Abstract
A facultative methylotroph, strain 2AzMo (VKM Ac-2782), was isolated from the coastal zone of the Sea of Azov. The cells of the isolate are aerobic gram-positive nonmotile rods. Optimal growth occurs at 1% NaCl, 28°C, and pH 7.5 with 1% СH3OH or 0.3% methylamine as the carbon and energy sources. A broad spectrum of polycarbon substrates is also utilized. Sequencing of the 16S rRNA gene of the strain revealed its similarity to Rhodococcus species: 99.9% to R. wratislaviensis IEGM 1112T (=NCIMB 13082T), 99.4% to R. imtechensis IEGM 940T (=RKJ300T), and 99.2% to R. koreensis IEGM 962T (=DNP505T). The level of DNA–DNA homology of strain 2AzMo with R. wratislaviensis IEGM 1112T (=NCIMB 13082T) was 76%, supporting its identification as a strain of this species. However, unlike strain 2AzMo, the type strain R. wratislaviensis IEGM 1112T, as well as other members of this genus (R. imtechensis IEGM 940T, R. koreensis IEGM 962T, and R. opacus IEGM 716T), do not grow on methanol and methylamine. Methanol oxidation by R.wratislaviensis 2AzMo is catalyzed by alcohol dehydrogenase, which uses 4-nitroso-N,N-dimethylaniline as an artificial electron acceptor. Methylamine is oxidized by methylamine dehydrogenase and the enzymes of the N-methylglutamate pathway. Formaldehyde is then assimilated via the fructose bisphosphate aldolase variant of the ribulose monophosphate pathway of C1 metabolism. Ammonium is assimilated by α-ketoglutarate reductive amination and via the glutamate cycle.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Terrestrial and marine ecosystems are sources of methanol, methylamine, and other C1 compounds, which are the substrates for a specialized group of aerobic methylotrophic bacteria (Trotsenko et al., 2010). Methanol is a widespread compound, which is formed as a result of demethylation of the cell wall pectin during active growth of plant cells; it is the main volatile organic metabolite of plants, since its emission into the atmosphere is over 100 Tg/year (Fedorov et al., 2011). Methanol is released during biomass combustion, lignin degradation, in hydrothermal seabed systems, and in the course of volcanic activity (Dorokhov et al., 2015). Methanol is also formed by methanotrophic bacteria during oxidation of methane, and it is one of archaeal metabolites (Trotsenko and Khmelenina, 2008; Chistoserdova and Lidstrom, 2013; Chistoserdova and Kalyuzhnaya, 2018; Enzmann et al., 2018). Not only gram-negative, but also gram-positive prokaryotes of the genera Amicilatopsis, Arthrobacter, Mycobacterium, and Bacillus possess the capacity for methylotrophy. Several pathways of methanol oxidation are known. In most gram-negative bacteria, PQQ-calcium-dependent ethanol/methanol dehydrogenases (MxaFI) play a key role in oxidation of methanol to formaldehyde (Trotsenko et al., 2010). However, methylotrophs possessing a lanthanide-containing methanol dehydrogenase (XoxF) have been recently revealed (Vu et al., 2016; Chu and Lidstrom, 2016). In gram-positive facultative methylotrophs, oxidation of methanol is carried out by either the NAD+-dependent form of this enzyme (Bacillus methanolicus C1) (Dijkhuizen and Arfman, 1990, Hektor et al., 2002), or by methanol: 4-nitroso-N,N-dimethylaniline (NDMA) oxidoreductases (nicotinoprotein methanol dehydrogenases), as is the case in Amicolatopsis methanolica and Mycobacterium gastri MB19 (Van Ophem et al., 1993; Bystrykh et al., 1993). The members of the genus Rhodococcus growing on ethanol are known to possess functioning NAD(P)+ and NDMA-dependent alcohol dehydrogenases as well (Van Ophem et al., 1993; Schenkels and Duine, 2000). At the same time, NDMA-dependent enzymes of rhodococci are able to oxidize both ethanol and methanol (Pirog et al., 2008).
Among the broad spectrum of C1 compounds of biogenic and abiogenic origin, methylated amines, reduced forms of organic nitrogen which are formed in nature as by-products of degradation of proteins, amino acids, some alkaloids, and nitrogen-containing pesticides, are present in plant and animal tissues as natural products of nitrogen metabolism. In marine environments, methylamine is released during degradation of quaternary amines, such as betaine, carnitine, choline, and trimethylamine N-oxide, which are used as osmolytes by many marine organisms (Nayak et al., 2015; Suleiman et al., 2016). Methylamine can be used via direct oxidation to formaldehyde by methylamine dehydrogenase (MADH) or methylamine oxidase (MAO), or via mediated pathways involving the transfer of methyl groups to amino acids (glutamate, alanine, or their keto analogs) followed by oxidation of the corresponding N-methylated amino acids. This two-step mechanism was named the N-methylglutamate (N-MG) pathway (Trotsenko et al., 2010).
We have recently isolated a new facultative methylotroph from the coastal zone of the Sea of Azov, which was assigned to the genus Rhodococcus. Methylotrophic members of this genus have been unknown until now, although rhodococci are widespread, and 63 species have been validly described (http://www. bacterio.cict.fr/qr/rhodococcus.htm). A detailed study of the metabolic pathways of methanol and methylamine in the new methylotrophic rhodococcus seems to be relevant.
The goal of the present work was taxonomic and metabolic characterization of the new gram-positive facultative methylotroph.
MATERIALS AND METHODS
Subjects of research. Strain 2AzMo (VKM Ac-2782) was isolated from the sand of the coastal zone of the Sea of Azov (Crimea, Russia) (45°27′25.1″ N, 36°27′59.7″ E). A sample of sand (5 g) was placed into an Erlenmeyer flask (750 mL) with 200 mL of the K medium and 0.5% methanol (vol/vol). The K medium contained the following (g/L): KH2PO4, 2.0; (NH4)2SO4, 2.0; NaCl, 0.5; MgSO4 · 7H2O, 0.1; FeSO4 · 7H2O, 0.002; pH 7.4. The enrichment and pure cultures were prepared as described previously (Kaparullina et al., 2017). The purity of the culture was tested by light microscopy and by homogeneity of the colonies on the agar K medium containing methanol and on Luria-Bertani medium. Type cultures of rhodococci used as reference cultures (R. wratislaviensis IEGM 1112T = NCIMB 13082T, R. imtechensis IEGM 940T = RKJ300T, R. koreensis IEGM 962T = DNP505T, and R. opacus IEGM 716T) were kindly provided by I.B. Ivshyna, Institute of Ecology and Genetics of Microorganisms, Ural Branch, Russian Academy of Sciences, Perm.
Study of the cultural, physiological, and biochemical properties of the isolate. To study morphology and motility of the cells, strain 2AzMo was grown on the agar K and Luria-Bertani media (2% Difco agar, United States). The presence of oxidase was determined using a 1% (wt/vol) solution of tetramethyl-p-phenylenediamine dihydrochloride. Catalase activity was detected by applying a 3% hydrogen peroxide solution to a culture streak grown on the agar medium.
The temperature range for growth was determined by growing the culture in the liquid K medium with methanol in hermetically sealed vials on a shaker (120 rpm) at temperatures from 4 to 43°C. The growth of the isolate at different concentrations of methanol (0.1–7.0%, vol/vol), salinity (0–7% NaCl), and pH was studied in the K medium. The pH values were adjusted by addition of 1 M NaOH and 5 N HCl; the pH optimum was determined by the specific growth rate of the strain at the initial pH values of the medium 5.0–10.0.
When studying ability of the isolate to use various organic compounds as a source of carbon and energy, the medium was supplemented with 0.05–0.3% (wt/vol) of the test substance instead of methanol, inoculated with an overnight culture, and incubated for 14 days on a shaker at the optimal temperature. All volatile substances were added in the amount of 0.5% (vol/vol medium).
API tests (API 20E and API 20NE; Biomerieux, France) were also used to determine the spectrum of utilized substrates and to identify some biochemical properties of the studied strain, according to the manufacturer’s instructions. Growth in the atmosphere of methane, dichloromethane, or H2/CO2/O2, as well as sensitivity to antibiotics, was analyzed as described (Kaparullina et al., 2017). Formation of indole from L-tryptophan was analyzed with Salkowski reagent (Gordon and Weber, 1951). The calibration curve was plotted with standard solutions of indoleacetic acid.
Microscopy. Studies of the morphology and motility of the cells were carried out using a Nikon Eclipse Ci optical microscope in the phase contrast mode (Nikon, Japan) equipped with a ProgRes SpeedXT core5 camera (Jenoptik, Germany).
Enzymological analysis was carried out in the cell-free extract, as described earlier (Doronina et al., 2018). The activity of nicotinoprotein methanol dehydrogenase was determined spectrophotometrically by reduction of 4-nitroso-N,N-dimethylaniline at 440 nm with methanol as an electron donor (Van Ophemet et al., 1993). The activity of the enzymes was expressed in nanomoles of the consumed substrate or formed product per min per 1 mg protein. Quantitative determination of protein was carried out by the Lowry method (Lowry et al., 1951).
RAPD analysis (random amplified polymorphic DNA method) was performed using the OPQ1 and OPQ6 primers (Balachandar et al., 2008).
MALDI-TOF/MS analysis. MALDI-TOF Autoflex Speed mass spectrometer (Bruker Daltonik GmbH, Germany) was used to obtain MALDI spectra of bacterial extracts according to the previously described procedure (Horneffer et al., 2004).
Isolation and analysis of DNA. DNA was isolated using the ZR Fungal/Bacterial DNA MiniPrep kit (Zymo Research, United States), according to the manufacturer’s recommendations. The 16S rRNA gene was amplified by PCR using the 27f and 1492r universal primers for prokaryotes (Lane, 1991). The reaction products were separated by electrophoresis in 1% agarose gel. Isolation and purification of DNA fragments from low-melting agarose were carried out on columns using the Zymoclean Gel DNA Recovery Kit (Zymo Research, United States), according to the manufacturer’s instructions. Sequencing of the PCR fragments was carried out using the CEQ Dye Terminator Cycle Sequencing kit (Beckman Coulter, United States) on a CEQ2000 XL analyzer (Beckman Coulter, United States). Determination of G + C DNA composition and DNA−DNA hybridization were carried out as described previously (Doronina et al., 2013).
Phylogenetic analysis. Preliminary analysis of the similarity of the 16S rRNA gene sequences was performed using the GenBank [NCBI] database and the BLAST software package [http://ncbi.nlm.nih.gov]. Nucleotide sequences of the 16S rRNA gene were aligned with the sequences of the reference strains using the CLUSTAL W software package for a more accurate determination of the phylogenetic position of the isolate (Thompson et al., 1997). Phylogenetic analysis was performed using the MEGA 5 software package (Tamura et al., 2011). Phylogenetic trees (phylograms) were constructed by the neighbor-joining method (Saitou and Nei, 1987). The statistical reliability of branching was estimated using bootstrap analysis of 100 alternative phylograms.
RESULTS AND DISCUSSION
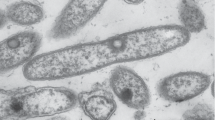
Morphological, cultural, physiological, and biochemical characteristics of the isolate. The cells of strain 2AzMo were gram-positive nonmotile straight or branching rods (Fig. 1). The colonies on the agar mineral medium containing methanol were yellow-brown (cream-colored on Luria-Bertani medium), pinpoint, with a convex profile, uneven edge, and rough surface. Strain 2AzMo grew in the liquid K medium containing methanol or in Luria-Bertani medium without cell aggregation and pigment formation. The strain was obligatory aerobic and catalase- and oxidase-positive; it did not require vitamins. It did not grow in the atmosphere of H2/O2/CO2, methane, or dichloromethane. Growth was observed in media with sucrose, raffinose, mannitol, alanine, citrate, inositol, fructose, maltose, glucose, acetate, phenylacetate, citrate, glutamate, succinate, gluconate, lactose, malate, α-ketoglutarate, pyruvate, methylamine, dimethylamine, methanol, ethanol, dimethyl sulfoxide, glycerol, formaldehyde, acetone, Tween 20, propanol, benzene, heptane, and toluene. However, strain 2AzMo did not use serine, arabinose, xylose, formate, acetamide, and trimethylamine. The strain was incapable of nitrate reduction. It had no activity of lysine and ornithine decarboxylases, arginine dihydrolase, and β-gluco- and β-galactosidase. It did not hydrolyze gelatin, did not form hydrogen sulfide and acetoin. The isolate was urease-positive. It synthesized 6 ± 1 μg/mL indole derivatives from L-tryptophan. It grew in the temperature range of 4–37°C and at pH 4.0–9.0 with an optimum at 28°C and pH 7.5 in the presence of 0.5–1% methanol and 1% NaCl; nevertheless, growth was inhibited by 7% NaCl. The strain was sensitive to gentamicin, streptomycin, lincomycin, neomycin, tetracycline, novobiocin, penicillin, kanamycin, chloramphenicol, and erythromycin; it was resistant to oxacillin and nalidixic acid.
Genotypic characteristics. Phylogenetic analysis of nucleotide sequences of the 16S rRNA gene indicated that strain 2AzMo had the highest similarity to Rhodococcus species: 99.9% to R. wratislaviensis IEGM 1112T, 99.4% to R. imtechensis IEGM 940T, and 99.2% to R. koreensis IEGM 962T (Fig. 2). Since a high level of similarity of the 16S rRNA was revealed, the method of randomly amplified polymorphic DNA (RAPD analysis) was also used to determine genotypic differences between the strain 2AzMo and R. wratislaviensis IEGM 1112T. The results of RAPD analysis showed that the strains had different patterns of amplification products; nevertheless, the results obtained using MALDI-TOF/MS revealed identity of the protein profiles. Moreover, the level of DNA‒DNA homology between the strain 2AzMo and R. wratislaviensis IEGM 1112T was 76%, which made it possible to assign our isolate to this species.
The phylogram showing the position of strain 2AzMo among members of the genus Rhodococcus and related genera of actinobacteria, on the basis of the results of comparison of the 16S rRNA nucleotide sequences. The scale corresponds to 10 nucleotide substitutions per each 100 nucleotides (evolutionary distances). The root was determined by the inclusion of the Turicella otitidis 234T (X73976) sequence as an external group. The figures show the statistical reliability of the order of branching, determined using the bootstrap analysis of 100 alternative trees.
Metabolic characteristics. The results of enzymological analysis of the pathways of the primary and intermediate metabolism in R. wratislaviensis 2AzMo cells grown on methanol are presented in Table 1. Activities of the classical PQQ-dependent and NAD+-dependent methanol dehydrogenases (MDG) were not detected in the cell extracts. At the same time, this strain was shown to oxidize methanol to formaldehyde with NDMA-dependent alcohol dehydrogenase using NDMA as an artificial electron acceptor, which seems to be a distinctive characteristic of these enzymes in actinobacteria. However, on the basis of genome analysis, the type strain R. wratislaviensis NBRC 100605T (NZ_BAWF01000000) proved to harbor an NDMA-dependent MDG (WP_037244517). We showed that R. wratislaviensis NBRC 100605T, R. imtechensis IEGM 940T, R. koreensis IEGM 962T, and R. opacus IEGM 716T did not grow on methanol and methylamine. According to the amino acid sequences, the NDMA-dependent methanol dehydrogenase of R. wratislaviensis NBRC 100605T showed more than 90% identity to the relevant enzyme in the members of the genera Rhodococcus and Mycobacterium and only 66% identity to the enzyme of Amycolatopsis methanolica (AAF21473). Strain 2AzMo showed high activity of formaldehyde and formate dehydrogenases with phenazine methosulphate (PMS) as an artificial electron acceptor. Enzymological analysis showed that the cell extracts did not possess activities of specific enzymes of the serine and ribulose bisphosphate pathways, although the sequences of the large subunit of ribulose-1,5-bisphosphate carboxylase were revealed in genomes of some rhodococci. The studied strain implemented the ribulose monophosphate pathway of C1 metabolism, which was evident from activity of the key enzyme of this pathway: hexulose phosphate synthase (HPS). The identity of amino acid sequences of the HPS of the type strain R. wratislaviensis NBRC 100605T (WP_037231807) with those of R. opacus (WP_005242427.1), R. jostii (WP_011600601), R. koreensis (WP_072949731), and Rh. erythropolis (WP_094617027) strains was more than 90%. Strain 2AzMo had a hexokinase active with ATP and enzymes of the oxidative pentose phosphate pathway (glucose-6-phosphate and 6-phosphogluconate dehydrogenases). The activity of 2-keto-3-deoxy-6-phosphogluconate aldolase (Entner−Doudoroff pathway) was absent. Since activity of fructose-1,6-bisphosphate aldolase was detected, this strain used the fructose-bisphosphate aldolase variant of the RMP pathway. Strain 2AzMo assimilated ammonium by reductive amination of α-ketoglutarate and by the functioning of the glutamate cycle enzymes.
Another C1 compound that plays an important biospheric role is methylamine. The results of enzymological analysis of activities of the enzymes of the pathways of the primary methylamine metabolism in the test strain grown on methylamine, as well as on methanol, are presented in Table 2. R. wratislaviensis 2AzMo was shown to implement a direct pathway of oxidation of methylamine to formaldehyde, since it had an PMS-dependent MADG, the activity of which was 1.5 times higher in the cells grown on methylamine than on methanol. The activity of another enzyme, methylamine oxidase, was extremely low, which probably excludes any significant contribution of this pathway to the oxidation of methylamine in the studied strain. We detected amine dehydrogenases in the genomes of some rhodococci, the amino acid sequences of which were only 30–42% identical to the MADG of methylotrophic members of the genera Methylopila and Methylobacterium. The activity of NDMA-dependent alcohol dehydrogenase with ethanol in the cells grown on both methanol and methylamine was found to be two times higher than the activity of the enzyme with methanol (Table 2). The activity of another enzyme of methylamine metabolism, N-methylglutamate dehydrogenase, catalyzing oxidative decomposition of N-methylglutamate to glutamate and formaldehyde, was higher with PMS than with NAD+. N-Methylglutamate oxidase activity was also revealed and was most pronounced in the cells grown on methylamine. The activity of γ-glutamylmethylamide lyase, which catalyzed the conversion of γ-glutamylmethylamide to formaldehyde and glutamate, was revealed in the studied strain. High activities of γ-glutamylmethylamide synthetase (GMAS) and N-methylglutamate synthase were also detected, which indicated implementation of the N-methylglutamate pathway. The search for the GMAS gene in the genomes of Rhodococcus strains showed that this enzyme was most likely annotated as a glutamate-ammonium ligase (type III) and had 43−45% identity with the GMAS proteins of members of the genus Methylobacterium. The enzymes assimilating trimethylamine and dimethylamine, as well as N-methylglutamate synthase, were revealed in the genome of R. jostii RHA1, although it is not clear how the subsequent conversion of N-methylglutamate can occur in this strain. Based on the results of our studies, a scheme of the metabolism of methanol and methylamine in R. wratislaviensis 2AzMo is presented (Fig. 3).
The scheme of methanol and methylamine oxidation in Rhodococcus wratislaviensis 2AzMo. 1, NDMA methanol dehydrogenase; 2, formaldehyde dehydrogenase; 3, formate dehydrogenase; 4, monoamine oxidase or methylamine dehydrogenase; 5, hexulose phosphate synthase; 6, γ-glutamylmethylamide synthetase; 7, N-methylglutamate synthase; 8, N-methylglutamate dehydrogenase; 9, γ-glutamylmethylamide lyase.
Thus, we provide the first evidence of the capacity of rhodococci for methylotrophy. The studied pathways of methanol and methylamine metabolism in the methylotrophic member of the genus Rhodococcus made it possible to reveal an important biospheric role of this microorganism and its promising application in various biotechnologies in future, including the development of biofilters and biosensors for decomposition and detection of pollutants.
COMPLIANCE WITH ETHICAL STANDARDS
Сonflict of interests. The authors declare that they have no conflict of interest.
Statement on the welfare of animals. This article does not contain any studies involving animals performed by any of the authors.
REFERENCES
Balachandar, D., Raja, P., and Sundaram, S.P., Genetic and metabolic diversity of pink-pigmented facultative methylotrophs in phyllosphere of tropical plants, Braz. J. Microbiol., 2008, vol. 39, pp. 68–73.
Bystrykh, L.V., Vonck, J., Van Bruggen, E.F., van Beeumen, J., Samyn, B., Govorukhina, N.I., Arfman, N., Duine, J.A., and Dijkhuizen, L., Electron microscopic analysis and structural characterization of novel NADP(H)-containing methanol: N,N'-dimethyl-4-nitrosoaniline oxidoreductases from the gram-positive methylotrophic bacteria Amycolatopsis methanolica and Mycobacterium gastri MB19, J. Bacteriol., 1993, vol. 175, pp. 1814−1822.
Chistoserdova, L. and Kalyuzhnaya, M.G., Current trends in methylotrophy, Trends Microbiol., 2018. pii: S0966-842X(18)30023-4. doi https://doi.org/10.1016/j.tim.2018.01.011
Chistoserdova, L. and Lidstrom, M.E., Aerobic methylotrophic prokaryotes, in The Prokaryotes, Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., and Thompson, F., Eds., Berlin: Springer, 2013, pp. 267–285.
Chu, F. and Lidstrom, M.E., XoxF acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense, J. Bacteriol., 2016, vol. 198, pp. 1317–1325.
Dijkhuizen, L. and Arfman, N., Methanol metabolism in thermotolerant methylotrophic Bacillus species, FEMS Microbiol. Rev., 1990, vol. 87, pp. 215–219.
Dorokhov, Y.L., Shindyapina, A.V., Sheshukova, E.V., and Komarova, T.V., Metabolic methanol: molecular pathways and physiological roles, Physiol. Rev., 2015, vol. 95, pp. 603–644.
Doronina, N.V., Kaparullina, E.N., and Trotsenko, Y.A., Methylopila musalis sp. nov., an aerobic facultatively methylotrophic bacterium isolated from banana fruit, Int. J. Syst. Evol. Microbiol., 2013, vol. 63, pp. 1847–1852.
Doronina, N.V., Kaparullina, E.N., Chemodurova, A.A., and Trotsenko, Yu.A., Paracoccus simplex sp. nov.—a new facultative methylotroph, utilizing methylamine, Microbiology (Moscow), 2018, vol. 87, pp. 541–550.
Enzmann, F., Mayer, F., Rother, M., and Holtmann, D., Methanogens: biochemical background and biotechnological applications, AMB Express, 2018, vol. 8, pp. 1–22.
Fedorov, D.N., Doronina, N.V., and Trotsenko, Y.A., Phytosymbiosis of aerobic methylobacteria: new facts and views, Microbiology (Moscow), 2011, vol. 80, pp. 443–454.
Gordon, S.A. and Weber, R.P., Colorimetric estimation of indoleacetic acid, Plant Physiol., 1951, vol. 26, pp. 192–195.
Hektor, H.J., Kloosterman, H, and Dijkhuizen, L., Identification of a magnesium-dependent NAD(P)(H)-binding domain in the nicotinoprotein methanol dehydrogenase from Bacillus methanolicus, J. Biol. Chem., 2002, vol. 277, pp. 46 966–46 973.
Horneffer, V., Haverkamp, J., Janssen, H.G., Steeg, P.F., and Notz, R., MALDI-TOF-MS analysis of bacterial spores:wet heat treatment as a new releasing technique for biomarkers and the influence of different experimental parameters and microbiological handling, J. Am. Soc. Mass. Spectrom., 2004, vol. 15, pp. 1444–1454. http://www.bacterio.cict.fr/qr/rhodococcus.htm.
Kaparullina, E.N., Trotsenko, Y.A., and Doronina, N.V., Methylobacillus methanolivorans sp. nov., a novel non-pigmented obligately methylotrophic bacterium, Int. J. Syst. Evol. Microbiol., 2017, vol. 67, pp. 425–431.
Lane, D.J., 16S/23S rRNA sequencing, in Nucleic Acid techniques in Bacterial Systematics, Stackebrandt, E. and Goodfellow, M., Eds., Chichester: Wiley, 1991, pp. 115–175.
Lowry, O.H., Rosebrough, N.J., Farr, A.L., and Randall, R.J., Protein measurement with the Folin phenol reagent, J. Biol. Chem., 1951, vol. 193, pp. 265–275.
Nayak, D.D. and Marx, C.J., Experimental horizontal gene transfer of methylamine dehydrogenase mimics prevalent exchange in nature and overcomes the methylamine growth constraints posed by the sub-optimal N-methylglutamate pathway, Microorganisms, 2015, vol. 3, pp. 60‒79.
Pirog, T.P., Korzh, Yu.V., Shevchuk, T.A., and Tarasenko, D.A., Peculiarities of C2 metabolism and intensification of the synthesis of surface-active substances in Rhodococcus erythropolis EK-1 grown in ethanol, Microbiology (Moscow), 2008, vol. 77, pp. 665–673.
Saitou, N. and Nei, M., The neighbour-joining method: a new method for reconstructing phylogenetic trees, Mol. Biol. Evol., 1987, vol. 4, pp. 405–425.
Schenkels, P. and Duine, J.A., Nicotinoprotein (NADH-containing) alcohol dehydrogenase from Rhodococcus erythropolis DSM 1069: an efficient catalyst for coenzyme-independent oxidation of broad spectrum of alcohols and interconversion of alcohols and aldehydes, Microbiology (UK), 2000, vol. 146, pp. 775–785.
Suleiman, M., Zecher, K., Yücel, O., Jagmann, N., and Philipp, B., Interkingdom cross-feeding of ammonium from marine methylamine-degrading bacteria to the diatom Phaeodactylum tricornutum, Appl. Environ. Microbiol., 2016, vol. 82, pp. 7113–7122.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S., MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods, Mol. Biol. Evol., 2011, vol. 28, pp. 2731–2739.
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G., The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools, Nucl. Acids Res., 1997, vol. 25, pp. 4876–4882.
Trotsenko, Yu.A. and Khmelenina, V.N., Ekstremophil’nye metanotrofy (Extremophilic Methanotrophs), Gal’-chenko, V.F., Ed., Pushchino: ONTI PNTs RAN, 2008.
Trotsenko, Yu.A., Doronina, N.V., and Torgon-skaya, M.L., Aerobnye metilobakterii (Aerobic Methylobacteria), Gal’chenko, V.F., Ed., Pushchino: ONTI PNTs RAN, 2010.
Van Ophem, P.W., van Beeumen, J., and Duine, J.A., Nicotinoprotein (NAD(P)-containing) alcohol/aldehyde oxidoreductases. Purification and characterization of a novel type from Amycolatopsis methanolica, Eur. J. Biochem., 1993, vol. 212, pp. 819–826.
Vu, H N., Subuyuj, G.A., Vijayakumar, S., Good, N.M., Martinez-Gomez, N.C., and Skovran, E. Lanthanide-dependent regulation of methanol oxidation systems in Methylobacterium extorquens AM1 and their contribution to methanol growth, J. Bacteriol., 2016, vol. 198, pp. 1250–1259.
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated by A. Panyushkina
Rights and permissions
About this article
Cite this article
Kaparullina, E.N., Trotsenko, Y.A. & Doronina, N.V. Characterization of Rhodococcus wratislaviensis, a New Gram-Positive Facultative Methylotroph, and Properties of Its C1 Metabolism. Microbiology 88, 46–53 (2019). https://doi.org/10.1134/S0026261718060103
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261718060103