Abstract
In recent years modified bacteriophage lysins have widely been investigated for the purposes of development of antibacterial therapy. Thus, effective and precise methods for the quantitative analysis of these enzymes are in high demand. The enzyme-linked immunosorbent assay (ELISA) method has been developed for the detection of recombinant modified endolysin LysAm24-SMAP in biological samples. The optimal parameters for protein detection were determined, in particular, the influence of salt and the composition of the buffer system for preparation of the samples was studied. The applicability of the immunodetection system of the genetically engineered endolysin LysAm24-SMAP in various biological samples with enzyme concentrations from 0.4 ng/mL was demonstrated. In addition, the influence of matrix effects in samples of animal organs and tissue homogenates and producer strain lysates and their individual components during the analysis was assessed and it was shown that 0.65 M NaCl addition in the ELISA buffer is crucial for achieving correct results and reduces nonspecific interactions in the case of LysAm24-SMAP. The effectiveness of the developed system in the immunochemical control of the bacteriolytic enzyme was confirmed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Lytic bacteriophages are obligate parasites causing death of infected bacteria; this has prompted further research of bacteriophages, and their lytic enzymes– lysins, as a treatment for bacterial infections, an alternative to conventional antibiotics [1, 2]. Bacteriophages secrete several types of lysins; including endolysins, released at the terminal stage of bacterial invasion, destroy the bacterial cell wall from within followed by release of newly assembled virions. These enzymes hydrolyze the peptide and/or glycosidic bonds of peptidoglycan, leading to turgor pressure disruption and cell death.
In recent years, the relevance of research and development in this area has increased due to the spreading antibiotic resistance in bacteria [3, 4]. Currently, there is active research into the use of endolysins for treatment of diseases caused by gram-positive and gram-negative pathogens that tend to develop multidrug resistance, including the ESKAPE pathogens – Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and other members of the Enterobacteriaceae family [5, 6]. The use of lysins against gram-negative bacteria has long been difficult due to the shielding effect of the outer membrane. In the last decade, however, the development of genetically modified molecules has made this possible. Methods of genetic engineering and targeted design make it possible to modify native phage endolysins, significantly increasing their bactericidal activity, improving the pharmacokinetic stability and physicochemical properties [7], and allow to biotechnologically obtain recombinant proteins.
Precise quantitative analysis is necessary at almost all stages of the lysins study. For example, during protein expression optimization, quantification becomes one of the main criteria for selecting optimal expression conditions. In addition, during pharmacokinetic studies of lysin-based drugs, it is necessary to assess the content and distribution of lysins in organs, tissues, and biological fluids of laboratory animals. Obviously, such studies require the development of a fast and highly specific screening method that allows the selection of highly efficient protein-producing strains, as well as the detection of low enzyme concentrations in biological samples.
There are several specifical and non-specifical ways to measure the quantitative content of protein: UV spectroscopy, colorimetry, IR spectroscopy, HPLC, and mass spectrometric methods of analysis. However, for complex mixtures, such as biological samples containing different proteins, immunodetection, e.g., enzyme-linked immunosorbent assay (ELISA), is more suitable. ELISA is characterized by high sensitivity, specificity and low time consumption, and does not require complex equipment [8]. The accuracy of this analysis is ensured by the specific antigen-antibody interaction, and the high sensitivity allows the measurement of proteins in nano- and picogram concentrations. However, in the case of endolysins, it is important to consider factors that may influence ELISA result: presence of homologous proteins in producer strains or animal organs homogenates. Also, interaction of lysins sites with components of ELISA buffers should be considered, since these enzymes often contain functional domains capable of binding to polysaccharides or prone to electrostatic interactions. In addition, in the case of biological samples, it is necessary to take into account the possible interaction of the analyte with enzymes, nucleic acids, and other biomolecules contained in the sample (the so-called matrix effect), leading to both proteolysis of endolysin and its aggregation with matrix components, that can significantly distort the measurement results. Optimal analysis conditions and, primarily, the composition of the dilution buffer, makes it possible to significantly neutralize the negative influence of the matrix on the measurement results.
LysAm24-SMAP is a genetically engineered protein with a molecular weight of 27.0 kDa, pI 9.95, obtained by fusion of the LysAm24 endolysin sequence [5] and the SMAP fragment. LysAm24 is a lysozyme-like N-acetylmuramidase of a bacteriophage that infects bacteria of the Acinetobacter genus, containing an additional cell wall binding domain at the N-terminus [5]. The C-terminus catalytic domain belongs to the glycoside hydrolase family 24 (GH24) and is common among lytic bacteriophages. Previously, LysAm24 showed activity against a wide range of gram-negative bacteria in vitro. In the future, its use will be the basis for the development of highly effective antimicrobial agents [5, 11]. The SMAP fragment is a sheep myeloid peptide used to enhance antibacterial activity [9, 10]. The addition of the SMAP peptide increases the calculated total charge of the protein to 19.8 at pH 7.5; its introduction into the molecule allows to effectively permeabilize the bacterial outer membrane. The development of an effective and accurate tool for the quantitative analysis of the LysAm24-SMAP will allow to draw conclusions from the results of the selection of protein expression conditions, as well as from experiments to determine the pharmacokinetic and pharmacodynamic properties of the drug.
The purpose of the work is to create and evaluate the applicability of an ELISA test system for the quantitative estimation of the endolysin in various biological samples, such as bacterial biomass lysates, organs and tissues homogenates, and the blood serum of experimental animals using the example of the modified endolysin LysAm24-SMAP, active against a wide range of gram-negative bacteria.
METHODOLOGY
Preparation of recombinant endolysin LysAm24-SMAP. Endolysin LysAm24-SMAP, obtained by recombinant expression, was used in the experiments. For this, the LysAm24-SMAP coding sequence, including the LysAm24 muramidase sequence (NCBI AN: APD20282.1) and a fragment of the antimicrobial peptide SMAP-29 (1–17, K2,7,13, RKLRRLKRKIAHKVKKY) on C-terminus of enzyme, was artificially synthesized in the pALTA-LysAm24-SMAP vector and integrated into the pET-42b expression vector (+). The correctness of the assembly of vector constructs was verified by Sanger sequencing.
Next, endolysin was obtained similarly to the protocol described in [12]. The constructs were introduced into the producer strain E. coli BL21(DE3)pLysS using the heat-shock transformation method that was cultured at 37°C in an incubator shaker in LB medium with the selective antibiotics (chloramphenicol and kanamycin) addition. Protein expression was induced with isopropyl-β-D-1-thiogalactopyranoside (AppliСhem, Germany). The producer biomass was harvested by centrifugation and disrupted. Then, two-stage chromatographic purification on an XK 16-600 column (GE Healthcare, United States) was carried out using a cation-exchange SP-sepharose resin (GE Healthcare, United States) and the gel-exclusion Superdex 75pg resin. The protein was eluted with phosphate-buffered saline (PBS tablets: 137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, pH 7.3-7.5, VWR, United States). The endolysin concentration was determined by measurement of the optical density at 280 nm wavelength (Implen NanoPhotometer, IMPLEN, Germany) and calculated using a predicted extinction coefficient of 0.852. The protein purity was determined by 16% SDS-PAGE.
Specific polyclonal antibodies purification. Rabbits were subcutaneously immunized with 140 μg of LysAm24-SMAP at least eight times, 14 days apart. The first immunization was carried out in Freund’s complete adjuvant (Sigma, United States), and all subsequent immunizations were carried out in Freund’s incomplete adjuvant. Five to seven days after the second, fourth, and subsequent even-numbered immunizations, 35–40 mL of blood was taken from the rabbits’ ear vein; after completion of the coagulation process, a blood clot was harvested, sodium azide was added to a 0.1% final concentration, and the serum was stored at 4°C.
Polyclonal antibodies (ABs) purification was carried out with immunoaffinity NHS Sepharose (GE Healthcare, Germany). To prepare the resin, LysАm24-SMAP was transferred into a conjugation buffer (0.2 M NaHCO3 (Amresco, United States), 0.5 M NaCl, pH 8.3) using PD-10 desalting columns (GE Healthcare, Germany), mixed with an equal volume of washed resin and incubated in a rotary shaker for 16–18 hours at room temperature. Free binding sites were blocked by Tris-HCl pH 8.0 solution added to a final concentration of 100 mM according to the manufacturer protocol of NHS Sepharose.
For ABs purification, NaCl was added to a concentration of 0.5 M to the immunized sera and loaded to the resin (LysАm24-SMAP-NHS-sepharose). Next, the resin was washed with a PBS solution with the addition of 0.5 M NaCl and eluted with an acetate buffer pH 2.5–2.8 (0.1 M CH3COOH, 0.15 M NaCl). Immediately after elution, Tris-HCl 1.0 M was added to pH 7.0–8.0. AB-containing fractions were transferred to a storage buffer (PBS with 0.1% NaN3, Amresco, United States) with Amicon Ultra-15 centrifugal concentrators (Millipore, Germany) 30 kDa, and the concentration of ABs in the sample was adjusted to 1–2 mg/mL. The specific activity of the obtained antibodies was determined by indirect ELISA.
Assessment of the antibody activity by indirect ELISA. The ELISA plate wells (96-well ELISA plates of high sorption ESP-96-D, Servicebio, China) were coated with endolysin by adding 100 μL of 1 μg/mL LysAm24-SMAP solution in a carbonate-bicarbonate buffer (CBB) at pH 9.3–9.6. Sorption was carried out overnight at a temperature of 4°C. The next day, free binding sites were blocked by 100 μL of blocking solution S002 (Xema, Russia) with the addition of sucrose up to 5% (Dia-m, Russia) and sorbitol up to 0.5% (Xema, Russia). The plates were incubated for 24 hours at 4°C. Then, the liquid was removed, the wells were dried for 24–48 hours at room temperature, packed in plastic bags, and stored at 4°C.
In the experiment, 100 μL of the test sera and purified antibodies were added to the wells in serial dilutions of 1 : 100, 1 : 1000, 1 : 10 000 in ELISA diluent S011 (PBS pH 7.2-7.4, casein, Tween-20, phenol-based preservative, dye, Xema, Russia), and incubated for one hour at 37°C, 600 rpm. The wells were washed with PBS and 0.1% Tween-20 to remove unbound antigens. Afterwards, 100 μL of anti-species antibodies (conjugate of goat polyclonal antibodies to rabbit IgG with horseradish peroxidase, HyTest, Russia) at a 1 : 25 000 dilution of was added, incubated for one hour at 37°C, 600 rpm, and washed.
To visualize the reaction, 100 μL of substrate buffer with tetramethylenebenzidine (R055, Xema, Russia) was added to the wells and incubated for 10 min. The reaction was stopped with 10% HCl solution. The optical density was measured at 450 nm (Multiscan FC, Thermo Scientific, United States).
Measurement of LysAm24-SMAP by sandwich ELISA. To measure the endolysin in bacterial lysates samples, in the presence of DNA and peptidoglycan, in samples of animals biological fluids, organs, and tissues, purified LysAm24-SMAP antibodies were conjugated with horseradish peroxidase similarly to the method described in [13] with the following modifications. To stop the peroxidase-sodium periodate reaction, the mixture was loaded to a PD-10 gel filtration column and the buffer was changed to a borate buffer (0.05 M H3BO3 pH 8.6, Helicon, Russia). Sodium borohydride (Merk, Germany) was added instead of sodium cyanoborohydride to stabilize the Schiff base during antibodies conjugation with horseradish peroxidase. Glycerol was added to the conjugate to 55% (vol/vol) concentration of and stored at -30°C.
For analysis, affinity-purified rabbit LysAm24-SMAP antibodies at 1 μg/ml concentration in CBB were loaded on ELISA plates. Sorption was carried out overnight at 4°C. Next, free binding sites were blocked by 100 μL of blocking solution S002 (Xema, Russia) with the addition of sucrose up to 5% and sorbitol up to 0.5%. The plates were incubated for 24 hours at 4°C. The liquid was removed from the wells, plates were dried for 24–48 hours at room temperature, packaged, and stored at 4°C.
In the experiment, endolysin solution of known concentration and the test samples were diluted in ELISA diluent S011 or in 50 mM Tris buffer pH 7.5 with 1.0% bovine serum albumin (BSA, Sigma-Aldrich, United States), 500 mM NaCl and 0.1% Tween-20 and added to the wells in 100 μL volume. The plates were incubated for one hour at 37°C, 600 rpm. The wells were washed three times with 300 μl of washing buffer (PBS with 0.1% Tween-20). Afterwards, 100 μL of affinity purified LysAm24-SMAP rabbit antibodies conjugated with horseradish peroxidase were added at 1 : 5000 dilution. The plates were incubated for one hour at 37°C, 600 rpm, then the wells were washed five times. Imaging was performed as described above.
For endolysins LysAp22-SMAP and LysECD7-SMAP, the analysis procedure was similar.
Measurement of LysECD7-SMAP and CFP-10 by sandwich ELISA. To compare the effect of additional salt in the S011 sample dilution buffer, a sandwich ELISA was performed for the LysECD7-SMAP and CFP-10 proteins. ABs purification, sorption, and blocking were carried out similarly to the LysAm24-SMAP test system procedure. For CFP-10, mouse monoclonal ABs to CFP-10 (sorption ABs) and a conjugate of mouse monoclonal ABs to CFP-10 with HRP (HyTest, Russia) were used. To set up the experiment, solutions of LysECD7-SMAP and CFP-10 with a known concentration and the test samples were diluted in ELISA diluent S011 and in S011 with the addition of 0.25, 0.5, and 1 M NaCl. Next, the analysis was carried out similarly to the sandwich ELISA method described above.
Sample preparation of bacterial lysate samples. To measure the concentration of endolysin in a negative lysate containing no endolysins, we used the strain E. coli BL21(DE3)pLysS (Evrogen, Russia).
Bacterial lysates were obtained using a standard culture disruption protocol for further protein purification [11]. For that, a E. coli culture was grown overnight at 37°C, 250 rpm in liquid LB medium with the addition of chloramphenicol, diluted with fresh medium in a 1 : 100 ratio and continued cultivation at 37°C, 250 rpm for eight hours. The biomass was harvested by centrifugation (15 min, 3000 g), and the cells were disrupted by sonication in a lysis buffer: 20 mM Tris-HCl pH 8.0, 250 mM NaCl, 0.1 mM disodium salt of ethylenediaminetetraacetic acid (Helicon, Russia). The bacterial lysate was centrifuged (30 min, 10 000 g), and the supernatant was saved.
Isolation of DNA and peptidoglycan. DNA and peptidoglycan were isolated from a culture of bacterial strain Acinetobacter baumannii Ts 50-16, deposited in the collection of the Gamaleya National Research Centre for Epidemiology and Microbiology, Ministry of Health of the Russian Federation. For this, the strain was grown in LB medium for 16–18 hours, the biomass was harvested by centrifugation (6000 g, 10 min). The CTAB method was used to isolate DNA from the biomass [14]. Peptidoglycan was isolated according to the method described in [15]. The DNA concentration was measured using a Qubit DNA HS Assay Kit and a Qubit 3.0 fluorometer (Thermo Fisher Scientific Eugene, United States), and peptidoglycan was measured using an Implen NanoPhotometer (Implen, Germany) using an OD206 calibration curve, that was based on peptidoglycan concentrations of Micrococcus luteus (Sigma, United States).
Obtaining homogenates of animal organs and tissues. All manipulations were performed on ice. Samples of mouse organs and tissues, stored in a low-temperature refrigerator at –80°C, were thawed at 4°C for the minimum time required to thaw the samples. Next, the homogenization buffer (PBS, 1.0 M NaCl, 1.0 mM EDTA-Na) was added to the test samples in a 1 : 4 ratio (400 μL of buffer per 100 mg of organ), phenylmethylsulfonyl fluoride (Dia-m, Russia) was added to the final concentration of 1 mM. One 5 mm steel ball was added to each tube, and the samples were homogenized on TissueLyser LT (QiaGen, Germany) for four minutes at a frequency of 50 Hz. Immediately after homogenization, samples were frozen and stored at –80°C. Before the assay the samples were thawed on ice, mixed, and harvested by centrifugation (16 000 g at 4°C, 10 min). The measurements were carried out in the supernatant.
Conclusion of the bioethical commission. The protocols for obtaining antibodies and samples using animals were approved by the Ethics Committee of N.F. Gamaleya National Research Center (Protocol No. 64 dated October 10, 2023).
Statistical analysis and data visualization. Data were processed using GraphPad Prism 9.5.0. We used two technical and two to five biological replicates in ELISA. The graphs represent mean values ± the standard deviation. The statistical significance in the optical density was calculated using analysis of variance (p < 0.05 was considered significant).
RESULTS AND DISCUSSION
When developing enzyme preparations for biomedical applications, issues assosiated with their detection arise. The ELISA method is one of the most accessible and convenient to use. Currently, few scientific works on the immunochemical determination of endolysins and other lytic enzymes have been published in the literature. For the antistaphylococcal endolysin SAL200, a pharmacokinetic study in monkeys used an enzyme-linked immunosorbent assay to quantify the molecule in serum [16]. The sandwich ELISA method is also known for quantitative analysis of lysostaphin, a staphylococcal bacteriocin, that is also a peptidoglycan-degrading enzyme, in the blood serum of rats [17]. However, currently there are no works describing the development of such test systems in the literature.
For LysAm24-SMAP, there are no specific quantitative methods with high sensitivity; therefore, in this work, a test system was developed for quantitative assay of the protein in various matrices. Also test system main parameters were estimated, such as sensitivity, specificity, dynamic range, and measurement accuracy.
After rabbits’ immunization with modified endolysin LysAm24-SMAP, hyperimmune sera were obtained and analyzed by indirect enzyme-linked immunosorbent assay (Fig. 1a). The results indicated the presence of specific antibodies in the immunized sera (the signal at 1 : 100 000 dilution exceeded 1 o.d. unit) and were suitable for their purification and conjugation with horseradish peroxidase.
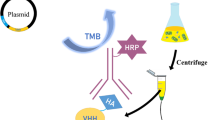
Test systems used in the work: (a) indirect ELISA, (b) sandwich ELISA. LysAm24-SMAP, antigen; Anti-LysAm24-SMAP, polyclonal antibodies isolated from the serum of immunized animals; Anti-Rabbit*HRP, anti-species conjugate of antibodies to rabbit IgG with horseradish peroxidase (E); TMB, substrate for visualization of the reaction. To visualize ELISA schemes, the free version of the website https://miro.com/ru/ was used.
Purified ABs specific to LysAm24-SMAP were used to develop a test system based on the “sandwich” ELISA principle (Fig. 1b) as ABs for plate sorption and conjugate with horseradish peroxidase obtaining. The method was chosen as two-step sandwich ELISA is optimal for the detection of antigens whose size allows simultaneous binding with two antibodies to different fragments of the molecule. Furthermore, the use of specific antigen capture by antibodies adsorbed on the plate at the first stage allows measurements in complex protein matrices, since the removal of unbound matrix components during washing allow to significantly reduce their influence on the reaction result, and to avoid the so-called “hook effect” [18]. This makes two-step ELISA preferable to one-step sandwich ELISA for measuring wide ranges of analytes in complex matrices.
Study of the range and specificity of the quantitative test system for LysAm24-SMAP measurement. LysAm24-SMAP solutions with concentrations ranged from 1.25 ng/mL to 100 ng/mL were measured using the sandwich ELISA method to determine the test system ranges for endolysin-containing samples. The cross-reactivity of the resulting antibodies was also assessed during the experiments. Muramidases are a common class of enzymes found in bacteriophages and bacteria [19, 20]. At the same time, they have significant homology in their catalytic and functional domains. Moreover, lysozyme-like proteins are also common in eukaryotic organisms, for example, in fish [21], mice, and cows [22]. In humans, such proteins are found in the blood, kidneys, intestine, and epithelium [23]. All abovementioned factors can affect the quantitative assessment results and lead to false positive results.
Two modified endolysins (LysAp22-SMAP and LysECD7-SMAP) were used to assess the specificity of the LysAm24-SMAP antibodies. These enzymes are also obtained from the genomes of Myoviridae family bacteriophages, infect gram-negative bacteria, and are additionally modified with SMAP peptide. At the same time, LysAp22-SMAP contains the lysozyme-like N-acetylmuramidase catalytic domain, similar to LysAm24, while LysECD7-SMAP has endopeptidase activity [5].
The expression and purification of the endolysins were carried out under similar conditions; therefore, the composition of the contaminant proteins are similar . Assessment of the test system specificity with two modified endolysins allows to analyze the presence of a sufficient number of antibodies to the contaminating proteins or to an identical SMAP fragment to give a signal that interferes with the interpretation of the results.
The analysis showed that cross-activity with other endolysins samples was not observed in the test system operating range (3–50 ng/mL). The optical density values did not exceed the background 0.1 o.d. units for heterologous antigens that indicates the high specificity of the obtained antibodies. The data suggest that ABs are produced for the LysAm24 endolysin sequence epitopes, while the SMAP peptide is not an immunodominant epitope. Simultaneously, the contribution of individual components of the hybrid molecule to its immunogenicity and their ability to ensure sandwich complexes formation is of interest for further research.
To sum up, study of the polyclonal antibodies showed that they were antigen-specific and did not have cross-reactivity with homologous proteins.
Effect of the salt content in a dilution buffer on LysAm24-SMAP measurement. The endolysin contains positively charged structures that can electrostatically interact with the sample matrix components. In this regard, the effect of NaCl different concentrations in the dilution buffer (S011) was studied.
It was shown that NaCl addition to the ELISA buffer during LysAm24-SMAP measurement led to a significant increase in the ELISA signal for low calibration concentrations (3.13 and 6.25 ng/mL) (Fig. 2a). At the same time, a dependence of the signal increase on the endolysin concentration was observed: even small concentrations of NaCl (0.25 M) added to buffer S011 led to signal increase for low (3.13–6.25 ng/mL) concentrations of endolysin, but had almost no effect for high (12.5–25.0 ng/mL) concentrations. In addition, this effect did not correlate with the added to the buffer solution salt concentration and the OD values at 1.0 M NaCl were lower than at 0.25 M NaCl.
In most cases, increasing the ionic strength leads to weaken non-specific and low affinity specific AG-AB interactions, resulting in decreased ELISA signal [24]. In our case, the opposite effect was observed. In order to assess how universal the effect of signal amplification by salt addition is in other “sandwich ELISA” test systems, this effect was investigated for two other proteins – endolysin LysECD7-SMAP (Fig. 2b) with polyclonal affinity isolated ABs, and Mycobacterium tuberculosis CFP-10 with commercial mouse monoclonal antibodies (Fig. 2c).
In both test systems, 0.25 or 1.0 M NaCl addition did not affect the ELISA signal over the entire range of specific antigen concentrations (Figs. 2b, 2c). It can therefore be concluded that the effect of signal increase with i NaCl concentration was characteristic of LysAm24-SMAP.
Effect of the dilution buffer composition on LysAm24-SMAP measurement. In order to assess whether the observed signal amplification effect is dependent on the buffer system used, a buffer of known composition was also used: 50 mM Tris-HCl pH 7.5 with the addition of 1% BSA, 0.1% Tween-20, and 0.15 M NaCl (TBS), in addition to the standard commercial ELISA buffer (S011), that has been successfully used in the development of test systems for antibodies to the SARS-CoV-2 nucleocapsid protein [25] and antibodies to the vaccinia virus [26]. To assess the salt effect, the working buffer was supplied with 0.5 M NaCl (the resulted NaCl concentration in buffer was 0.65 M).
Dilution of samples in the TBS buffer without the additional salt (0.5 M NaCl) led to increase in the measured signal relative to dilution in S011 (Fig. 3a). Moreover, the effect was more pronounced in the 0.4–12.5 ng/mL range of enzyme concentration, and the signal was 4.5–2.3 times increased. At 25.0–50.0 ng/ml protein concentrations, the ratio of the TBS/S011 measured optical densities was only 1.4–1.1. With the additional 0.5 M NaCl, the signal was also higher in TBS, compared to commercial ELISA buffer (Fig. 3b); however, the dependence on the endolysin concentration was not maintained and the OD of TBS/S011 increase was 1.2–1.4 times over the entire range of measured concentrations.
This may be due to the interaction of the endolysin with the low concentration ELISA diluent component that inhibits the binding of the enzyme to the ABs, resulting in an underestimation of the signal at low protein concentrations. Particularly, we can speculate that this component could be polysaccharide, since LysAm24-SMAP contains peptidoglycan binding domain (pfam01471) [27] and is potentially capable to interact with glycosidic compounds. In addition, residual nucleic acids interacting with endolysin through ionic bonds can be considered as inhibitors. The salt addition to the buffer partially destroyed this bond, “releasing” endolysin from the complex and leading to the signal increase.
As a result, the developed detection method using TBS with the additional 0.5 M NaCl allows to determine LysAm24-SMAP in samples in the 0.4–25.0 ng/ml concentration range.
Measurement of LysAm24-SMAP in bacterial lysates. The accuracy of the enzyme detection added to the bacterial cell lysate was assessed to study the applicability of the ELISA method for the quantitative endolysin assay during its production and optimization of the recombinant expression system. For this purpose, samples of three independently obtained biomass lysates of a laboratory E. coli BL21(DE3) pLysS LysAm24-SMAP expression strain were used in the experiment.
The signal excess over calibration was 500% in S011 when 2 ng/mL of LysAm24-SMAP was added to E. coli biomass lysates. It should be mentioned that there was no non-specific direct lysate components binding to the antibodies on the plate and in the conjugate, since the measured OD remained at the ELISA buffer background level in the samples without LysAm24-SMAP. Overestimation of the measured endolysin concentrations in bacterial lysates decreased to 300-200% in TBS and buffer solutions with the 0.5 M NaCl addition (Fig. 4). The OD of negative lysate samples of E. coli without endolysin also did not exceed the background values; therefore, no cross-activity with producer proteins was observed.
One of the hypotheses to explain this effect is the interaction of endolysin with matrix components. This is a result of the protein binding to polysaccharides through the domain described above. In particular, such results can be explained by non-covalent aggregation of LysAm24-SMAP with polymeric matrix components. This process presumably led to the Antigen-Antibody (AG-AB) complex formation that is characterized by an excess amount of binding to the conjugate antigen per antibody molecule on the plate. This happened because the necessary epitopes became available when aggregating. Thus, at the moment, this test system is suitable for endolysin detection in bacterial lysates, but not quantitative analysis, and requires further optimization.
The influence of matrix components on the results of ELISA analysis. Components of lysed bacterial cells that may contribute to protein measurements include biopolymers such as peptidoglycan (PG) and DNA, released in large quantities into the extracellular environment during the destruction of bacterial cells and potentially capable to bind LysAm24-SMAP. For example, DNA (and other nucleic acids), can interact electrostatically with endolysin due to their charge, making its detection difficult. Similarly, peptidoglycan released during the lysis of bacterial cells can irreversibly interact with the enzyme due to the peculiar LysAm24 domain organization, preventing its correct detection.
Total DNA and cell wall PG were obtained for A. baumannii. DNA and PG were added before measurements to the 3.1 ng/mL endolysin solution at concentrations of 1.22–9.75 and 1.19–9.50 µg/mL, respectively.
It was shown that the addition of DNA to an endolysin solution resulted in smaller and non-linear fluctuations in the concentration ratios: from 80 to 97% (Fig. 5a), that is within the acceptable error limits of this analytical method [28, 29]. The PG addition had little effect on the endolysin measurements (Fig. 5b). The ratios of the added and measured enzyme concentrations varied linearly from 93 to 120%. At the same time, the linear dependence of the measured to the added concentration ratio on the matrix components concentration was preserved. Thus, the molecules studied do not appear to be a significant cause of the results obtained overestimation and should not introduce significant bias in the endolysin measurement in lysates. However, further study of other components presented in large quantities in matrix is required, including various types of polysaccharides.
Measurement of LysAm24-SMAP in animal the serum and organs. One of the critical stages in the drug development is the pharmacokinetic study in animals, that involves distribution of the compound in the organs, tissues, and biological fluids. ELISA remains one of the most accessible and convenient methods of quantitative analysis for protein drugs, and particularly LysAm24-SMAP. However, in this case, the characteristics of biological samples, such as the presence of endogenous proteases and other interfering substances of a protein nature, as well as the pH value, may affect the results of the experiment. During our study, the possibility of using the developed method on animals’ samples was evaluated.
Liver, lung, and serum samples were collected from mice and homogenized to evaluate the matrix effects of animal biological samples. After homogenization, endolysin was added at 200 ng/mL concentration to the samples and then diluted in TBS and TBS with the 500 mM NaCl addition (Fig. 6).
The influence of the buffer composition on the endolysin concentration measurement (% of measured concentration from applied concentration) in, I, liver homogenates; II, lungs; III, blood serum. 1, TBS; 2, TBS + 0.5 M NaCl. The results are presented in the range diagrams: lines, medians; boxes, interquartile range; whiskers, min–max.
As in the case of bacterial lysates, the use of TBS without additional salt resulted in the overestimation of the measured concentration compared to the added concentration. Thus, in blood serum the increase ranged from 259 to 441%, and for organs (liver and lungs), 180–330%. When 0.5 M NaCl was added in the buffer to a final concentration of 0.65 M NaCl, the measured concentration values reached the added concentration both in the organ homogenates and in the blood serum: 101–120% for liver homogenates, 55–112% for the blood serum, and 70–110% for lung homogenates.
Therefore, for homogenates, close to the expected results were obtained using TBS with 0.5 M NaCl as a buffer for samples dilution. It should be mentioned that values vary from organ to organ. Concentrations most comparable to the added endolysin level were found in liver and lung homogenates. The underestimation of the serum values is presumably due to the proteases effect, since protease inhibitors were not used during sample preparation.
Thus, in our study the ELISA test system was developed. Its applicability was assessed for the immunodetection of the engineered endolysin LysAm24-SMAP in various biological samples with an enzyme concentration of 0.4 ng/mL. At the same time, the 0.5 M NaCl addition turned out to be critical for correct results obtaining in animal organs and tissues samples.
Our results showed that for bacteriophage lytic enzymes, matrix effects as well as the buffer composition, can significantly affect the measurement. Therefore, their careful selection is necessary in each specific case. The developed test system will be used for future studies on the biological and pharmacokinetic properties of the modified endolysin LysAm24-SMAP.
REFERENCES
Gerstmans, H., Rodríguez-Rubio, L., Lavigne, R., and Briers, Yv., Biochem. Soc. Trans., 2016, vol. 44, no. 1, pp. 123–128. https://doi.org/10.1042/bst20150192
Love, M.J., Bhandari, D., Dobson, R.C.J., and Billington, C., Antibiotics (Basel), 2018, vol. 7, no. 1, p. 17. https://doi.org/10.3390/antibiotics7010017
Huemer, M., Mairpady Shambat, S., Brugger, S.D., and Zinkernagel, A.S., EMBO Rep., 2020, vol. 21, no. 12, p. e51034. https://doi.org/10.15252/embr.202051034
Baquero, F., Int. Microbiol., 2021, vol. 24, no. 4, pp. 499–506. https://doi.org/10.1007/s10123-021-00184-y
Antonova, N., Vasina, D., Lendel, A., Usachev, E., Makarov, V., Gintsburg, A., Tkachuk, A., and Gushchin, V., Viruses, 2019, vol. 11, no. 3, p. 284. https://doi.org/10.3390/v11030284
Gutiérrez, D. and Briers, Yv., Curr. Opin. Biotechnol., 2021, vol. 68, pp. 15–22. https://doi.org/10.1016/j.copbio.2020.08.014
Fursov, M.V., Abdrakhmanova, R.O., Antonova, N.P., Vasina, D.V., Kolchanova, A.D., Bashkina, O.A., Rubalsky, O.V., Samotrueva, M.A., Potapov, V.D., Makarov, V.V., Yudin, S.M., Gintsburg, A.L., Tkachuk, A.P., Gushchin, V.A., and Rubalskii, E.O., Viruses, 2020, vol. 12, no. 5, p. 545. https://doi.org/10.3390/v12050545
Tabatabaei, M.S. and Ahmed, M., Methods Mol. Biol., 2022, vol. 2508, pp. 115–134. https://doi.org/10.1007/978-1-0716-2376-3_10
Antonova, N., Vasina, D., Rubalsky, E., Fursov, M., Savinova, A., Grigoriev, I., Usachev, E., Shevlyagina, N., Zhukhovitsky, V., Balabanyan, V., Potapov, V., Aleshkin, A., Makarov, V., Yudin, S., Gintsburg, A., Tkachuk, A., and Gushchin, V., Biomolecules, 2020, vol. 10, no. 3, p. 440. https://doi.org/10.3390/biom10030440
Dawson, R.M. and Liu, Ch., Drug Dev. Res., 2009, vol. 70, no. 7, pp. 481–498. https://doi.org/10.1002/ddr.20329
Vasina, D.V., Antonova, N.P., Grigoriev, I.V., Yakimakha, V.S., Lendel, A.M., Nikiforova, M.A., Pochtovyi, A.A., Remizov, T.A., Usachev, E.V., Shevlyagina, N.V., Zhukhovitsky, V.G., Fursov, M.V., Potapov, V.D., Vorobev, A.M., Aleshkin, A.V., Laishevtsev, A.I., Makarov, V.V., Yudin, S.M., Tkachuk, A.P., and Gushchin, V.A., Front. Microbiol., 2021, vol. 12. https://doi.org/10.3389/fmicb.2021.748718
Arshinov, I.R., Antonova, N.P., Grigoriev, I.V., Pochtovyi, A.A., Tkachuk, A.P., Gushchin, V.A., and Vasina, D.V., Appl. Biochem. Microbiol., 2022, vol. 58, no. S1, pp. S65–S74. https://doi.org/10.1134/s0003683822100027
Alves, N.J., Antibody Ther., 2019, vol. 2, no. 1, pp. 33–39. https://doi.org/10.1093/abt/tbz002
Minas, K., Mcewan, N.R., Newbold, Ch.J., and Scott, K.P., FEMS Microbiol. Lett., 2011, vol. 325, no. 2, pp. 162–169. https://doi.org/10.1111/j.1574-6968.2011.02424.x
Li, G. and Peter Howard, S., Methods Mol. Biol., 2017, vol. 1615, pp. 143–149. https://doi.org/10.1007/978-1-4939-7033-9_11
Jun, S.Yo., Jung, G.M., Yoon, S.J., Youm, S.Yo., Han, H., Lee, J., and Kang, S.H., Clin. Exp. Pharmacol. Physiol., 2016, vol. 43, no. 10, pp. 1013–1016. https://doi.org/10.1111/1440-1681.12613
Grishin, A.V., Lavrova, N.V., Lyashchuk, A.M., Strukova, N.V., Generalova, M.S., Ryazanova, A.V., Shestak, N.V., Boksha, I.S., Polyakov, N.B., Galushkina, Z.M., Soboleva, L.A., Vetchinin, S.S., Pavlov, V.M., Karyagina, A.S., and Lunin, V.G., Molecules, 2019, vol. 24, no. 10, p. 1879. https://doi.org/10.3390/molecules24101879
Ross, G.M.S., Filippini, D., Nielen, M.W.F., and Salentijn, G.I., Anal. Chem., 2020, vol. 92, no. 23, pp. 15587–15595. https://doi.org/10.1021/acs.analchem.0c03740
Adhya, S., Merril, C.R., and Biswas, B., Cold Spring Harbor Perspect. Med., 2014, vol. 4, no. 1, p. a012518. https://doi.org/10.1101/cshperspect.a012518
Höltje, J.-V., Arch. Microbiol., 1995, vol. 164, no. 4, pp. 243–254. https://doi.org/10.1007/bf02529958
Chen, T., Rao, Yi., Li, J., Ren, Ch., Tang, D., Lin, T., Ji, J., Chen, R., and Yan, A., Int. J. Mol. Sci., 2020, vol. 21, no. 2, p. 501. https://doi.org/10.3390/ijms21020501
Callewaert, L. and Michiels, Ch.W., J. Biosci., 2010, vol. 35, no. 1, pp. 127–160. https://doi.org/10.1007/s12038-010-0015-5
Liu, R., Meng, Q., Dai, Y., and Zhang, Y., Chin. J. Biotechnol., vol. 39, pp. 4482–4496. https://doi.org/10.13345/j.cjb.230241
Xu, H., Lu, J.R., and Williams, D.E., J. Phys. Chem. B, 2006, vol. 110, no. 4, pp. 1907–1914. https://doi.org/10.1021/jp0538161
Generalova, L.V., Grigoriev, I.V., Vasina, D.V., Tkachuk, A.P., Kruzhkova, I.S., Kolobukhina, L.V., Burgasova, O.A., and Guschin, V.A., Bull. Russ. State Med. Univ., 2022, pp. 14–21. https://doi.org/10.24075/brsmu.2022.005
Gushchin, V.A., Ogarkova, D.A., Dolzhikova, I.V., Zubkova, O.V., Grigoriev, I.V., Pochtovyi, A.A., Iliukhina, A.A., Ozharovskaia, T.A., Kuznetsova, N.A., Kustova, D.D., Shelkov, A.Y., Zrelkin, D.I., Odintsova, A.S., Grousova, D.M., Kan, V.Y., Davtyan, S.A., Siniavin, A.E., Belyaeva, E.D., Botikov, A.G., Bessonova, A.A., Vasilchenko, L.A., Vasina, D.V., Kleymenov, D.A., Slutskiy, E.A., Tkachuk, A.P., Burgasova, O.A., Loginova, S.Y., Rozhdestvensky, E.V., Shcheblyakov, D.V., Tsibin, A.N., Komarov, A.G., Zlobin, V.I., Borisevich, S.V., Naroditsky, B.S., Logunov, D.Y., and Gintsburg, A.L., Front. Immunol., 2022, vol. 13, p. 1023164. https://doi.org/10.3389/fimmu.2022.1023164
Antonova, N., Vasina, D., Lendel, A., Usachev, E., Makarov, V., Gintsburg, A., Tkachuk, A., and Gushchin, V., Viruses, 2019, vol. 11, no. 3, p. 284. https://doi.org/10.3390/v11030284
Stiller, J., Jasensky, A.-K., Hennies, M., Einspanier, R., and Kohn, B., J. Vet. Diagn. Invest., 2016, vol. 28, no. 3, pp. 235–243. https://doi.org/10.1177/1040638716634397
Biswas, S. and Saha, M.K., Immunochem. Immunopathol., 2015, vol. 1, no. 2, p. 1000109. https://doi.org/10.4172/icoa.1000109
Funding
This study was supported by the Russian Science Foundation, project no. 23-74-01068, https://rscf.ru/project/23-74-01068/.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Protocols for obtaining antibodies and samples using animals were approved by the Ethics Committee of N.F. Gamaleya National Research Center (Protocol No. 64 dated October 10, 2023).
CONFLICT OF INTEREST
The authors of this work declare that they have no conflicts of interest.
Additional information
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Klimova, A.A., Grigoriev, I.V., Vasina, D.V. et al. Development of Immunoassay for Detection of Engineered Endolysin LysAm24-SMAP. Appl Biochem Microbiol 60, 765–775 (2024). https://doi.org/10.1134/S0003683824604384
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683824604384