Abstract
New zinc complexes, [ZnBr2(HL)] (1), [ZnBr(HL)(NCS)]·0.5H2O (2), [Zn(HL)I2] (3), and [ZnL2] (4), where L is the monoanionic form of 2-amino-N′-(pyridin-2-ylmethylene)benzohydrazide (HL), have been prepared and characterized by elemental analysis, IR and UV-Vis spectra, as well as single crystal X-ray diffraction. The Zn atoms in complexes 1-3 are in distorted trigonal-bipyramidal coordination, and that in complex 4 is in octahedral coordination. The crystal structures of the complexes are stabilized by hydrogen bonds. The antibacterial and antifungal activities of the compounds were evaluated.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Hydrazones and their derivatives are a special type of Schiff bases, which have interesting and fascinating pharmaceutical and biological activities like antibacterial, antifungal, anti-inflammatory, antiviral and antitumor [1-4]. Metal complexes derived from hydrazone ligands are important in bioinorganic chemistry [5-11]. In particular, when coordinated to transition metals, they significantly expand the lipophilic character that contributes to increasing potential biological complexes [12]. In order to obtain metal complexes, it is necessary to use active metals that have low toxicity to organisms [13]. Zinc complexes with hydrazone ligands have shown various biological activities [14-17]. In the past few years, a number of zinc complexes with hydrazone ligands have been reported for their synthesis, crystal structures and biological application. The coordination geometries of zinc complexes are usually tetrahedral, square pyramidal, trigonal bipyramidal and octahedral [18-23]. The hydrazones adopt either ketone or enol form during the coordination. However, it is hard to predict which form they will present. As a continuation of our work on the hydrazone complexes [24-26], and to control the coordination mode of hydrazone ligands, as well as in pursuit of new biological agents, we reported here a series of new zinc complexes, [ZnBr2(HL)] (1), [ZnBr(HL)(NCS)]·0.5H2O (2), [Zn(HL)I2] (3) and [ZnL2] (4), where HL is 2-amino-N′-(pyridin-2-ylmethylene)benzohydrazide (Scheme 1).
EXPERIMENTAL
Materials and measurements. The starting materials 2-pyridinecarboxaldehyde and 2-aminobenzohydrazide (AR grade) were purchased from Bailingwei Chemicals Co. Ltd. of China. Zinc chloride, zinc bromide, zinc iodide, zinc acetate, ammonium thiocyanate and solvents with AR grade were purchased from Xiya Chemicals Co. Ltd. of China. The ligand HL was prepared according to the literature method [27]. CHN elemental analyses were performed on a PerkinElmer 240C elemental analyzer. IR spectra were recorded on a Jasco FTIR-4000 spectrometer as KBr pellets in the 4000-400 cm–1 region. UV-Vis spectra were recorded on a LAMBDA 35 spectrometer. Single crystal X-ray diffraction was carried out on a Bruker SMART 1000 CCD diffractometer.
Synthesis of [ZnBr2(HL)] (1). To a 15 mL methanolic solution of HL (0.24 g, 1.0 mmol) a methanolic solution (15 mL) of zinc bromide (0.26 g, 1.0 mmol) was added slowly. The mixture was stirred for 30 min and filtered. The colorless solution was left undisturbed in air for slow evaporation. After 5 days colorless single crystals of the complex that separated, were washed with cold methanol and dried in vacuo over silica gel indicator. Yield: 0.22 g (47%). Anal. calcd for C13H11Br2N4OZn (%): C 33.62; H 2.39; N 12.06. Found (%): C 33.41; H 2.50; N 11.87. IR data (cm–1): 3473, 3358 (NH2), 3210 (NH), 1632 (C=N), 1577, 1552, 1522, 1471, 1446, 1357, 1285, 1247, 1162, 937, 776, 742, 534, 492. UV-Vis data (nm, L/(mol·cm)): 306, 6.72·103; 325, 4.51·103; 380, 1.72·103.
Synthesis of [ZnBr(HL)(NCS)]·0.5H2O (2). To a 15 mL methanolic solution of HL (0.24 g, 1.0 mmol) and ammonium thiocyanate (0.076 g, 1.0 mmol) a methanolic solution (15 mL) of zinc bromide (0.26 g, 1.0 mmol) was added slowly. The mixture was stirred for 30 min and filtered. The colorless solution was left undisturbed in air for slow evaporation. After 5 days colorless single crystals of the complex that separated, were washed with cold methanol and dried in vacuo over silica gel indicator. Yield: 0.31 g (67%). Anal. calcd for C14H13BrN5O1.5SZn (%): C 37.15; H 2.89; N 15.47. Found (%): C 36.86; H 3.07; N 15.32. IR data (cm–1): 3477, 3359 (NH2), 3210 (NH), 2088 (NCS), 1633 (C=N), 1580, 1551, 1521, 1463, 1442, 1399, 1355, 1328, 1290, 1276, 1245, 1163, 1102, 935, 775, 739, 696, 669, 530. UV-Vis data (nm, L/(mol·cm)): 239, 5.59·103; 325, 2.87·103; 390, 3.13·103.
Synthesis of [Zn(HL)I2] (3). Complex 3 was prepared by metathesis of complex 1, with zinc bromide replaced by zinc iodide (0.32 g, 1.0 mmol). Yield: 0.35 g (62%). Anal. calcd for C13H12I2N4OZn (%): C 27.91; H 2.16; N 10.01. Found (%): C 28.16; H 2.25; N 9.89. IR data (cm–1): 3468, 3358 (NH2), 3214 (NH), 1632 (C=N), 1581, 1547, 1518, 1471, 1446, 1352, 1327, 1298, 1247, 1158, 1102, 928, 780, 742, 670, 526, 492. UV-Vis data (nm, L/(mol·cm)): 303, 6.81·103; 325, 4.32·103; 368, 3.87·103.
Synthesis of [ZnL2] (4). To a 15 mL methanolic solution of HL (0.24 g, 1.0 mmol) and 2,2′-bipyridine (0.016 g, 1.0 mmol) a methanolic solution (15 mL) of zinc bromide (0.26 g, 1.0 mmol) was added slowly. The mixture was stirred for 30 min and filtered. The colorless solution was left undisturbed in air for slow evaporation. After 5 days colorless single crystals of the complex that separated, were washed with cold methanol and dried in vacuo over silica gel indicator. Yield: 0.25 g (46%). Anal. calcd for C26H22N8O2Zn (%): C 36.58; H 2.63; N 15.24. Found (%): C 36.71; H 2.55; N 15.05. IR data (cm–1): 3451, 3392 (NH2), 3277 (NH), 1603 (C=N), 1561, 1488, 1463, 1445, 1340, 1285, 1251, 1145, 1073, 920, 759, 687, 632, 593, 539, 462. UV-Vis data (nm, L/(mol·cm)): 238, 5.35·103; 295, 3.87·103; 383, 5.56·103.
X-ray diffraction. Diffraction intensities for the complexes were collected at 298(2) K using a Bruker SMART 1000 CCD area-detector diffractometer with MoKα radiation (λ = 0.71073 Å). The collected data were reduced with the SAINT [28], and multi-scan absorption correction was performed using the SADABS [29]. The structures of the complexes were solved by direct method and refined against F2 by full-matrix least-squares method using the SHELXL-2018/3 package [30]. All of the non-hydrogen atoms were refined anisotropically. The amino H atoms were located from difference Fourier maps and refined isotropically, with N–H and H⋯H distances restrained to 0.90(1) Å and 1.45(2) Å, respectively. The water H atoms were located from difference Fourier maps and refined isotropically, with O–H and H⋯H distances restrained to 0.96(1) Å and 1.47(2) Å, respectively. The remaining hydrogen atoms were placed in calculated positions and constrained to ride on their parent atoms. There is no acceptor for O3–H3A and O3–H3B bonds in complex 2, which is due to the disorder of the water molecule, and it is difficult to add the hydrogen atoms at the right positions. The crystallographic data for the complexes are summarized in Table 1. Selected bond lengths and angles are given in Tables 2 and 3.
Antimicrobial assay. The antibacterial activities of the compounds were tested against B. subtilis, S. aureus, E. coli, and P. fluorescens using MH (Mueller–Hinton) medium. The antifungal activities of the compounds were tested against C. albicans and A. niger using RPMI-1640 medium. The MIC values of the tested compounds were determined by a colorimetric method using the dye MTT [31]. A stock solution of the aroylhydrazone compound (150 μM) in DMSO was prepared and graded quantities (75 μM, 37.5 μM, 18.8 μM, 9.4 μM, 4.7 μM, 2.3 μM, 1.2 μM, 0.59 μM) of the tested compounds were incorporated in specified quantity of the corresponding sterilized liquid medium. A specified quantity of the medium containing the compound was poured into microtitration plates. Suspension of the microorganism was prepared to contain approximately 105 cfu/mL and applied to microtitration plates with serially diluted compounds in DMSO to be tested and incubated at 37 °C for 24 h and 48 h for bacteria and fungi, respectively. Then the MIC values were visually determined on each of the microtitration plates, 50 μL of PBS (phosphate buffered saline 0.01 M, pH = 7.4) containing 2 mg of MTT/mL was added to each well. Incubation was continued at room temperature for 4-5 h. The content of each well was removed, and 100 μL of isopropanol containing 5% 1 M HCl was added to extract the dye. After 12 h of incubation at room temperature, the optical density was measured with a microplate reader at 550 nm.
RESULTS AND DISCUSSION
Chemistry. The hydrazone HL was facial prepared by the reaction of 2-pyridinecarboxaldehyde and 2-aminobenzohydrazide with 1:1 molar ratio in methanol. The zinc complexes 1-3 with tetrahedral coordination were synthesized from the reaction of HL with zinc bromide, zinc bromide and ammonium thiocyanate, and zinc iodide, respectively in methanol. The zinc complex 4 with octahedral coordination was synthesized from the reaction of HL with zinc bromide in the presence of 2,2′-bipyridine (Scheme 2). Interestingly, when 2,2′-bipyrdine was replaced by other Lewis bases like 4,4′-bipyridine, pyridine, or 1,10-phenanthroline, the same structures as complex 4 can be obtained. Thus, the presence of Lewis bases contributes to the enol form of the hydrazone compound. While no Lewis bases, the hydrazone presents in ketone form. All the complexes are soluble in methanol and ethanol. Single crystals of the complexes were obtained by slow evaporation of the methanolic solution of the complexes. The crystals of the complexes are stable in air at room temperature.
The coordination of HL to Zn cation affects the bond lengths with adjacent atoms in the structure of organic ligand when compared with the structure of individual HL. In complexes 1-3, the ligands are in neutral ketone form. The C6=N2 bond length in HL is shorter than those in complexes 1 and 2, and comparable to that in complex 3. The N2–N3 bond length in HL is longer than those in complexes 1-3. The C7–N3 bond length in HL is shorter than those in complexes 1 and 3, and longer than that in complex 2. The C7–O1 bond length in HL is shorter than those in complexes 2 and 3, and comparable to that in complex 1. While in complex 4, the ligands are in monoanionic enolate form. The C6=N2 and C7–O1 bond lengths in HL are shorter than those in complex 4. The N2–N3 and C7–N3 bond lengths in HL are longer than those in complex 4.
Structure description of complexes 1-3. The molecular structures of complexes 1-3 are shown in Figs. 1-3, respectively. The complexes crystallize in the monoclinic space group P21/c. The Zn atom in each complex is in distorted trigonal-bipyramidal coordination, with the equatorial plane defined by the imino nitrogen (N2) of the hydrazone ligand, and two halide or pseudohalide ligands, viz. Br1 and Br2 for 1, Br1 and N5 for 2, and I1 and I2 for 3, and with the axial positions occupied by the pyridine nitrogen (N1) and carbonyl oxygen (O1) atoms of the hydrazone ligand. The Zn–O and Zn–N bond lengths in the complexes are similar, and are comparable to those observed in zinc complexes with hydrazone ligands [32-37]. The trigonal-bipyramidal coordination in each complex is distorted from ideal geometry, as indicated by the bond lengths and angles. The equatorial bond angles of 114.67(13)-129.47(13)° for 1, 114.4(4)-132.0(2)° for 2, and 114.44(13)-130.57(13)° for 3, as well as the angles among the axial and equatorial donor atoms of 71.14(15)-102.22(13)° for 1, 73.1(4)-100.1(3)° for 2, and 71.13(15-103.84(12)° for 3, indicate the distortion of the trigonal-bipyramidal coordination. The distortion can be further observed from the perpendicular angles between the two axial bonds of 145.25(15)° for 1, 147.9(4)° for 2, and 144.93(16)° for 3.
The pyridine ring and the benzene ring form a dihedral angle of 6.2(4)° for 1, 5.7(4)° for 2, and 7.5(5)° for 3. The dihedral angle between the benzene ring and the ring C8–C9–N4–H4B⋯O1–C7 is 1.2(4)° for 1, 5.4(4)° for 2, and 3.9(5)° for 3, indicating they are approximately coplanar. This is caused by the presence of N4–H4B⋯O1 hydrogen bond. In the crystal structure of complex 1, the molecules are linked through N–H⋯Br hydrogen bonds (Table 4), to form two-dimensional network along the bc plane (Fig. 4). In the crystal structure of complex 2, the complex molecules are linked by water molecules through N–H⋯O and O–H⋯Br hydrogen bonds (Table 4), to form one-dimensional chains along the b axis. The amino H atom forms an intramolecular N–H⋯O hydrogen bond (Fig. 5). In the crystal structure of complex 3, the molecules are linked through N–H⋯I hydrogen bonds (Table 4), to form two-dimensional network along the ab plane (Fig. 6).
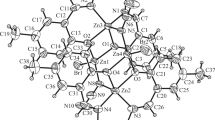
Structure description of complex 4. The molecular structure complex 4 is shown in Fig. 7. The complex crystallizes in the monoclinic space group P21/n. The Zn atom in the complex is in octahedral coordination, with the equatorial plane defined by the pyridine nitrogen (N1), imino nitrogen (N2) and enolate oxygen (O1) and the other imino nitrogen (N6) from two hydrazone ligands, and with the two axial positions occupied by the pyridine nitrogen (N5) and enolate oxygen (O2) of the hydrazone ligand. The octahedral coordination is distorted from octahedral geometry, as indicated by the bond lengths and angles. The Zn–O and Zn–N bond lengths in the complex are 2.0758(18)-2.1566(18)° and 2.040(2)-2.257(2)°, respectively, which are comparable to those observed in zinc complexes with hydrazone ligands [38-42]. The cis and trans coordinate bond angles are 73.96(8)-109.41(8)° and 149.31(8)-175.37(8)°, indicate the distortion of the octahedral coordination.
The pyridine ring and the benzene ring form a dihedral angle of 7.3(4)° for one hydrazone ligand, and 22.7(5)° for the other one. The dihedral angle between the C8–C13 benzene ring and the ring C8–C13–N4–H4A⋯N3–C7 is 1.1(4)°, and that between the C21–C26 benzene ring and the ring C21–C22–N8–H8A⋯N7–C20 is 1.8(4)°. There present N4–H4A⋯N3 and N8–H8A⋯N7 hydrogen bonds in the ligands. In the crystal structure of the complex, the molecules are linked through N–H⋯O hydrogen bonds (Table 4), to form two-dimensional network along the bc plane (Fig. 8).
IR and UV spectra. The IR spectra of the free hydrazone and the complexes were recorded in the region of 4000-400 cm–1 using KBr pellets. The infrared absorption bands at 3482 cm–1 and 3373 cm–1 are assigned to the stretching vibrations of NH2, and that at 3205 cm–1 is assigned to NH. The free hydrazone exhibits strong band at 1641 cm–1 due to the C=O group, and the medium band at 1611 cm–1 can be attributed to azomethine group, μ(C=N) [43]. In the complexes 1-3, these bands are observed at 1632-1633 cm–1 for the conjugated groups –CH=N–N=C(O)–. While in the spectrum of complex 4, the C=N vibration is located at 1603 cm–1 [44]. The wave numbers of the azomethine groups in complexes 1, 2 and 3 are located at higher frequencies, while that in complex 4 is located at lower frequency when compared with that of HL. This agrees with the different types of the hydrazone ligands, viz. ketone for 1, 2 and 3, and enol for 4. The complexes show characteristic absorptions in the region 3200-3500 cm–1 that can be assigned to NH stretching bands. In the spectrum of complex 2, the intense and typical band at 2088 cm–1 is assigned to the vibration of the thiocyanate ligand [45].
The UV spectra of the hydrazone and the complexes were recorded in methanol. In the spectrum of the free hydrazone, the bands at 350 nm are attributed to the azomethine chromophore π–π* transition. The bands at higher energies (300 nm) are associated with the benzene π–π* transition. In the spectra of the complexes, the peaks in the region 295-325 nm can be assigned to intraligand transitions, and those in the region 360-390 nm may correspond to the ligand-to-metal charge transfer [46].
Antimicrobial activity. The hydrazone and the complexes were screened for antibacterial activity against two Gram (+) bacterial strains (Bacillus subtilis and Staphylococcus aureus) and two Gram (–) bacterial strains (Escherichia coli and Pseudomonas fluorescens) by MTT method. The MIC (minimum inhibitory concentration) values of the compounds against four bacteria are listed in Table 5. Kanamycin and Penicillin G were used as the standard agents. The free hydrazone has medium activity against B. subtilis, and weak activity against S. aureus and E. coli, while inactive against P. fluorescens. In general, the complexes have better activities against the bacteria than the free hydrazone. Interestingly, complexes 1-3 have similar activities against all the bacteria strains. Thus, the Br, NCS and I ligands have no obvious influence on the antibacterial activity. The three complexes showed strong activity against B. subtilis and S. aureus, medium activity against E. coli, and weak activity against P. fluorescens. Complex 4 showed good activity against B. subtilis, and weak activity against the remaining bacteria strains. Interestingly, complexes 1-3 have effective activity against B. subtilis and S. aureus, which are comparable to the reference drug Penicillin G.
The antifungal activity of the compounds was also evaluated against two fungal strains (Candida albicans and Aspergillus niger) by MTT method. Ketoconazole was used as a reference agent. Complexes 1-3 have weak activity against P. fluorescens, while no activity against C. albicans and A. niger. Complex 4 has very weak activity against P. fluorescens, which is weaker than complexes 1-3.
CONCLUSIONS
In summary, the present paper reports the synthesis, characterization and crystal structures of four new zinc complexes with the ligand 2-amino-N′-(pyridin-2-ylmethylene)benzohydrazide. The hydrazone ligand coordinates to the Zn atom through the pyridine nitrogen, imino nitrogen and carbonyl oxygen. When Lewis bases were added to the reaction mixture, the hydrazone ligand coordinates to the Zn atom through the pyridine nitrogen, imino nitrogen and enolate oxygen. Thus, the presence of Lewis bases contributes to the enolate form of the hydrazone compound. The complexes have effective antibacterial activity against S. aureus.
ACKNOWLEDGMENTS
This work was financially supported by the high-level scientific research achievement funding program of Liaoning Normal University (grant number 21GDL003).
ADDITIONAL INFORMATION
CCDC - 2116737 (1), 2116738 (2), 2116739 (3) and 2116740 (4) contain the supplementary crystallographic data for this article. These data can be obtained free of charge at http://www.ccdc.cam.ac.uk/const/retrieving.html or from the Cambridge Crystallographic Data Centre (CCDC), 12 Union Road, Cambridge CB2 1EZ, UK; Fax: +44(0)1223-336033 or E-mail: deposit@ccdc.cam.ac.uk.
REFERENCES
K. Pyta, A. Janas, M. Szukowska, P. Pecyna, M. Jaworska, M. Gajecka, F. Bartl, and P. Przybylski. Eur. J. Med. Chem., 2019, 167, 96-104. https://doi.org/10.1016/j.ejmech.2019.02.009
I. Shabeeb, L. Al-Essa, M. Shtaiwi, E. Al-Shalabi, E. Younes, R. Okasha, and M. Abu Sini. Lett. Org. Chem., 2019, 16, 430-436. https://doi.org/10.2174/1570178616666181227122326
Y. C. Wu, X. D. Ding, L. Ding, Y. S. Zhang, L. Cui, L. Sun, W. Li, D. Wang, and Y. F. Zhao. Eur. J. Med. Chem., 2018, 158, 247-258. https://doi.org/10.1016/j.ejmech.2018.09.004
G.-X. He and L.-W. Xue. Acta Chim. Slov., 2021, 68, 567-574. https://doi.org/10.17344/acsi.2020.6333
A. Thirugnanasundar, M. P. Kesavan, S. M. Kumar, L. Ravi, R. Bhaskar, G. Rajagopal, and J. Rajesh. Inorg. Chim. Acta, 2021, 526, 120543. https://doi.org/10.1016/j.ica.2021.120543
J. Szklarzewicz, A. Jurowska, D. Matoga, K. Kruczala, G. Kazek, B. Mordyl, J. Sapa, and M. Papiez. Polyhedron, 2020, 185, 114589. https://doi.org/10.1016/j.poly.2020.114589
P. Bera, P. Brandao, G. Mondal, H. Jana, A. Jana, A. Santra, and P. Bera. Polyhedron, 2017, 134, 230-237. https://doi.org/10.1016/j.poly.2017.06.024
R. Fekri, M. Salehi, A. Asadi, and M. Kubicki. Inorg. Chim. Acta, 2019, 484, 245-254. https://doi.org/10.1016/j.ica.2018.09.022
H.-Y. Qian. Acta Chim. Slov., 2019, 66, 995-1001. https://doi.org/10.17344/acsi.2019.5247
Z.-Q. Sun, S.-F. Yu, X.-L. Xu, X.-Y. Qiu, and S.-J. Liu. Acta Chim. Slov., 2020, 67, 1281-1289. https://doi.org/10.17344/acsi.2020.6236
L.-W. Xue, Y.-J. Han, and X.-Q. Luo. Acta Chim. Slov., 2019, 66, 622-628. https://doi.org/10.17344/acsi.2019.5039
D. A. Paixão, I. M. Marzano, E. H. L. Jaimes, M. Pivatto, D. L. Campos; F. R. Pavan, V. M. Deflon, P. Maia, A. M. da Costa Ferreira, I. A. Uehara, M. J. B. Silva, F. V. Botelho, E. C. Pereira-Maia, S. Guilardi, and W. Guerra. J. Inorg. Biochem., 2017, 172, 138-146. https://doi.org/10.1016/j.jinorgbio.2017.04.024
J. D. Siqueira, S. F. de Pellegrin, S. S. dos Santos, B. A. Iglesias, P. C. Piquini, L. P. Arantes, F. A. Soares, O. A. Chaves, A. Neves, and D. F. Back. J. Inorg. Biochem., 2020, 204, 110950. https://doi.org/10.1016/j.jinorgbio.2019.110950
B. B. Hollo, J. Magyari, S. Armakovic, G. A. Bogdanovic, M. V. Rodic, S. J. Armakovic, J. Molnar, G. Spengler, V. M. Leovac, and K. M. Szecsenyi. New J. Chem. 2016, 40, 5885-5895. https://doi.org/10.1039/C6NJ00560H
V. Singh, A. Katiyar, and S. Singh. J. Coord. Chem., 2009, 62, 1336-1346. https://doi.org/10.1080/00958970802546016
H. L. Zhu, J. H. Zhu, C. Chen, Z. Wei, and L. Tian. Asian J. Chem., 2013, 25, 8444-8446. https://doi.org/10.14233/ajchem.2013.14779
Y.-L. Sang, X.-S. Lin, and W.-D. Sun. Acta Chim. Slov., 2020, 67, 581-585. https://doi.org/10.17344/acsi.2019.5595
I. Demir, M. Bayrakci, K. Mutlu, and A. I. Pekacar. Acta Chim. Slov., 2008, 55, 120-124. https://doi.org/10.1353/dss.2008.0061
A. Sousa-Pedrares, N. Camina, J. Romero, M. L. Duran, J. A. Garcia-Vazquez, and A. Sousa. Polyhedron, 2008, 27, 3391-3397. https://doi.org/10.1016/j.poly.2008.08.011
Y. Luo, J. Wang, X. Ding, R. Ni, M. Li, T. Yang, J. Wang, C. Jing, and Z. You. Inorg. Chim. Acta, 2021, 516, 120146. https://doi.org/10.1016/j.ica.2020.120146
R. A. Gossage and H. A. Jenkins. Acta Chim. Slov., 2009, 56, 329-333.
P. H. O. Santiago, M. B. Santiago, C. H. G. Martins, and C. C. Gatto. Inorg. Chim. Acta, 2020, 508, 119632. https://doi.org/10.1016/j.ica.2020.119632
N. Kitanovski and M. Pockaj. Acta Chim. Slov., 2021, 68, 475-482. https://doi.org/10.17344/acsi.2020.6634
G.-H. Sheng, X. Han, Z. You, H.-H. Li, and H.-L. Zhu. J. Coord. Chem., 2014, 67, 1760-1770. https://doi.org/10.1080/00958972.2014.916795
Y. Huo, Y.-T. Ye, X.-S. Cheng, and Z.-L. You. Inorg. Chem. Commun., 2014, 45, 131-134. https://doi.org/10.1016/j.inoche.2014.04.008
Z. L. You, H. Y. Yu, B. Y. Zheng, C. L. Zhang, C. W. Lv, K. Li, and L. Pan. Inorg. Chim. Acta, 2018, 469, 44-50. https://doi.org/10.1016/j.ica.2017.09.011
L. Mazur, K. N. Jarzembska, R. Kaminski, K. Wozniak, E. Pindelska, and M. Zielinska-Pisklak. Cryst. Growth Des., 2014, 14, 2263-2281. https://doi.org/10.1021/cg401866x
Bruker, SMART and SAINT. Madison: Bruker AXS Inc., 2002.
G. M. Sheldrick. SADABS. Göttingen, Germany: University of Göttingen, 1996.
G. M. Sheldrick. Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3-8. https://doi.org/10.1107/S2053229614024218
J. Meletiadis, J. F. Meis, J. W. Mouton, J. P. Donnelly, and P. E. Verweij. J. Clin. Microbiol., 2000, 38, 2949-2954. https://doi.org/10.1128/JCM.38.8.2949-2954.2000
A. A. R. Despaigne, J. G. da Silva, A. C. M. Do Carmo, O. E. Piro, E. E. Castellano, and H. Beraldo. J. Mol. Struct., 2009, 920, 97-102. https://doi.org/10.1016/j.molstruc.2008.10.025
A. A. R. Despaigne, J. G. da Silva, A. C. M. do Carmo, F. Sives, O. E. Piro, E. E. Castellano, and H. Beraldo. Polyhedron, 2009, 28, 3797-3803. https://doi.org/10.1016/j.poly.2009.07.059
M. Bakir, R. R. Conry, O. Green, and W. H. Wulder. J. Coord. Chem., 2008, 61, 3066-3079. https://doi.org/10.1080/00958970801993672
A. A. R. Despaigne, J. G. da Silva, A. C. M. do Carmo, F. Sives, O. E. Piro, E. E. Castellano, and H. Beraldo. Polyhedron, 2009, 28, 3797-3803. https://doi.org/10.1016/j.poly.2009.07.059
M. Bakir, O. Green, and W. H. Mulder. J. Mol. Struct., 2008, 873, 17-28. https://doi.org/10.1016/j.molstruc.2007.03.001
C. E. Kyriakidis, P. C. Christidis, P. J. Rentzeperis, and I. A. Tossidis. Z. Kristallogr., 1990, 193, 261-269. https://doi.org/10.1524/zkri.1990.193.3-4.261
A. Ray, S. Banerjee, S. Sen, R. J. Butcher, G. M. Rosair, M. T. Garland, and S. Mitra. Struct. Chem., 2008, 19, 209-217. https://doi.org/10.1007/s11224-007-9274-7
C. M. Armstrong, P. V. Bernhardt, P. Chin, and D. R. Richardson. Eur. J. Inorg. Chem., 2003, 1145-1156. https://doi.org/10.1002/ejic.200390146
P. Barbazan, R. Carballo, and E. M. Vazquez-Lopez. CrystEngComm, 2007, 9, 668-675. https://doi.org/10.1039/b703442c
A. A. R. Despaigne, J. G. da Silva, A. C. M. do Carmo, O. E. Piro, E. E. Castellano, and H. Beraldo. Inorg. Chim. Acta, 2009, 362, 2117-2122. https://doi.org/10.1016/j.ica.2008.09.040
B. Samanta, J. Chakraborty, S. Shit, S. R. Batten, P. Jensen, J. D. Masuda, and S. Mitra. Inorg. Chim. Acta, 2007, 360, 2471-2484. https://doi.org/10.1016/j.ica.2006.12.019
M. Zhang, D.-M. Xian, H.-H. Li, J.-C. Zhang, and Z.-L. You. Aust. J. Chem., 2012, 65, 343-350. https://doi.org/10.1071/CH11424
Z.-L. You, D.-H. Shi, J.-C. Zhang, Y.-P. Ma, C. Wang, and K. Li. Inorg. Chim. Acta, 2012, 384, 54-61. https://doi.org/10.1016/j.ica.2011.11.039
S. Basak, S. Sen, S. Banerjee, S. Mitra, G. Rosair, and M. T. G. Rodriguez. Polyhedron, 2007, 26, 5104-5112. https://doi.org/10.1016/j.poly.2007.07.025
S. Das and S. Pal. J. Mol. Struct., 2005, 753, 68-79. https://doi.org/10.1016/j.molstruc.2005.05.037
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interests.
Additional information
Text © The Author(s), 2022, published in Zhurnal Strukturnoi Khimii, 2022, Vol. 63, No. 8, 97415.https://doi.org/10.26902/JSC_id99095
Rights and permissions
About this article
Cite this article
Zhang, L., Feng, X., Gu, Y. et al. SYNTHESES, CRYSTAL STRUCTURES, AND ANTIMICROBIAL ACTIVITIES OF ZINC COMPLEXES DERIVED FROM 2-AMINO-N′- (PYRIDIN-2-YLMETHYLENE)BENZOHYDRAZIDE. J Struct Chem 63, 1358–1370 (2022). https://doi.org/10.1134/S0022476622080170
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0022476622080170