng B., Chekanova J.A., Ma J., Ren G. 2018. Degradation of unmethylated miRNA/miRNA*s by a DEDDy-type 3' to 5' exoribonuclease Atrimmer 2 in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A.115, E6659–E6667.
Kamminga L.M., Luteijn M.J., den Broeder M.J., Redl S., Kaaij L.J., Roovers E.F., Ladurner P., Berezikov E., Ketting R.F. 2010. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J.29, 3688–3700.
Szczepinska T., Kalisiak K., Tomecki R., Labno A., Borowski L.S., Kulinski T.M., Adamska D., Kosinska J., Dziembowski A. 2015. DIS3 shapes the RNA polymerase II transcriptome in humans by degrading a variety of unwanted transcripts. Genome Res.25, 1622–1633.
Tisseur M., Kwapisz M., Morillon A. 2011. Pervasive transcription: Lessons from yeast. Biochimie.93, 1889–1896.
Thiebaut M., Kisseleva-Romanova E., Rougemaille M., Boulay J., Libri D. 2006. Transcription termination and nuclear degradation of cryptic unstable transcripts: A role for the Nrd1–Nab3 pathway in genome surveillance. Mol. Cell.23, 853–864.
Thompson D.M., Parker R. 2007. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae.Mol. Cell. Biol.27, 92–101.
Belostotsky D. 2009. Exosome complex and pervasive transcription in eukaryotic genomes. Curr. Opin. Cell Biol.21, 352–358.
Chekanova J.A., Gregory B.D., Reverdatto S.V., Chen H., Kumar R., Hooker T., Yazaki J., Li P., Skiba N., Peng Q., Alonso J., Brukhin V., Grossniklaus U., Ecker J.R., Belostotsky D.A. 2007. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell.131, 1340–1353.
Davidson L., Francis L., Cordiner R.A., Eaton J.D., Estell C., Macias S., Caceres J.F., West S. 2019. Rapid depletion of DIS3, EXOSC10, or XRN2 reveals the immediate impact of exoribonucleolysis on nuclear RNA metabolism and transcriptional control. Cell Rept.26, 2779–2791 e5.
Preker P., Nielsen J., Kammler S., Lykke-Andersen S., Christensen M.S., Mapendano C.K., Schierup M.H., Jensen T.H. 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science.322, 1851–1854.
van Dijk E.L., Chen C.L., d’Aubenton-Carafa Y., Gourvennec S., Kwapisz M., Roche V., Bertrand C., Silvain M., Legoix-Ne P., Loeillet S., Nicolas A., Thermes C., Morillon A. 2011. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature.475, 114–117.
Wery M., Descrimes M., Vogt N., Dallongeville A.S., Gautheret D., Morillon A. 2016. Nonsense-mediated decay restricts lncRNA levels in yeast unless blocked by double-stranded RNA structure. Mol. Cell.61, 379–392.
Muhlrad D., Parker R. 1999. Aberrant mRNAs with extended 3' UTRs are substrates for rapid degradation by mRNA surveillance. RNA.5, 1299–1307.
Ustianenko D., Pasulka J., Feketova Z., Bednarik L., Zigackova D., Fortova A., Zavolan M., Vanacova S. 2016. TUT-DIS3L2 is a mammalian surveillance pathway for aberrant structured non-coding RNAs. EMBO J.35, 2179–2191.
Funding
This work was supported by the Russian Foundation for Basic Research (project no. 18-34-00247mol_a) and the Russian Science Foundation (project no. 19-14-00327).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This work does not contain any studies involving animals or human subjects performed by any of the authors.
Additional information
Translated by T. Tkacheva
Abbreviations: 3′-UTR, 3′-untranslated region; AMD, AU-rich element-mediated decay; ARE, AU-rich element; CUT, cryptic unstable transcript; Dis3L2, Dis3-like 3′–5′ exoribonuclease; GMD, GU-rich element-mediated decay; GRE, GU-rich element; hTRAMP, human TRAMP; NEXT, nuclear exosome targeting complex; NGD, no-go decay; NMD, nonsense-mediated decay; NRD, nonfunctional rRNA decay; NSD, nonstop decay; PABP, poly(A)-binding protein; PAP, poly(A) polymerase; PPD, PABPN1 and PAPα/γ-mediated decay; PTC, premature termination codon; RTD, rapid tRNA decay; SINE, short interspersed element; siRNA, small interfering RNA; SKI, Superkiller (complex, a cytoplasmic exosome cofactor); SMD, Staufen1-mediated decay; TRAMP, Trf4/5–Air1/2–Mtr4 polyadenylation complex; snoRNA, small nucleolar RNA; snRNA, small nuclear RNA; ncRNA, non-coding RNA; pre-mRNA, precursor mRNA; pre-microRNA, precursor microRNA; pri-microRNA, primary microRNA transcript (pre-microRNA precursor); TUTase, terminal uridylyl transferase.
Rights and permissions
About this article
Cite this article
Tatosyan, K.A., Ustyantsev, I.G. & Kramerov, D.A. RNA Degradation in Eukaryotic Cells. Mol Biol 54, 485–502 (2020). https://doi.org/10.1134/S0026893320040159
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893320040159