Abstract
A nonmotile gram-negative, short, rod-shaped strain, BGMRC 2024T, was isolated from the sediment of Rhizophora stylosa collected in the Beibu Gulf, China. The strain BGMRC 2024T exhibited optimal growth at 25 to 28°C, pH 7.0, and in the presence of 1–4% (wt/v) NaCl. Analysis of the 16S rRNA gene sequences indicated that BGMRC 2024T was closely related to Pseudooceanicola antarcticus CGMCC 1.12662T (96.23% sequence similarity), P. algae Lw-13eT (96.17%), P. lipolyticus 157T (96.16%), and P. pacificus 216_PA32_1T (96.09%). The digital DNA-DNA hybridization (dDDH) values and the average nucleotide identity (ANI) of the strain Pseudooceanicola antarcticus CGMCC 1.12662T were 24.7 and 77.52%, respectively. The DNA G+C content was 67.4 mol %. The main respiratory quinone was ubiquinone-10. The polar lipids of strain BGMRC 2024T included four unidentified phospholipids, two unidentified ninhydrin-positive lipids, one unidentified ninhydrin-positive phospholipid, and three unidentified lipids. The major fatty acids were C19:0 cyclo ω8c, summed feature 8 (C18:1ω7c and/or C18:1ω6c), and C16:0. Based on these results, BGMRC 2024T represents a novel species of the genus Pseudooceanicola, for which the name Pseudooceanicola albus sp. nov. is proposed. The type strain is BGMRC 2024T (=KCTC52117T = DSM102086T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The genus Pseudooceanicola, with Pseudooceanicola atlanticus 22II-s11gT as the type strain belongs to the family Rhodobacteraceae. It was proposed by Lai et al. (2015). Based on their genotypic and phenotypic characteristics, six misclassified species were subsequently transferred from the Oceanicola genus to the Pseudooceanicola genus (Lai et al., 2015). Currently, there are 13 Pseudooceanicola species with validly published names: Pseudooceanicola aestuarii (Li et al., 2020), P. algae (Wolter et al., 2021), P. antarcticus (Huo et al., 2014; Lai et al., 2015), P. atlanticus (Lai et al., 2015), P. batsensis (Cho et al., 2004; Lai et al., 2015), P. endophyticus (Zheng et al., 2021), P. flagellates (Huo et al., 2014; Lai et al., 2015), P. lipolyticus (Huang et al., 2018), P. marinus (Lin et al., 2007; Lai et al., 2015), P. nanhaiensis (Gu et al., 2007; Lai et al., 2015), P. nitratireducens (Zheng et al., 2010; Lai et al., 2015), P. onchidiid (Yin et al., 2020), and P. pacificus (Lina et al., 2020), (https://lpsn.dsmz.de/genus/ pseudooceanicola). All of them were isolated from ocean environmental resources such as seawater, sediments, or invertebrates.
In the course of the investigation of the bacterial community from offshore areas of the Beibu Gulf, China, a novel bacterium, designated BGMRC 2024T, was isolated from the soil of Rhizophora stylosa. In the present study, we conducted a polyphasic taxonomic study to clarify the taxonomic position of strain BGMRC 2024T.
MATERIALS AND METHODS
Bacterial Isolation and Cultivation
The sediment sample from the Rhizophora stylosa rhizosphere was obtained from the Beibu Gulf, Beihai, PR China (21°44′53″ N, 108°36′14″ E). To obtain bacterial isolates, sediment samples were homogenized in sterile seawater for 1 h. After 1 h, 200-μL aliquots of the suspension were plated on ISP2 medium (2.0 g yeast extract, 2.0 g malt extract, 2.0 g D-(+)-glucose anhydrous, 15.0 g agar powder, and 1 L seawater). The plates were incubated at 28°C for 7 days. An ivory colony was used to isolate the bacterial strain BGMRC 2024. The pure culture of the strain was preserved in glycerol suspension (30%, v/v) at –80°C (Li et al., 2021). For comparative analyzes, P. atlanticus MCCC1A09160T and P. lipolyticus MCCC 1K03317T were purchased from the Marine Culture Collection of China (MCCC), and P. antarcticus CGMCC1.12662T was obtained from the China General Microbiological Culture Collection Center (CGMCC).
Phenotypic and Biochemical Characteristics
The cells of strain BGMRC 2024T were grown in ISP2 at 28°C for 48 h for morphological observation and physiological tests. Scanning electron microscopy (FEI Quanta 250 environmental scanning electron microscope) was used to observe the morphology and size of the cells. The presence of flagella was observed using transmission electron microscopy (HITACHI transmission electron microscope HT7700). Gram staining was tested using a Gram stain kit (Difco) according to the manufacturer’s instructions. Hydrolysis tests for casein, gelatin, starch, urea, and Tweens 20 and 40 were carried out by conventional methods (Dong and Cai, 2001). Strain BGMRC 2024T was incubated in ISP2 medium at different temperatures (4, 10, 15, 20, 25, 28, 37, 40, and 45°C), different concentrations of NaCl (0–5%, w/v, at a 0.5% interval), and different pH values (pH 4.0–12.0 at 1 pH unit intervals) to measure its growth (OD600) using an UV-1800 spectrometer (Lai et al., 2015). Oxidase activity was determined with 1% (w/v) N,N,N ′,N ′-tetramethyl-p-phenylenediamine, and catalase activity was determined by formation of bubbles upon addition of 3% H2O2 (Choi et al., 2014). The GEN III microplate (Biolog, Inc., United States) was used to test the utilization of acid production from different carbon sources. Other enzyme activities and physiological tests were determined using the API ZYM, API 20E, and API 50 CH system strips (bioMérieux), which were used to test the biochemical characteristics of the strain.
Phylogenetic Analysis
Genomic DNA was extracted using a bacterial genomic DNA Mini kit following the manufacturer’s recommendations. The 16S rRNA gene was amplified using AccuPower PCR PreMix (Bioneer, Daejeon, Korea) and the bacteria-specific primers 27F and 1492R (Yang et al., 2018). The 16S rRNA gene sequence was used for pairwise sequence alignment performed by the EzBioCloud server (http://www.ezbiocloud.net) (Kim et al., 2012). Multiple sequence alignment was performed with the CLUSTAL W program of the MEGA 7 package (Tamura et al., 2011). Evolutionary distances for the phylogenetic trees were calculated with the Kimura two-parameter model (Kimura et al., 1980). Phylogenetic trees were constructed by using the neighbor-joining (Saitou and Nei, 1987), maximum-likelihood (Felsenstein, 1981), and maximum-parsimony (Fitch et al., 1971), algorithms using the MEGA7.0 version (Tamura et al., 2011). The topologies of the phylogenetic trees were assessed by bootstrap analysis based on 1000 replicates (Felsenstein, 1985).
Genome Analysis
The draft genome sequence of strain BGM-RC 2024T was determined using the Illumina Hiseq 4000 system (Illumina, San Diego, CA, United States) and was sequenced by BGI (Wuhan, China). Genomic information of Pseudooceanicola antarcticus Ar-45T (GCA_002786285.1), P. atlanticus 22II-s11gT (GCA_000768315.1), P. lipolyticus 157T (GCA_002786325.1), P. nanhaiensis DSM 18065T (GCA_000688295.1), P. batsensis HTCC2597T, P. marinus CECT7751T (GCA_900172385.1), and P. nitratireducens DSM 29619T (GCA_900112545.1) was downloaded from GenBank, and the new strain BGMRC 2024T (WUMU00000000) was sequenced in BGI. The G + C DNA contents of strain BGM-RC 2024T and closely related type strains were calculated from their whole genome sequence. The average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) values between strain BGMRC 2024T and closely related type strains were calculated using the ANI calculator tool (https:// www.ezbiocloud.net/tools/ani) and the genome-to-genome distance calculator tool (https://ggdc. dsmz.de/ggdc.php) (Meier-Kolthoff et al., 2013), respectively. Gene function prediction was performed using the Rapid Annotation using Subsystem Technology (RAST) server (https://rast.nmpdr.org) (Aziz et al., 2008). The codon Tree method selects single-copy PATRIC PGFams and analyzes aligned proteins and coding DNA from single-copy genes using the program RAxML (Stamatakis et al., 2008).
Chemotaxonomic Analysis
The strain BGMRC 2024T and the reference strains P. atlanticus MCCC1A09160T, P. lipolyticus MCCC 1K03317T, and P. antarcticus CGMCC1.12662T were grown in ISP2 medium supplemented with 3% sea salt at 28°C for three days and then the biomass was collected for chemotaxonomic analysis.
The composition of cellular fatty acids was determined at Yunnan University (Yunnan, China) using the Sherlock Microbial Identification System (Version 6.0). Fatty acids were analyzed using gas chromatography and identified using the microbial identification software package based on the TSBA6 6.0 database (Kämpfer et al., 1996). Polar lipids were extracted with chloroform/methanol (1 : 2, v/v) solvent (Komagata and Suzuki, 1987) and analyzed by the method described previously (Cao et al., 2014). The respiratory quinones were extracted from freeze dried cells of strain BGMRC 2024T with chloroform/methanol (2 : 1, vol/vol) (Komagata and Suzuki, 1987) and analyzed by ultra-performance liquid chromatography (Nakagawa et al., 1993).
RESULTS AND DISCUSSION
Phenotypic and Physiological Characteristics
Cells of strain BGMRC 2024T were short nonmotile rods, 0.8 μm wide and 0.5–1.0 μm long (Fig. S1), and stained gram-negative. The colonies were white. The growth of strain BGMRC 2024T was observed at 1.0–7.0% (w/v) NaCl, (optimum at 3% w/v), the temperature range of 20–37°C (optimum at 28°C), and pH 6.0–9.0 (optimum at pH 7.0) (Table 1). The activities of oxidase and catalase were positive. The comparison of the morphological, physiological, and biochemical characteristics between strain BGMRC 2024T and the reference strains P. atlanticus MCCC1A09160T, P. lipolyticus MCCC 1K03317T, and P. antarcticus CGMCC1.12662T is presented in Tables 1 and S1.
The strain BGMRC 2024T shared most of the features with other members of the genus Pseudooceanicola. However, there were some differences between the novel isolate and the closely related species. For example, strain BGMRC 2024T was negative for Tween 20 and Tween 40 hydrolysis (Table 1). In the API ZYM, strain BGMRC 2046T showed positive activity for α-glucanase and β-glucosidase, unlike P. atlanticus MCCC1A09160T and P. lipolyticus MCCC 1K03317T. In API 50CH, strain BGMRC 2024T produced acid from L-arabinose, D-fructose, D-sorbitol, D-melibiose, D-turanose, and D-mannose, unike P. atlanticus MCCC1A09160T, P. antarcticus CGMCC1.12662T, and P. lipolyticus MCCC 1K03317T (Table 1 and Table S1).
Phylogenetic Analysis
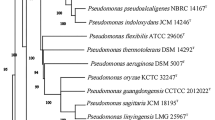
The sequenced length of the 16S rRNA gene of the strain BGMRC 2024T was 4 588 940 bp. Phylogenetic analysis of strain BGMRC 2024T based on 16S rRNA gene sequences indicated that the novel strain belonged to the genus Pseudooceanicola and exhibited 16S rRNA gene sequence similarities with the type strains Pseudooceanicola antarcticus CGMCC 1.12662T (96.23% sequence identity), P. algae Lw-13eT (96.17%), P. lipolyticus 157T (96.16%), and P. pacificus 216_PA32_1T (96.09%), respectively. In the analysis of the neighbor-joining tree, strain BGMRC 2024T formed a distinct clade with P. antarcticus CGMCC 1.12662T, P. marinus CECT 7751T (96.17%), and P. lipolyticus 157T (Fig. 1). The maximum-parsimony and maximum-likelihood trees also showed the closely phylogenetic relationship of the strain BGMRC 2024T and the reference strains (Figs. S3 and S4).
Neighbor-joining phylogenetic tree based on the 16S rRNA gene sequences showing the relationships between strain BGMRC 2024T and related taxa. Amaricoccus kaplicensis Ben101T was used as the outgroup. The numerals at the nodes represent the percentage bootstrap values (>50%) of 1000 bootstrap replicates from NJ algorithms. Bar, 0.01 substitutions per nucleotide position.
The ANI and dDDH values of strain BGMRC 2024T to its closely related species (P. antarcticus Ar-45T, P. atlanticus 22II-s11gT, P. lipolyticus 157T, P. nanhaiensis DSM 18065T, P. batsensis HTCC2597T, P. marinus CECT7751T, and P. nitratireducens DSM 29619T) ranged from 74.3 to 77.5% and 23.3 to 24.7%, the recommended cutoff for genus delineation at genome level ( Goris et al., 2007; Richter et al., 2009) (Table 2). However, the protein-based core genome phylogeny indicated that the strain BGMRC 2024T formed a subgroup with P. endophyticus CBS1P-1T, P. marinus CECT 7751T, and P. antarcticus CGMCC 1.12662T and a robust clade with genus Pseudooceanicola (Fig. S5). Thus, strain BGMRC 2024T represents a novel species of the genus Pseudooceanicola.
Genomic Characteristics
The draft genome size of strain BGMRC 2024T (4.6 Mb) was much smaller than that of P. atlanticus 22II-s11gT (4.9 Mb). The draft genome of strain BGMRC 2024T was deposited in DDBJ/ENA/GenBank under the accession number WUMU00000000.
The G + C content of genomic DNA is 67.4 mol %, which is close to the values for P. nanhaiensis (67.9%) and P. marinus (66.8%). Gene prediction allowed the annotation of 5068 protein-coding genes. The genome of BGMRC 2024T is much larger than those of P. antarcticus Ar-45T (4.3 Mb), P. batsensis HTCC2597T (4.4 Mb), P. marinus CECT7751T (4.5 Mb), and P. nitratireducens DSM 29619T (4.1 Mb). The numbers of genes putatively involved in various aspects of the stress response, iron acquisition and metabolism, regulation and cell signaling, protein metabolism, respiration, amino acids, and derivatives in the genome of BGMRC 2024T was higher than those of P. antarcticus Ar-45T and P. atlanticus 22II-s11gT, while the numbers of genes putatively involved in nitrogen metabolism, lipids, fatty acids, motility and chemotaxis, isoprenoids, membrane transport, and virulence, disease, and defense in the genome of BGMRC 2024T was lower than those of P. antarcticus Ar-45T and P. atlanticus 22II-s11gT (Supplementary Table S2). Furthermore, denitrification genes were present in P. antarcticus Ar-45T and P. atlanticus 22II-s11gT, but not in strain BGMRC 2024T. Based on the strain annotation, the genome sequences of BGMRC 2024T unraveled the presence of genes involved in ectoine biosynthesis (Supplementary Table S3). Moreover, the new strain and the other three types strains P. atlanticus MCCC1A09160T, P. antarcticus CGMCC1.12662T, and P. lipolyticus MCCC 1K03317T possessed the genes putatively encoding riboflavin synthesis clusters.
Chemotaxonomic Analysis
The main cellular fatty acids (>5%) of strain BGMRC 2024T were identified as C19:0 cyclo ω8c (37.14%), summed feature 8 (C18:1ω7c and/or C18:1ω6c (28.32%)), C16:0 (17.92%), and C18:1ω7c 11-methyl (6.18%) (Table 2). Overall, strain BGMRC 2024T had a fatty acid profile similar to those of reference strains P. atlanticus MCCC1A09160T, P. lipolyticus MCCC 1K03317T, and P. antarcticus CGMCC1.12662T. However, strain BGMRC 2024T possessed relatively higher amounts of C19:0 cyclo ω8c and summed feature 8 (C18:1ω7c and/or C18:1ω6c), and lower amounts of C14:0, C18:0, C10:0 3-OH, and C12:0 3-OH (Table 2). The polar lipids profile of strain BGMRC 2024T consisted of two unidentified phospholipids (PL1,6), one unidentified ninhydrin-positive phospholipids (NPL), four unidentified ninhydrin-positive lipids (AL1–4), and three unidentified lipids (L3,4,11) (Fig. S2). However, the presence of one unidentified ninhydrin-positive phospholipid (NPL), one unidentified phospholipid (PL6), one unidentified ninhydrin-positive lipid (AL4), and one unidentified lipid (L11) differentiates the novel strain from those reference strains (Fig. S2). The predominant respiratory quinone was the ubiquinone Q-10, which was identical to that of other type strains of the genus Pseudooceanicola.
Description of Pseudooceanicola albus sp. nov.
Pseudooceanicola albus. (al’bus. L. masc. adj. albus white, referring to the color of the colonies).
The cells are short nonmotile rods with a size 0.4–0.8 μm wide and 0.5–1.0 μm long, and stain gram-negative. The colonies are white. The oxidase and catalase activities are positive. Growth occurs at 20 to 37°C (optimum at 28°C), pH 6.0 to 9.0 (optimum at pH 7.0) and 1.0 to 7.0% (w/v) NaCl (optimum, 1–3%). The main fatty acids are C19:0 cyclo ω8c, summed feature 8 (C18:1ω7c and/or C18:1ω6c), and C16:0. The main respiratory quinone is ubiquinone-10. The DNA G+C content is 67.4 mol %. In the API 20E tests, the strain was positive for O-nitrophenyl-β-D-galactopyranoside, urease test, and the VP test. In the API ZYM system, positive reactions are observed with leucine arylamidase, esterase (C4), esterase lipase (C8), naphthol-ASBI-phosphohydrolase, alkaline phosphatase, acid phosphatase, α-glucanase, and β-glucosidase. The results for D-fructose, D-sorbitol, D-melibiose, D-sucrose, D-turanose, L-arabinose and D-mannose, and ferric citrate of esculin are positive in the API 50CH system.
The type strain, BGMRC 2024T (=KCTC52117T = DSM102086T), was isolated from the rhizosphere soil of Rhizophora stylosa collected from the Beibu Gulf of China. The GenBank/EMBL/DDBJ accession numbers for the draft genome and 16S rRNA gene sequences of strain BGMRC 2024T are WUMU00000000 and MN067774, respectively.
REFERENCES
Aziz, R.K., Bartels, D., Best, A.A., DeJongh, M., Disz, T., Edwards, R.A., Formsma, K., Gerdes, S., Glass, E.M., Kubal, M., Meyer, F., Olsen, G.J., Olson, R., Osterman, A.L., Overbeek, R.A., et al., The RAST Server: rapid annotations using subsystems technology, BMC Genomics, 2008, vol. 9, p. 75.
Blin, K., Shaw, S., Kloosterman, A.M., Charlop-Powers, Z., Van Wezel, G.P., Medema, M.H., and Weber, T., AntiSMASH 6.0: improving cluster detection and comparison capabilities, Nucleic. Acids Res., 2021, vol. 49, pp. W29–W35.
Cao, J., Lai, Q., Li, G., and Shao, Z., Pseudopedobacter beijingensis gen. nov., sp. nov., isolated from coking wastewater activated sludge, and reclassification of Pedobacter saltans as Pseudopedobacter saltans comb. nov., Int. J. Syst. Evol. Microbiol., 2014, vol. 64, pp. 1853–1858.
Choi, J.H., Seok, J.H., Cha, J.H., and Cha, C.J., Lysobacter panacisoli sp. nov., isolated from ginseng soil, Int. J. Syst. Evol. Microbiol., 2014, vol. 64, pp. 2193–2197.
Cho, J.C. and Giovannoni, S.J., Oceanicola granulosus gen. nov., sp. nov. and Oceanicola batsensis sp. nov., poly-beta-hydroxybutyrate-producing marine bacteria in the order “Rhodobacterales,” Int. J. Syst. Evol. Microbiol., 2004, vol. 54, pp. 1129–1136.
Dong, X. and Cai, M., Determinative Manual for Routine Bacteriology, Beijing: Scientific Press, 2001.
Felsenstein, J., Evolutionary trees from DNA sequences: a maximum likelihood approach, J. Mol. Evol., 1981, vol. 17, pp. 368–376.
Fitch, W.M., Toward defining the course of evolution: minimum change for a specific tree topology, Syst. Zool., 1971, vol. 20, pp. 406–416.
Felsenstein, J., Confidence limits on phylogenies: an approach using the bootstrap, Evolution., 1985, vol. 39, pp. 783–791.
Gu, J., Guo, B., Wang, Y.N., Yu, S.L., Inamori, R., Qu, R., Ye, Y.G., and Wu, X.L., Oceanicola nanhaiensis sp. nov., isolated from sediments of the South China Sea, Int. J. Syst. Evol. Microbiol., 2007, vol. 57, pp. 157–160.
Huo, Y.Y., Li, Z.Y., You, H., Wang, C.S., Post, A.F., Oren, A., and Xu, X.W., Oceanicola antarcticus sp. nov. and Oceanicola flagellatus sp. nov., moderately halophilic bacteria isolated from seawater, Int. J. Syst. Evol. Microbiol., 2014, vol. 64, pp. 2975–2979.
Huang, M.M., Guo, L.L., Wu, Y.H., Lai, Q.L., Shao, Z.Z., Wang, C.S., Wu, M., and Xu, X.W., Pseudooceanicola lipolyticus sp. nov., a marine alphaproteobacterium, reclassification of Oceanicola flagellatus as Pseudooceanicola flagellatus comb. nov. and emended description of the genus Pseudooceanicola, Int. J. Syst. Evol. Microbiol., 2018, vol. 68, pp. 409–415.
Kämpfer, P. and Kroppenstedt, R.M., Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa, Can. J. Microbiol., 1996, vol. 42, pp. 989–1005.
Komagata, K. and Suzuki, K., Lipid and cell-wall analysis in bacterial systematics, Methods Microbiol., 1987, vol. 19, pp. 161–207.
Kimura, M., A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences, J. Mol. Evol., 1980, vol. 16, pp. 111–120.
Kim, O.S., Cho, Y.J., Lee, K., Yoon, S.H., Kim, M., Na, H., Park, S.C., Jeon, Y.S., Lee, J.H., Yi, H.N., Won, S.H., and Chun, J.S., Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species, Int. J. Syst. Evol. Microbiol., 2012, vol. 62, pp. 716–721.
Li, R., Li, Y., Kristiansen, K., and Wang, J., SOAP: short oligonucleotide alignment program, Bioinformatics, 2008a, vol. 24, pp. 713–714.
Li, D., Liu, C.M., Luo, R., Sadakane, K., and Lam, T.W., MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph, Bioinformatics, 2015b, vol. 31, pp. 1674–1676.
Lin, K.Y., Sheu, S.Y., Chang, P.S., Cho, J.C., and Chen, W.M., Oceanicola marinus sp. nov., a marine alphaproteobacterium isolated from seawater collected off Taiwan, Int. J. Syst. Evol. Microbiol., 2007, vol. 57, pp. 1625–1629.
Li, Y.Y., Wang, Y., Wang, Y.N., Lin, F., Zhu, H.M., Lai, Q.L., Shao, Z.Z., and Zhou, H.T., Pseudooceanicola aestuarii sp. nov., isolated from the Jiulong River Estuary in PR China, Int. J. Syst. Evol. Microbiol., 2020, vol. 70, pp. 6220–6225.
Lai, Q., Li, G., Liu, X., Du, Y., Sun, F., and Shao, Z., Pseudooceanicola atlanticus gen. nov. sp. nov., isolated from surface seawater of the Atlantic Ocean and reclassification of Oceanicola batsensis, Oceanicola marinus, Oceanicola nitratireducens, Oceanicola nanhaiensis, Oceanicola antarcticus and Oceanicola flagellatus, as Pseudooceanicola batsensis comb. nov., Pseudooceanicola marinus comb. nov., Pseudooceanicola nitratireducens comb. nov., Pseudooceanicola nanhaiensis comb. nov., Pseudooceanicola antarcticus comb. nov., and Pseudooceanicola flagellatus comb. nov., Antonie van Leeuwenhoek, 2015, vol. 107, pp. 1065–1074.
Lina, L.Y., Lai, Q.L., Li, J.Y., Shao, Z.Z., and Yu, Z.Q., Pseudooceanicola pacificus sp. nov., isolated from deep-sea sediment of the Pacific Ocean, Int. J. Syst. Evol. Microbiol., 2020, vol. 70, pp. 4372–4377.
Li, M., Liu, K., Liu, Y.H., Gao, C.H., and Yi, X.X., Bruguierivorax albus gen. nov. sp. nov. isolated from mangrove sediment and proposal of Bruguierivoracaceae fam. nov., Curr. Microbiol., 2021, vol. 78, pp. 856–866.
Meier-Kolthoff, J.P., Auch, A.F., Klenk, H.P., and Göker, M., Genome sequence-based species delimitation with confidence intervals and improved distance functions, BMC Bioinformatics, 2013, vol. 14, p. 60.
Nakagawa, Y. and Yamasato, K., Phylogenetic diversity of the genus Cytophaga revealed by 16S rRNA sequencing and menaquinone analysis, J. Gen. Microbiol., 1993, vol. 139, pp. 1155–1161.
Rüger, H.J. and Höfle, M.G., Marine star-shaped-aggregate-forming bacteria: Agrobacterium atlanticum sp. nov.; Agrobacterium meteori sp. nov.; Agrobacterium ferrugineum sp. nov., nom. rev.; Agrobacterium gelatinovorum sp. nov., nom. rev. and Agrobacterium stellulatum sp. nov., nom. rev., Int. J. Syst. Bacteriol., 1992, vol. 42, pp. 133–143.
Stamatakis, A., Hoover, P., and Rougemont, J., A rapid bootstrap algorithm for the RAxML Web servers, Syst. Bi-ol., 2008, vol. 57, pp. 758–771.
Saitou, N. and Nei, M., The neighbor-joining method: a new method for reconstructing phylogenetic trees, Mol. B-iol. Evol., 1987, vol. 4, pp. 406–425.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S., MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods, Mol. Biol. Evol., 2011, vol. 28, pp. 2731–2739.
Wolter, L.A., Wietz, M., Ziesche, L., Sven, B., Janina, L., Anja, P., Rolf, D., Stefan, S., and Thorsten, B., Pseudooceanicola algae sp. nov., isolated from the marine macroalga Fucus spiralis, shows genomic and physiological adaptations for an algae-associated lifestyle, Syst. Appl. Microbiol., 2021, vol. 44, p. 126166.
Yang, Q., Jiang, Z.W., Huang, C.H., Zhang, R.N., Li, L.Z., Yang, G., Feng, L.J., Yang, G.F., Zhang, H., Zhang, X., and Mu, J., Hoeflea prorocentri sp. nov., isolated from a culture of the marine dinoflagellate Prorocentrum mexicanum PM01, Antonie. Van. Leeuwenhoek., 2018, vol. 111, pp. 1845–1853.
Yin, Q., Song, Z.M., Liang, J.Y., Wang, Y., Zheng, X.L., Li, S.F., and Xu,Y., Pseudooceanicola onchidii sp. nov., isolated from a marine invertebrate from the South China Sea, Int. J. Syst. Evol. Microbiol., 2020, vol. 70, pp. 1224–1230.
Zheng, Q., Chen, C., Wang, Y.N., and Jiao, N.Z., Oceanicola nitratireducens sp. nov., a marine alphaproteobacterium isolated from the South China Sea, Int. J. Syst. Evol. Microbiol., 2010, vol. 60, pp. 1655–1659.
Zheng, Z.Q., Chen, M.S., Yan, X.R., and Tuo, L., Pseudooceanicola endophyticus sp. nov., a novel endophytic bacterium isolated from bark, Int. J. Syst. Evol. Microbiol., 2021, vol. 71, pp. 1–7.
Funding
The work was supported by the Joint Fund Project for Regional Innovation and Development set up by the National Natural Science Foundation (U20A20101), the Natural Science Foundation of Guangxi (2020GXNS-FGA297002), Middle-Aged Teachers Scientific Research Project of Guangxi Universities (2022KY0283), Youth Fund Project of Guangxi University of Chinese Medicine (2021QN009), Special Fund for Bagui Scholars of Guangxi and the Innovation Project of Guangxi Graduate Education (YCSW2022356), Special Fund for Bagui Scholars of Guangxi, High-level Talent Inheritance and Innovation Team of Guangxi Traditional Chinese Medicine (2022A007), Guangxi University of Chinese Medicine “Guipai Traditional Chinese Medicine Inheritance and Innovation Team” Project (2022B005), Guangxi University of Chinese Medicine-Guipai Xinglin Top Talent Funding Project (2022C008).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflicts of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Supplementary Information
Rights and permissions
About this article
Cite this article
Li, M., Li, F.T., Gao, C.H. et al. Pseudooceanicola albus sp. nov., Isolated from Mangrove Sediment within the Beibu Gulf. Microbiology 92, 622–629 (2023). https://doi.org/10.1134/S0026261723601070
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261723601070