Abstract
MicroRNA-219-1 (miR-219-1) acts as a tumor suppressor in a variety of cancers but, the regulatory epigenetic mechanism involved in its gene expression level has not been studied. Using real-time polymerase chain reaction (real-time PCR) and bisulfite genomic sequencing technology, promoter methylation level of miR-219-1 and gene expression levels of miR-219-5p and miR-219-1-3p were determined respectively, in glioblastoma multiforme (GBM) (n = 31), their adjacent normal tissues (n = 31), and GBM U87 cell line. Following treatment of GBM U87 cells with 5-aza-2′-deoxycitidine (5-aza-dC), miR-219-1 promoter methylation, their target mRNA, and protein levels were determined by genomic bisulfite modification, real-time-PCR, and ELISA techniques, respectively. Our results showed that gene expression levels of miR-219-5p and miR-219-1-3p were significantly lower in GBM patients relative to their adjacent normal tissues (p < 0.01). MiR-219-1 promoter had a high level of methylation in GBM tissues (p < 0.01) and a negative correlation was observed between the miRNAs gene expression and methylation levels in GBM tissues (p < 0.01). Treatment of GBM U87 cells by 5-aza-dC decreased the level of miR-219-1 methylation, amount of target mRNA, and levels of cyclin A2 and mucin 4 (MUC4) proteins, and increased the expression levels of miR-219-5p and miR-219-1-3p (p < 0.01). Using external miR-219-5p and miR-219-1-3p, the expression of cyclin A2 and MUC4 were suppressed and proliferative activity of the U87MG cell line was reduced (p < 0.01). These findings suggested that DNA methylation has a crucial role in the regulation of miR-219-1 gene expression and that hypermethylated miR-219-1 may be involved in GBM pathogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Annual incidence of central nervous system (CNS) cancer is about 3.5 in 100,000 people which represents 1.9% of all newly diagnosed cancer cases and 2.3% of cancer deaths worldwide [1]. Gliomas are the most common and lethal neuroepithelial tumors and comprise majority of the malignant brain tumors in humans [2]. Gliomas are clinicopathologically classified by the World Health Organization (WHO) into four different grades (I-IV) [3] and glioblastoma multiforme (GBM) is the most aggressive form (grade IV) of these malignant brain tumors [2]. Median survival of the newly diagnosed GBM is 12-15 months and the five-year survival rate is less than 3% [4]. Consequently, elucidating molecular mechanisms underlying GBM development and progression is crucial for improving conventional therapeutic strategies and facilitating development of new therapies for this aggressive disease.
MicroRNAs (miRNAs) are non-coding, small RNA molecules (19 to 24 nucleotides) that post-transcriptionally regulate gene expression in both plants and animals [5]. They are involved in regulation of a variety of biological processes during normal development including cell cycle, proliferation, differentiation, apoptosis, and many more molecular pathways [6]. It has been shown that the altered gene expression of miRNAs is involved in different pathological processes, such as cancer [7]. Besides genetic mechanisms, epigenetic modifications such as DNA methylation, and covalent histone modification play an important role in controlling miRNAs expression [8]; furthermore, CpG island hyper-methylation of tumor suppressor miRNAs contributes to carcinogenesis [8, 9].
Processing of the miR-219-1 precursor by dicer enzyme generates two mature miRNAs: miR-219-5p from the 5′-end, and miR-219-1-3p from the 3′-end of the pre-miR-219-1. Since these two mature products have unique seed regions, each miRNA targets different mRNAs (http://www.mirbase.org/). A recent study demonstrated that miR-219-5-p has low level of gene expression in glioma and functions as a tumor suppressor in GBM cells [10]. Although, the study confirmed the Sal-like protein 4 as a target mRNA of miR-219-5p [10], cyclin A2 was identified as a direct target of miR-219-5p in esophageal squamous cell carcinoma that may also have an oncogenic effect on glioma [11]. Similarly, the tumor suppressor function of miR-219-1-3p by targeting mucin 4 (MUC4) gene has been reported in pancreatic cancer [12]. While the tumor suppressor functions of miR-219-1 has been studied in a variety of cancers, there is no evidence of epigenetic modulation of miR-219-5p and miR-219-1-3p in GBM.
Despite the fact that miR-219-1 is one of the most significantly down-regulated miRNAs in various tumor types [10, 12], its gene expression level underlying epigenetic modification, and whether this regulatory mechanism may be associated with the miR-219-1 functional role have not been clearly elucidated in GBM. In this work we studied relationship between the level of miR-219-1 gene expression and its promoter methylation levels in attempt to answer the question whether epigenetic modification of this miRNA could result in down-regulation of its targets.
MATERIALS AND METHODS
Primary tumor samples. Analyses of 31 fresh GBM tumor samples and their histologically normal adjacent tissues was performed in this study. A total of 17 males and 14 females (1.4 : 1) ranging in age from 25 to 79 years (median 59 years) participated in the present study. The detailed clinicopathological features of GBM samples are described in the table. All samples were obtained from patients undergoing surgical resection at the Department of Neurosurgery, Imam Reza hospital, Tabriz, Iran, in accordance with the Committee on Human Research approved procedure.
Two pathologists reviewed all specimens and only primary GBM samples were included in the study. No patients had received blood transfusion, radiotherapy, or chemotherapy before surgery. Patients were excluded if they had a prior history of diabetes, hepatic fibrosis, multiple sclerosis, schizophrenia, other cancers such as hepatocellular carcinoma, papillary thyroid carcinoma, ovarian cancer, colorectal cancer, etc. Moreover, the surrounding normal tissues with any neoplastic tumors or necrotic lesions were also excluded from the study. All specimens were snap-frozen in liquid nitrogen immediately after surgery and then stored at –80°C until used for investigation. Written informed consent was obtained from all the patients for the use of their tumor tissues. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Cell line, cell cultivation and 5-aza-dC treatment. Human GBM cancer cell line, U87 (ATCC® HTB-14™), was purchased from the Pasteur Institute of Iran and was cultured no longer than one month following purchase. Cell line was cultured under sterile condition at 37°C, 5% CO2, in Dulbecco’s Modified Eagle’s Medium (DMEM; Life Technologies, Inc., USA) supplemented with streptomycin (100 µg/ml), penicillin (100 U/ml) (Life technologies, Inc.), 2 mM glutamine (Life Technologies, Inc.) and 10% fetal bovine serum (FBS, Life Technologies, Inc). In order to determine correlation of the promoter hyper-methylation of miR219-1 and miR-219-5p and miR-219-1-3p gene expression levels, bisulfite genomic sequencing and real-time polymerase chain reaction (real-time PCR) were used to evaluate the promoter CpG island methylation and mRNA expression levels of miRNA, respectively, following incubation of U87 cell line in the presence or absence of 5-aza-2′-deoxycitidine (5-aza-dC). Briefly, GBM U87 cells were seeded at a density of 1 × 105 cells per well in a six-well plate, incubated for 24 h, and then supplemented with fresh medium containing 1 or 5 µM 5-aza-dC (purity ≥ 98%) (Sigma-Aldrich, USA) for 72 h. At the end of the treatment, the medium was replaced with fresh medium without 5-aza-dC, and the cells were cultured for additional 48 h. Stock solutions of 5-aza-dC were dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich) and baseline was established in mock treatment of cells with the same volume of DMSO in triplicate.
Methylation analyses. Methylation of the promoter CpG island of miR-219-1 was determined by the bisulfite genomic sequencing technique using sodium bisulfite-treated genomic DNA. In particular, DNA was extracted from the U87 cells and the frozen tissues by a DNeasy Blood & Tissue Kit (Qiagen, Germany). To perform bisulfite conversion of DNA, 1 µg of extracted DNA was treated with sodium bisulfite according to the EpiTect® Fast Bisulfite Conversion kit (Qiagen) protocol and subjected to polymerase chain reaction (PCR) using the following primer sets: forward primer; 5′-GTGATTTTTGATTTTTGTTTTTTTT and reverse primer; 5′-TTCACCTACACTTATTCCAACAAAC-3′. Thermal cycling conditions were: 95°C for 5 min, 60°C for 10 min; 95°C for 5 min, 60°C for 10 min. Samples were then purified using EpiTect spin-column and eluted with 15 µl of Buffer EB supplied in the kit. To observe distribution of the methylation patterns in an individual molecule we used the pGEM-T Easy Vector System II (Promega, USA), and cloned the purified PCR products into pGEM-T Easy Vector. Using blue/white screening and ABI 3730 XL DNA Analyzer (Applied Biosystem, USA), 8-10 clones from each samples were sequenced. To control complete bisulfite conversion of DNA, cytosines (Cs) that are not followed by guanine (non CpG Cs) were reviewed after sequencing of the analyzed region. Since such non CpG Cs should be converted to thymine (T) following bisulfite treatment and PCR, the presence of Cs in these positions served as an indicator for incomplete bisulfite conversion. Methylation levels for each CpG site within the DNA amplicon was quantified by measuring the ratio between the peak height values of C and T, using basic equation for the methylation percentage: (C/(C + T) × 100).
RNA extraction and real-time PCR. Total RNA including miRNAs was extracted from all tissues and cells using a MirVanaTM miRNA isolation kit (Ambion, USA) following the manufacturer’s instructions. To study expression of miR-219-5p and miR-219-1-3p, a qScript™microRNA cDNA Synthesis Kit (Quanta, USA) was used to synthesize complementary DNA (cDNA) and gene expression levels of miR-219-5p and miR-219-1-3p were determined using PerfeCTa® microRNA Assays (Quanta) according to manufacturer’s instructions with a Rotor-Gene 6000 real-time PCR cycler (Qiagen) system. U6 small nuclear RNA (U6 snRNA), an endogenous control, was used to normalize expression levels of miR-219-5p and miR-219-1-3p by the comparative Ct (ΔΔCt) method and relative miRNA gene expression level corresponded to the 2–ΔCt.
Universal primer provided in the kit served as reverse primer and miRNA was amplified by a specific forward primer. Our specific forward primers for detection of miR-219-5p and miR-219-1-3p were 5′-TGA TTG TCC AAA CGC AAT TCT-3′ and 5′-AGAGTTGAGTCTGGACGTCCCG-3′, respectively. Also, U6 snRNA forward primer was 5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse primer was 5′-GGAACGCTTCACGAATTTG-3′. Each qPCR reaction was performed in duplicate and U6 snRNA was used to normalize expression level of miR-219.
To study gene expression levels of MUC4 and cyclin A2, 1 µg of the total RNA was reverse transcribed to complementary DNA and subsequently diluted 1/10. Real-time PCR was performed using specific primers and SYBR Green detection technology as previously described [12, 13]. Expression levels relative to miR-219-1 were determined by ΔΔCt method.
Enzyme-linked immunosorbent assay (ELISA). MUC4 and cyclin A2 protein levels were measured with a Human MUC4 ELISA Kit (My BioSource, USA 2504728) and Rat Cyclin-A2 ELISA Kit (My BioSource), respectively, based on the standard quantitative enzyme immunoassay technique.
Cell transfection and proliferation assay. Generally, 10 nM of miR-219-5-p and miR-219-1-3p precursor molecules, mimicking miR-219-5p and miR-219-1-3p, respectively, and negative control non-specific miRNA (NC) were transfected into U87MG cell line using Lipofectamine TM RNAiMAX (Invitrogen, USA). The day before transfection, cells were seeded to an antibiotic free medium. MiR-219-5p and miR-219-1-3p transfected U87MG cells were cultured, and then incubated with 20 µl of BrdU label for 2 h. The cells were fixed, and incorporated BrdU was detected using a BrdU cell proliferation kit (Abcam, USA) according to the manufacturer’s instruction.
Statistical analysis. Student’s t-test was used to compare mean methylation and gene expression levels of miR-219-1 in GBM and adjacent normal tissues. To compare the mean methylation and gene expression levels of miR-219-1, MUC4 and cyclin A2 levels in cells before and after 5-Aza-dC treatment, the ANOVA test was performed. Linear Pearson correlation was used to study relationship between the miR-219-1 promoter methylation and its gene expression level. Statistical analysis was performed using SPSS version 22.0, and p < 0.05 was considered as statistically significant.
RESULTS
MiR-219-1 and clinicopathological features of GBM patients. Associations between the miR-219-1 gene expression, methylation levels, and clinicopathological characteristics were assessed in all 31 GBM patients. Since our patient group consisted of a relatively small number of GBM tumor samples (n = 31), to get more accurate results the miR-219-5p (n = 31) and miR-219-1-3p (n = 31) gene expression levels as miR-219-1 were pooled together and the relationships between the miR-219-1 gene expression (n = 62), methylation levels (n = 31), and clinicopathological features of GBM patients were examined. Based on the mean value of miR-219-1 expression and methylation levels, all GBM patients were divided into four groups; patients with low (n = 37) and high (n = 25) miR-219-1 gene expression levels and patients with low (n = 13) and high miR-219-1 methylation levels (n = 18). As it is shown in the table no statistically significant differences were observed between the miR-219-1 gene expression, methylation levels and age (<50 and ≥50), gender (male/female), tumor location (frontal lobe, temporal lobe, parietal lobe and occipital lobe) in the patient group. However, our statistical analysis demonstrated that the low gene expression and high methylation levels of miR-219-1 were associated with the tumor size (< 5 cm vs. ≥ 5 cm) and Karnofsky Performance Scale (KPS; < 80 vs. ≥ 80) in the patient samples.
MiR-219-1 and target gene expression levels in GBM tissues. We determined the expression levels of miR-219-5p, miR-219-1-3p, cyclin A2, and MUC4 in the tumor-derived tissues from GBM patients and compared them with the normal adjacent tissues using real-time PCR technique. Comparative Ct method was used to determine the relative expression ratio of miRNA and target genes in the tumor samples compared to the normal adjacent tissues. We found lower gene expression levels of miR-219-5p and miR-219-1-3p in 27 (87%) and 25 (80%) samples of the GBM tissues, respectively compared with the normal adjacent tissues.
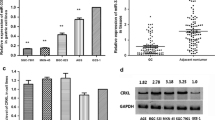
As one can see in Fig. 1, a and b, there were significant reductions in the miR-219-5p and miR-219-1-3p gene levels in the tumor tissues from GBM patients compared to the normal adjacent tissues (Student’s t-test, p < 0.01). As shown in Fig. 1, c and d we also observed high gene expression levels of cyclin A2 (1.60 vs 0.39; Student’s t-test, p < 0.01) and MUC4 (1.71 vs 0.62; Student’s t-test p < 0.05) in GBM tumor tissues compared to the adjacent normal tissues. High expression levels of cyclin A2 and MUC4 in GBM tissues indicated that these oncogenes may be involved in glioma carcinogenesis.
Gene expression levels of MiR-219-5p (a), miR-219-1-3p (b), cyclin A2 (c), and MUC4 (d) in GBM tumor tissues (n = 31) and normal adjacent tissues (n = 31) determined by real-time PCR using 2−ΔCT method. Expression of miRNA in individual tumor samples were measured relative to U6 snRNA (* p < 0.01). Cyclin A2 (c) and MUC4 (d) protein gene expression levels were normalized to the levels of B actin (ACTB) mRNA. Gene expressions data for individual human tissues are presented as mean ± standard deviation (SD). * p < 0.01 for all GBM tumors compared to non-GBM specimens.
Methylation status of MiR-219-1 promoter in GBM tumor tissues. A CpG island in the promoter region of miR-219-1 (from –2802 to –2952 bp) containing 22 CpGs was found using methprimer (http://www.urogene.org/methprimer/) and inserting 3000 nucleotides from the upstream region of miR-219-1 (Fig. 2a). Analysis of relative methylation levels by subcloning of PCR products and genomic bisulfite sequencing technique revealed higher level of the miR-219-1 DNA methylation in the GBM tumor tissues (70.7%) compared to the adjacent normal tissues (16.4%; Student’s t-test, p < 0.01; Fig. 2, b and c).
Methylation upstream of miR-219-1 and its association with gene expression level. a) Map of the CpG island showing the number and positions of CpGs in relation to the miR-219-1 promoter. CpG island of 151 bp (from –2802 to –2952 bp) downstream of the pre-miR-219-1 start codon, covering 22 CpG sites) was used for methylation analysis. b) Scheme of distribution of CpGs in the promoter region of miR-219-1 in a random GBM tumor sample, its corresponding normal adjacent tissue, and GBM U87 cell line. For each sample 8 separate clones were sequenced. Each row represents methylation level of the individual CpG (ranging between 0 and 1) in a separate clone, except the last row that represents position of each CpG on chromosome 6. To simplify visualization, the first four figures of each CpG position number (3320) have been removed from all CpG site numbering so that the position of 4867 of a CpG represents 33204867th nucleotide on chromosome 6. c) Difference in miR-219-1 promoter methylation between the GBM tumor tissues and adjacent normal tissues (* p < 0.01). Results are presented as a percent of methylated cytosines in CpG island (C/(C + T) × 100). d) Negative correlation between methylation of the miR-219-1 promoter and miR-219-5p (R = –0.82) and miR-219-1-3p gene expressions (R = –0.75) in GBM tumor tissue (p < 0.01).
To elucidate whether the downregulation of miR-219-1 in GBM tumors is a result of its promoter hypermethylation, we additionally performed correlation analysis between the miR-219-1 promoter methylation and the gene expression levels of miR-219-5p and miR-219-1-3p in GBM tumor tissue samples. It can be seen in Fig. 2d that according to the results of Pearson’s correlation analysis the expression levels of miR-219-5p (R = –0.82, p < 0.01) and miR-219-1-3p (R = –0.75, p < 0.01) genes correlated negatively with the miR-219-1 promoter hypermethylation. These results suggested that the miR-219-1 promoter hypermethylation in GBM tissues may cause downregulation of the gene expression.
Effects of 5-aza-dC on methylation and gene expression levels of miR-219-1. In order to assess the effects of 5-aza-dC on the level of miR-219-1 promoter methylation, mean methylation level of the miRNA promoter was examined before and after treatments of U87MG cell line with 1 or 5 µM of 5-aza-dC for 72 h. Before treatment of cells with 5-aza-dC, genomic bisulfite sequencing technique showed a 64.5% mean methylation level in the promoter region of the mir-219-1 gene. However, the miR-219-1 methylation level decreased by 26% and 51% following 72-h treatment of the cells with 1 or 5 µM 5-aza-dC, respectively. These results suggested that 5-Aza-dC was capable to decrease the mean methylation level of miR-219-1 at the promoter region (Fig. 3a). Furthermore, the levels of expression of miR-219-5p and miR-219-1-3p genes before and after treatment of cells with 1 or 5 mM of 5-azadC were investigated using real-time PCR. The results shown in Fig. 3b demonstrated that demethylation of DNA using 1 or 5 µM 5-aza-dC resulted in the 1.9 and 4.1-fold increase of the level of miR-219-5p gene expression in the U87MG cells, respectively. It was also shown that the treatment of cells with concentrations of 1 or 5 µM 5-aza-dC increased the levels of expression of miR-219-1-3p gene by 1.8- and 3.2-fold, respectively (Fig. 3c).
Effects of 5-aza-dC on methylation and gene expression levels of miR-219-1. Cells of glioblastoma cell line, U87, were treated with 5-aza-dC at 1 or 5 µM for 72 h. a) Effects of treatment with different concentrations of 5-Aza-dC for 72 h on promoter methylation level of miR-219-1. b and c) Effects of the dose- and time dependent of treatment with 5-Aza-dC on the levels of expression of miR-219-5p and miR-219-1-3p genes. Error bars represent SD of the results of triplicate polymerase chain reactions from a single experimental set, and P represents statistical significance of the difference between cell groups (* p < 0.05, ** p < 0.01).
Effects of 5-aza-dC on cyclin A2 and MUC4 gene expression and protein levels. To study the effects of 5-aza-dC treatment on gene expression and protein levels of the cyclin A2 and MUC4, U87 cells were treated with 1 or 5 µM of 5-aza-dC for 72 h and real-time PCR and ELISA techniques were used, respectively. Our results showed reduction in the level of expression of the cyclin A2 gene when the cells were treated with 1 or 5 µM 5-aza-dC (0.29- and 0.72-fold, respectively; Fig. 4a). Furthermore, demethylation of DNA by 1 or 5 µM 5-aza-dC decreased expression level of the MUC4 gene in U87 cells (0.11- and 0.81-fold, respectively Fig. 4b). However, as shown in Fig. 4, c and d treatment of the cells with 1 µM of 5-Aza-dC did not decrease protein levels of cyclin A2 and MUC4 and reduction of the protein level was observed only following treatment with 5 µM of 5-Aza-dC. Protein level of cyclin A2 in the untreated U87MG cells using ELISA was 310.8 pg/ml. Treatment of the cells with 1 or 5 µM 5-aza-dC reduced protein levels of cyclin A2 by 12.1 pg/ml (p > 0.05) and 112.7 pg/ml (p < 0.01), respectively. Protein level of MUC4 in untreated U87MG cells was 12 ng/ml and reduction by 1.2 ng/ml (p > 0.05) and 6.3 ng/ml (p < 0.01) were observed after treatment of the cell with concentration of 1 and 5 µM of 5-aza-dC, respectively (Fig. 4d).
Suppressive effects of ectopic miR-219-1 expression on target genes and cell proliferation rate. Since our study provides an evidence that miR-219-5p and miR-219-1-3p have a low gene expression levels in U87MG cell line and that their promoter region is hypermethylated, we investigated whether the restored miR-219-5p and miR-219-1-3p expression (mediated by transient transfection of dsRNA mimicking miR-219-5p and miR-219-1-3p into U87MG cell line) could suppress cell proliferation. Our results showed that transfection of the cells by dsRNA mimicking miR-219-5p and miR-219-1-3p induced expression of the candidate miRNA gene by 4- and 5-fold, respectively, and restoration of these miRNA resulted in a significant reduction of the U87MG cell proliferation rate (Fig. 5, a and b; p < 0.01). These results suggest that miR-219-15p and miR-219-1-3p may have an anti-proliferative effect in the U87MG cells. To verify that cyclin A2 and MUC4 are miR-219-5p and miR-219-1-3p targets, we performed real-time PCR with U87MG cells transfected with miR-219-5p and miR-219-1-3p mimics. Our results showed that the cyclin A2 and MUC4 mRNA levels reduced 48 h after transfection with mir-219-5p and miR-219-1-3p by 43% and 51%, respectively (Fig. 5c; p < 0.01) as compared with the cells transfected with miRNA negative control.
Tumor suppressive effects of miR-219-1 in U87 cell line. Cells were transfected with 10 nM of pre-miR miRNA precursor molecule mimicking miR-219-5p, miR-219-1-3p or control non-specific dsRNA (Pre-miR NC #1) using Lipofectamine TM RNAiMAX. a) Number of viable cells at 24-96 h after transfection was assessed by Brdu cell proliferation assay. Results are presented as means of triplicate determinations in these experiments; * p < 0.05 versus Pre-miR NC #1-transfected cells, in a statistical analysis with the Mann–Whitney U-test. b) Effect of transfection of 10 nM miR-219-5p and miR-219-1-3p on transcript levels of cyclin A2 and MUC4 mRNA in U87MG cells at 48 h relative to the cells transfected with 10 nM control oligonucleotide. c) Cyclin A2 and MUC4 gene expression levels were determined by real time PCR relative to the reference gene (actin β). Values represent mean ± SD of independent experiments (* p < 0.05, ** p < 0.01).
DISCUSSION
In recent years, several expression profiling studies have been carried out to determine the miRNAs expression patterns in GBM [14, 15]. These studies, demonstrated different gene expression patterns of miRNAs in GBM compared to the normal tissues and concluded that deregulated miRNAs could be involved in the pathogenesis of GBM. In the present study we observed low expression levels of miR-219-5p and miR-219-1-3p genes in GBM compared to normal adjacent tissues. The result of low gene expression level of miR-219-5p is in agreement with the previous studies [16]. Although, the decreased expression level of miR-219-1-3p gene has been reported in pancreatic cancer [12] there was no data available about the expression level of miR-219-1-3p gene in GBM.
We also found that the low gene expression levels of miR-219-5p and miR-219-1-3p were associated with the GBM tumor size and KPS (Karnofsky Performance Scale) index. Previously association of the low gene expression level of miR-219-1 (miR-219-5p) and clinicopathological characteristics of the patients such as advanced WHO grade and KPS Index was established [10]. Since our test group consisted from the patient with the most aggressive grade of glioma (grade IV or GBM) and not with gliomas of different grades (I, II, III, and IV) we were not able to include the WHO grades as a variable in the present study of gene expression. KPS is a simple approach to determine functional status of the patient. This can be used to assess effectiveness of cancer therapies, quality of life, and prognosis for individual patients. The Karnofsky Performance Score (KPS) ranges from 100 to 0, where 100 is “no evidence of disease” and 0 is “death”. Due to association of the miR-219-5p and miR-219-1-3p gene expression levels and KPS Index, it seems that the decreased gene expression of our candidate miRNAs could be related to the patient’s prognosis.
The observed CpG island hypermethylation in the promoter region of miR-219-1 in GBM as compared with the normal adjacent tissues could explain, at least in part, epigenetic silencing of miR-219-1 in the GBM tissues. There is a growing amount of data supporting the suggestion that DNA hyper-methylation at the promoter region may be involved in the silencing of tumor suppressor genes in several cancer cells [17]. More interestingly, several studies confirmed that a number of tumor suppressor miRNAs are down-regulated by hypermethylation of the promoter CpG islands in cancer cells [18-20]. Therefore, it seems that DNA methylation-mediated silencing of the tumor suppressor miRNAs could be a novel pathogenic mechanism in different stages of cancer. Aberrant DNA methylation of miR-219-1 has been observed in noncancerous cases including chronic inflammation pain and long-term night shift workers [21, 22]. However, there has been no study to identify DNA methylation of miR-219-1 (miR-219-5p and miR-219-1-3p) in GBM and at present, this is the only study of miR-219-1 methylation status to include patients with GBM. The reason that we selected miR-219-1 as a candidate gene to study epigenetic modification is that the promoter analysis by methprimer (http://www.urogene.org/methprimer/) revealed a large CpG island within its promoter region. Therefore, it was hypothesized that hypermethylation of the promoter of this miRNA could be involved in its down-regulation. Since regulation of miRNA expression by DNA methylation is a complex process, further studies on the methylation patterns of miRNA in GBM are warranted.
In our study treatment of glioblastoma multiforme cell line, U87MG, with 5-Aza-dC led to the increased expression levels of miR-219-5p and miR-219-1-3p genes and decreased gene expression levels of cyclin A2 and MUC4, respectively. Close relationship between the deregulated cyclin A2, chromosomal instability, and cancer cell proliferation has been predicted and increased gene expression of cyclin A2 has been implicated in a variety of cancer types [23]. Similarly, it has been reported that overexpression of MUC4, the membrane-bound glycoprotein, could result in epithelial cancer progression, aggressive behavior, poor outcomes and therapy resistance [24]. Since the aberrant methylation of DNA could be involved in miRNA downregulation, it is reasonable to suggest that the demethylating agent, 5-Aza-dC, is involved in miR-219-1 restoration, which could, in turn, lead to the decreased levels of target mRNAs, cyclin A2 as a target of miR-219-5p, and MUC4 as a target of miR-219-1-3p. 5-Aza-dC, a nucleoside analog, is incorporated into the DNA of rapidly growing tumor cells during replication and inhibits DNA methylation by trapping DNA methyltransferases on the DNA leading to their depletion inside the cell [25].
In the present study cyclin A2 and MUC4 protein levels were reduced only following treatment with high concentrations of 5-aza-dc (5 µM) and low concentrations of demethylating agent (1 µM) had no effect on the target protein levels. Also, there was no proportional correlation between the effects of different concentrations of 5-aza-dC on the levels of reductions of target proteins. In other words, cyclin A2 and MUC4 protein levels were not reduced 5-fold as compared to 1-fold reduction of protein levels by 1 µM of 5-Aza dC. In general, correlation between the cellular protein levels and expression levels of their corresponding mRNAs is not strong. They often show correlation of ~0.40, which implies that only ~40% of the variation in protein concentration can be explained by the changes in mRNA amounts [26]. To explain the remaining ~60% of the variation, some combination of post-transcriptional regulation and measurement noise must be considered.
Our results provided evidence that miR-219-5p and miR-219-1-3p exhibited antiproliferative effect on the U87 cell line. Using the gain-of-function approach, Jiang et al. transfected glioma cells with miR-219-5p mimics and determined cell proliferation, migration, and invasion. The authors found that the ectopic miR-219-5p reduced the rate of cell growth and concluded that miR-219-5p was involved in the negative regulation of cell growth [10]. In a similar study Lahdoui et al. observed suppression of the cell proliferation rate associated with the decreased cell migration by following overexpression of miR-219-1-3p in the pancreatic cancer cell lines [12]. MicroRNAs (miRNAs) have crucial roles in the cancer development and progression including repression of oncogene and/or tumor suppressor mRNAs or blocking of protein translation. Based on the oncogene or tumor suppressor targets, miRNAs act as oncogenic (by targeting tumor suppressor) or tumor suppressive molecules (by targeting oncogene) [27]. Since the oncogenic functions of miR-219-5p and miR-219-1-3p mRNA targets, cyclin A2 (in esophageal squamous cells) and MUC4 (in pancreatic cancer cells), have been documented in previous studies [23, 24] it seems likely that our candidate miRNAs as tumor suppressor molecules are capable of inducing cell proliferation arrest in cancerous cells including GBM. The most obvious differences between our study and the previous studies are: (i) previous studies provided no evidence about underlying mechanism of low expression level of miR-219-5p and miR-219-1-3p, and (ii) there were no data about restoration of the candidate miRNA suppressed by epigenetic factors and following effects on its tumor-suppressive functions.
There are several limitations in our study that could be addressed in future studies. Firstly, this study was based on a relatively small sample size that limited our possibility to extract more accurate results especially with respect to the relationship between the patient characteristics and miRNA gene expression. Therefore, other studies of a larger sample sizes, will be necessary to confirm our findings. Secondly, we studied only oncogenic targets of the candidate miRNAs affected by DNA methylation. According to the multiple oncogenic or tumor suppressor targets of an individual miRNA, extensive analysis is needed to map the communication network between the epigenetic modification of miRNA and the level of their target mRNAs expression. Finally, we used normal adjacent tissue as a control in the present study. Considering that there are no comprehensive and conclusive studies addressing limitations and that little is known about the global gene expression profile of this type of control subjects in GBM, the results may be suboptimal. Therefore, to obtain more accurate results, transcriptomic profiling of normal adjacent tissue in GBM should be conducted in future studies.
Role of epigenetic regulation of miR-219-1 in pathogenesis of human GBM was investigated in this work. To the best of our knowledge, it was demonstrated for the first time that (i) mir-219-5p and miR-219-1-3p are methylation-sensitive miRNA in glioblastoma multiforme development; (ii) restoring expression of the candidate miRNA by demethylating agents decreased the levels of its oncogenic target mRNA and protein. These results indicate that the miRNA-based signatures might serve as a basis for the development of novel potential therapies for glioblastoma multiforme cancer and further research on epigenetic drug development will be conducted in the near future.
Abbreviations
- 5-aza-dC:
-
5-aza-2′-deoxycitidine
- DMED:
-
Dulbecco’s Modified Eagle’s Medium
- ELISA:
-
enzyme-linked immunosorbent assay
- GBM:
-
glioblastoma multiforme
- miRNA:
-
microRNA
- MUC4:
-
mucin 4
- real-time PCR:
-
real time polymerase chain reaction
References
International Agency for Research on Cancer (IARC). GLOBOCAN 2008. Estimated Incidence, Mortality and 5-Year Prevalence: Both Sexes. Available online: http://globocan.iarc.fr (accessed on 16 October 2013).
Dolecek, T. A., Propp, J. M., Stroup, N. E., and Kruchko, C. (2012) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005-2009, Neuro Oncol., 14, v1-v49.
Louis, D. N., Ohgaki, H., Wiestler, O. D., Cavenee, W. K., Burger, P. C., et al. (2007) The 2007 WHO classification of tumours of the central nervous system, Acta Neuropathol., 114, 97-109.
Wen, P. Y., and Kesari, S. (2008) Malignant gliomas in adults, N. Engl. J. Med., 359, 492-507.
Bartel, D. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function, Cell, 116, 281-297.
Vidigal, J. A., and Ventura, A. (2015) The biological functions of miRNAs: lessons from in vivo studies, Trends Cell. Biol., 25, 137-147.
Reddy, K. B. (2015) MicroRNA (miRNA) in cancer, Cancer Cell Int., 15, 38.
Liu, X., Chen, X., Yu, X., Tao, Y., Bode, A. M., et al. (2013) Regulation of microRNAs by epigenetics and their interplay involved in cancer, J. Exp. Clin. Cancer. Res., 32, 96.
Konno, M., Koseki, J., Asai, A., Yamagata, A., Shimamura, T., et al. (2019) Distinct methylation levels of mature microRNAs in gastrointestinal cancers, Nat. Commun., 10, 3888.
Jiang, B., Li, M., Ji, F., and Nie, Y. (2017) MicroRNA-219 exerts a tumor suppressive role in glioma via targeting Sal-like protein 4, Exp. Ther. Med., 14, 6213-6221.
Ma, Q. (2019) MiR-219-5p suppresses cell proliferation and cell cycle progression in esophageal squamous cell carcinoma by targeting CCNA2, Cell. Mol. Biol. Lett., 24, 4.
Lahdaoui, F., Delpu, Y., Vincent, A., Renaud, F., Messager, M., et al. (2015) miR-219-1-3p is a negative regulator of the mucin MUC4 expression and is a tumor suppressor in pancreatic cancer, Oncogene, 34, 780-788.
Lee, R. S., Sohn, S., Shin, K. H., Kang, M. K., Park, N. H., and Kim, R. H. (2017) Bisphosphonate inhibits the expression of cyclin A2 at the transcriptional level in normal human oral keratinocytes, Int. J. Mol. Med., 40, 623-630.
Piwecka, M., Rolle, K., Belter, A., Barciszewska, A. M., Żywicki, M., et al. (2015) Comprehensive analysis of microRNA expression profile in malignant glioma tissues, Mol. Oncol., 9, 1324-1340.
Ondracek, J., Fadrus, P., Sana, J., Besse, A., Loja, T., et al. (2017) Global microRNA expression profiling identifies unique microRNA pattern of radioresistant glioblastoma cells, Anticancer Res., 37, 1099-1104.
Rao, S. A. M., Arimappamagan, A., Pandey, P., Santosh, V., Hegde, A. S., et al. (2013) miR-219-5p inhibits receptor tyrosine kinase pathway by targeting EGFR in glioblastoma, PLoS One, 8, e63164, https://doi.org/10.1371/journal.pone.0063164.
Guo, M., Peng, Y., Gao, A., Du, C., and Herman, J. G. (2019) Epigenetic heterogeneity in cancer, Biomark Res., 7, 23.
Ghasemi, A., Fallah, S., and Ansari, M. (2016) MiR-153 as a tumor suppressor in glioblastoma multiforme is downregulated by DNA methylation, Clin. Lab., 62, 573-580.
Wang, L. Q., and Chim, C. S. (2015) DNA methylation of tumor-suppressor miRNA genes in chronic lymphocytic leukemia, Epigenomics, 7, 461-473.
Serra, P. L., and Esteller, M. (2012) DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer, Oncogene, 31, 1609-1622.
Pan, Z., Zhu, L. J., Li, Y. Q., Hao, L. Y., Yin, C., et al. (2014) Epigenetic modification of spinal miR-219 expression regulates chronic inflammation pain by targeting CaMKIIγ, J. Neurosci., 16, 9476-9483.
Shi, F., Chen, X., Fu, A., Hansen, J., Stevens, R., et al. (2013) Aberrant DNA methylation of miR-219 promoter in long-term night shiftworkers, Environ. Mol. Mutagen, 54, 406-413.
Malumbres, M., and Barbacid, M. (2009) Cell cycle, CDKs and cancer: a changing paradigm, Nat. Rev. Cancer, 9, 153-166.
Xia, P., Choi, A. H., Deng, Z., Yang, Y., Zhao, J., and Wang, Y. (2017) Cell membrane-anchored MUC4 promotes tumorigenicity in epithelial carcinomas, Oncotarget, 8, 14147-14157.
Egger, G., Liang, G., Aparicio, A., and Jones, P. A. (2004) Epigenetics in human disease and prospects epigenetic therapy, Nature, 429, 457-463.
De Sousa Abreu, R., Penalva, L. O., Marcotte, E., and Vogel, C. (2009) Global signatures of protein and mRNA expression levels, Mol. Biosyst., 5, 1512-1526.
Svoronos, A. A., Engelman, D. M., and Slack, F. J. (2016) OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer, Cancer Res., 76, 3666-3670.
Funding
This work was supported by the IRNA University of Medical Sciences (IUMS) [project no. 24756].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare no conflicts of interest in financial or any other sphere. All procedures performed in present study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Additional information
Published in Russian in Biokhimiya, 2021, Vol. 86, No. 4, pp. 496-510, https://doi.org/10.31857/S0320972521040047.
Rights and permissions
About this article
Cite this article
Ghasemi, A., Mohammadi, A. & Fallah, S. Epigenetic Modification of MicroRNA-219-1 and Its Association with Glioblastoma Multiforme. Biochemistry Moscow 86, 420–432 (2021). https://doi.org/10.1134/S0006297921040040
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297921040040