Abstract
Hli proteins of the multicellular cyanobacterium Arthrospira platensis have been studied in this work. According to the NCBI database, three Hli genes found in the A. platensis genome encode proteins 47, 64, and 69 amino acid (aa). A. platensis cells were incubated under light stress (500 µmol photons/m2 s, 1 h). The association of Hli proteins with pigment–protein complexes of thylakoid membranes was then studied by two-dimensional electrophoresis and subsequent mass spectrometry. Only Hli 47 aa was detected in the composition of pigment–protein complexes by matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) mass spectrometry. The identified Hli47 protein was shown to be associated with photosystem II and a homolog of the HliC protein of Synechocystis sp. Bioinformatic analysis has shown that the amino acid sequence of the identified protein shows a higher homology with proteins of multicellular cyanobacteria and a lower of homology with the amino acid sequence of Hli proteins of unicellular cyanobacteria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Much attention has recently been paid to cyanobacteria due to the possibility of their widespread use in biofuel production and bioremediation [1]. They adapt well to a variety of conditions, including extreme ones; they synthesize poorly studied stress proteins and secondary metabolites [2]. The identification and characterization of such proteins is necessary both to study the adaptation of cells to stress and to increase their resistance to stress in the practical application of cyanobacteria. Light-stress proteins are of particular interest, since excess light is one of the main stress effects on the cell. Light is necessary for photosynthesis, but an excess of light energy can disrupt the functioning of the photosynthetic apparatus as a result of photoinhibition and photodestruction [3]. When the absorbed energy cannot be fully utilized, an increase in the formation of singlet oxygen and other ROS occurs in electron transport reactions. Various protective mechanisms have formed to dissipate excessive absorbed energy in photosynthetic organisms [4]. Among the protective mechanisms of photosynthesis, stress proteins induced by intense light deserve separate consideration. These include the stress proteins Hlips (high light inducible proteins) of cyanobacteria with one transmembrane helix. Four Hli proteins (HliA, HliB, HliC, and HliD) found in the model cyanobacterium Synechocystis. Hlips proteins are essential for the maintenance of normal cell activity. It is assumed that Hli proteins are involved in such important processes as the regulation of chlorophyll biosynthesis; the transport and binding of chlorophyll molecules; singlet oxygen quenching; the assembly and repair of photosystem 2; and the non-photochemical dissipation of absorbed light energy [5]. Hli proteins are associated with photosystem II and, apparently, with photosystem I [6, 7]. However, the main function of these proteins is not yet clear, and there are practically no data in the literature on the Hli proteins of other cyanobacterial species.
Cyanobacteria with long-wavelength forms of chlorophyll are of great interest. This is due to the assumption of their participation in the protection of the photosynthetic apparatus from photodestruction [8]. We used a filamentous, multicellular, extremophilic cyanobacterium, Arthrospira platensis, that contains these forms of chlorophyll [8]. Long-wavelength chlorophyll with a fluorescence band at 760 nm was also found in other filamentous cyanobacteria, e.g., Pseudoanabaena sp., Phormidium uncinatum, and Nostoc muscorum, but not in the unicellular cyanobacteria Synechocystis sp. and Synechococcus elongatus, which are often used for research on the physiology and biochemistry of photosynthesis [8]. In this regard, we hypothesized that the difference in the photoprotection of the photosynthetic apparatus of unicellular and multicellular cyanobacteria, which differ in the presence of long-wavelength chlorophylls, was also reflected in the composition of Hli stress proteins.
The goal of this work is to identify and study the localization of Hli proteins in the pigment–protein complexes of thylakoids of the multicellular, extremophilic cyanobacteria A. platensis and to determine their structural and functional features with bioinformatics programs.
EXPERIMENTAL
Object of study. The multicellular cyanobacterium Arthrospira platensis (Nordst.) Geitl. IPPAS B-256 and unicellular cyanobacteria Synechocystis sp. PCC 6803 (hereinafter referred to as Synechocystis) served as the study objects. Cyanobacterial cells were grown at an illumination intensity of 50 μmol photons/m2 s, a temperature of 30°С, and aeration with air until the middle of the logarithmic growth phase [6, 7]. Some of the cells were exposed to light stress (500 μmol photons/m2 s, for 1 h).
Isolation of thylakoid membranes and the fractionation of chlorophyll-protein complexes. Thylakoid membranes were isolated from cyanobacterial cells with a method described earlier [6, 7]. A mild, nonionic detergent, n-dodecyl-β-D-maltoside (β-DM), was used to extract native photosystem complexes from thylakoid membranes. The detergent was added to thylakoid membranes in a detergent–chlorophyll ratio of 15 : 1 and incubated at 4°C for 30 min. The membrane lysate was centrifuged at 18 000 g for 10 min. The thylakoid membrane precipitate was used for further analysis.
The content of chlorophyll a in the samples was determined in an ethanol extract with the formula [Chla] (mg/mL) = 1.21A664 – 0.17 × A625 [9], where A664 is the absorption at a wavelength of 664 nm and A625 is the absorption at a wavelength of 625 nm.
Fractionation of pigment–protein and protein complexes of thylakoid membranes. Fractionation was performed via electrophoresis in native, unstained, polyacrylamide gel (PAAG) [10]. A tenfold buffer (200 mM BisTris pH 7.0; 75% sucrose, 1.0 M 6-aminocaproic acid) was added to the lysate of A. platensis thylakoid membrane in a ratio of 10 : 1. Gradient PAAG (4–12%) was used for electrophoresis in the first direction, which was performed on a Hoefer device (United States) at a voltage of 100 V for 2.5 h. After electrophoresis, the gel plate was photographed, and one band was then used for electrophoresis in the second direction under denaturing conditions in the presence of Na dodecyl sulfate (SDS-Na). Protein electrophoresis was carried out in Tris-glycine buffer (25 mM Tris, 250 mM glycine, 0.1% SDS-Na, pH 7.5) at a constant current of 120 mA for 2 h in 12.5% PAAG. The gel was then stained with Coomassie R-250 overnight.
Protein identification via MALDI-TOF mass spectrometry. Hydrolysis of tryptic protein in polyacrylamide gel stained with Coomassie Brilliant Blue was carried out as follows. A piece of gel 3–4 mm3 in size was washed twice to remove the dye in 100 μL of a 40% acetonitrile solution in 0.1 M NH4HCO3 for 20 min at 37°C. After the removal of the gel-dehydration solution, 100 μL of acetonitrile was added. The acetonitrile was then removed, a piece of the gel was dried, and 3.5 μL of a solution of modified trypsin (Promega, United States) in 0.05 M NH4HCO3 was added to it at a concentration of 15 μg/mL. The hydrolysis was carried out for 3 h at 37°C; then, 5.25 μL of 0.5% trifluoroacetic acid (TFA) in a 50% solution of aqueous acetonitrile was added, and the solution was thoroughly mixed. The hydrolysate was used to obtain MALDI mass spectra. The samples were prepared for mass spectrometry as follows: 1.5 μL of a sample solution and 0.5 μL of a solution of 2,5-dioxybenzoic acid (Aldrich, United States, 10 mg/mL in 20% aqueous acetonitrile, 0.5% TFA) were mixed on the target, and the resulting mixture was dried in air. The proteins were identified with the Mascot software (www.matrixscience.com). The mass spectra were processed with the FlexAnalysis 3.3 software package (Bruker Daltonics, Germany). The equipment and software for mass spectrometry at the Industrial Biotechnologies Center for Collective Use of the Federal Research Center of Biotechnology of the Russian Academy of Sciences were used.
RESULTS AND DISCUSSION
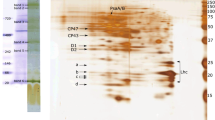
Fractionation of chlorophyll–protein complexes of cyanobacterial thylakoid membranes. Chlorophyll–protein complexes of the cyanobacteria Synechocystis and A. platensis were fractionated via native electrophoresis in PAAG (Fig. 1). Fractionation of pigment–protein complexes in native gel revealed the following components of thylakoid membranes: trimers and monomers of photosystem I, dimers and monomers of the photosystem II complex, NADP-oxidoreductase, the cytochrome complex b6-f, and proteins not included in the complexes (free protein zone). It was shown that the compositions of the pigment–protein complexes of the unicellular cyanobacteria Synechocystis and the multicellular A. platensis are similar.
Localization of Hli proteins in chlorophyll–protein complexes of A. platensis thylakoid membranes. Gels with fractionated complexes were subjected to electrophoresis in the second direction under denaturing conditions. Figure 2 shows the fractionation of proteins of A. platensis chlorophyll–protein complexes under denaturing conditions. To determine the localization of proteins in chlorophyll–protein complexes of the second-direction gel, Coomassie-stained regions of the gel with detected Hli proteins were selected. The staining of Coomassie proteins, the molecular weight of the Hli protein, and its most likely localization in the chlorophyll–protein complexes of cyanobacteria were taken into account in the selection. The selected gel samples were analyzed via MALDI-TOF mass-spectrometric analysis. In samples 1, 2, and 3, the Hli protein was identified with a length of 47 by. In these samples, in addition to the Hli47 protein, photosystem-II proteins were found. It follows from the obtained data that the identified protein Hli47 A. platensis is associated with photosystem II.
The A. platensis protein Hli, which was identified via mass spectrometry, was found in the NCBI database (https://www.ncbi.nlm.nih.gov/protein/WP_ 00661948923.02.19, accessed on February 23, 2019) [11]. Bioinformatic analysis was used to describe some of the structural and functional features of the identified Hli47 protein.
The degree of phylogenetic relationship of the identified protein with Hli proteins of other organisms was assessed with the Smart Blast program [https://blast.ncbi.nlm.nih.gov/blast/smartblast/]. It was shown that the amino acid sequence of the identified A. platensis protein Hli has a high degree of identity with Hli proteins of other multicellular cyanobacteria (Hlip 47 aar, Leptolyngbya sp., 89%; Hlip 47 aa, Geitlerinema sp., 87%). At the same time, the identified protein has much less similarity, only 55%, with the amino acid sequence of the Hli protein of the unicellular Synechocystis sp. (shown below in Fig. 4).
The closest phylogenetic relations to Hlip 47 aa A. platensis are the Hli proteins of other multicellular (filamentous) cyanobacteria. The nearest neighbors, Limnospira indica and Planktothrix agardhii, contain Hli proteins with an amino acid sequence that is almost similar to Hlip 47 aar A. platensis. The result is consistent with both the data obtained with the Smart Blast program and the literature data [12]. The genus Limnospira was recently placed into a separate group from the genus Arthrospira, but Planktothrix is closely related to Arthrospira [12]. The Hli proteins of the unicellular cyanobacteria Synechocystis are phylogenetically less similar to Hli 47 aar A. platensis. When searching for amino acid sequences identical to Hlip 47 aa A. platensis, the Smart Blast program finds almost none in unicellular cyanobacteria.
To determine the similarity of Hli proteins of Synechocystis sp. and A. platensis with the UniProt program (https://www.uniprot.org/), their amino acid sequences were aligned. It is shown that their common identity is only 15.5% (Fig. 3). Pairwise comparison of each of the Hli protein sequences of Synechocystis sp. with the sequence of the identified protein Hli 47 aa A. platensis showed that the amino acid sequence of the Synechocystis sp. HliC protein is most homologous to the sequence of the identified A. platensis 47 aa Hli protein. Their common identity is 55% (Fig. 4a).
Alignment of amino acid sequences of Synechocystis. and A. platensis proteins: (a) Synechocystis HliC and the identified protein A. platensis Hli 47 aa; (b) Synechocystis HliD and A. platensis Hli 64 aa; (c) Synechocystis HliA and A. platensis Hli 69 aa. Dark gray (*) shows a complete aar match; gray (:) indicates close aar properties; and light gray (.) denotes similar aar properties.
The HliC protein was previously isolated from the cyanobacteria Synechocystis sp., and some of its structural and physicochemical properties have been characterized [13]. The obtained structural data suggested that at least four chlorophyll molecules and two β-carotene molecules bind to the HliC protein. This indicated the participation of these proteins in photoprotection and their ability to bind harmful, free chlorophyll molecules, which cause photodegradation.
To gain a better understanding of the degree of homology of the primary structure of Synechocystis sp. and A. platensis Hli proteins, a pairwise comparison of each of the Synechocystis sp. Hli with sequences of A. platensis Hli proteins 64 and 69 aa. This comparison showed that A. platensis Hli 64 aa had the highest percentage of similarity to the HliD protein sequence of Synechocystis sp. (55%, Fig. 4b). The amino acid sequence of A. platensis Hlip 69 aar had the greatest similarity to HliA Synechocystis sp. (56%, Fig. 4c). The homology between the presented protein pairs directly depends on the length of their amino acid sequence. It should also be noted that even the largest percentage of amino acid sequence similarity between Synechocystis sp. and A. platensis Hli did not exceed 56%. Thus, the Hli proteins of A. platensis contain homologs of the previously characterized Synechocystis proteins: Hli64 is a homolog of HliD; Hli69 is a homolog of HliA, and Hli47 is a homolog of HliC. There was no HliB homolog in the A. platensis genome.
A graph of the hydrophobicity of the A. platensis Hli 47 aa protein was plotted with the EMBOSS Pepwindow program (https://www.ebi.ac.uk/Tools/seqstats/emboss_pepwindow/, accessed on November 28, 2019) (Fig. 5). It follows from the obtained graph that the identified protein Hli 47 aa contained a rather extended hydrophobic region that is characteristic of transmembrane domains and was a membrane protein.
To clarify the functions of the identified A. platensis protein Hli 47 aa and to understand the spatial arrangement of the molecule and its localization in the cell, a three-dimensional model of the protein was built with the I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER, accessed on October 29, 2019) (Fig. 6). I-TASSER modeling [14–16] begins with a search for structure templates in the Protein Data Bank (PDB) library. As the most successful template for the construction of a model of A. platensis Hli 47 aa, the program selected a model of the crystal structure of the photoprotective protein PsbS of spinach (Spinacia oleracea). It is a chloroplast membrane protein localized in photosystem II of spinach with a molecular weight of 22 kDa and contains four transmembrane helices. This PsbS protein of photosystem II plays an important role in non-photochemical quenching [17], which protects plants from photodamage under conditions of excessive illumination [18].
Structural model of the low molecular weight, light-induced A. platensis protein Hli 47 aa. The model was calculated based on the crystal structure of the photoprotective protein PsbS of spinach (Spinacia oleracea, http://www.rcsb.org/). The spatial arrangement of the protein molecule in the thylakoid membrane is shown relative to the cell stroma and thylakoid lumen.
Thus, the data obtained with bioinformatics methods indicate that A. platensis Hlip 47 aa is a transmembrane, single-stranded protein and a component of the cyanobacterial cell membrane. Analysis of the protein structure confirmed the presence of ligand binding sites, of which chlorophyll a is the most significant. It is possible that the protein is involved in the non-photochemical quenching of excessively absorbed light energy.
The biological processes involving the identified protein supposedly include the metabolism of nitrogenous compounds. In studies conducted earlier on Synechocystis, it was found that the content of the HliC protein in the cell increases significantly upon a nitrogen deficiency. At the same time, the content of other Hli proteins remains at a low level [19]. The primary HliC structure has been noted to have significant similarities to A. platensis Hli 47 aa.
Based on the similarity of the amino acid sequences of A. platensis Hli 47 aa and Synechocystis sp. HliC, it can be assumed that they perform a similar photoprotective function, the binding of phototoxic free chlorophyll molecules, in cyanobacterial cells. By binding free chlorophyll, Hli proteins prevent the formation of reactive oxygen species [20].
The family of light-induced Hli stress proteins of cyanobacteria is considered to be an evolutionary precursor of chlorophyll a/b binding proteins of the light harvesting complex (LHC) of plants and algae [4]. Most of the research on these important proteins has been done on Synechocystis cyanobacterial cells. The A. platensis Hli proteins of multicellular cyanobacteria were studied for the first time in this work. According to the NCBI database three Hli genes that encode proteins of 47, 64 and 69 aa were found in the A. platensis genome. Four genes encoding Hli proteins have been identified in the fully sequenced Synechocystis cyanobacterial genome: HliB, HliC, and HliD. In addition, the Hli domain was found at the C terminus of the ferrochelatase enzyme [21, 22]. The A. platensis genome does not contain a gene for a protein homologous to the Synechocystis sp. HliB protein.
A homolog of the HliC protein is involved in the association with the pigment–protein complexes in A. platensis Hli47. Evidence of the binding of other A. platensis Hli proteins with PSI and PSII pigment–protein complexes requires further research. Bioinformatic analysis showed that the amino acid sequence of the Hli47 protein exhibits a higher degree of homology with the proteins of multicellular cyanobacteria and a lower degree of homology with the amino acid sequence of the Hli proteins of unicellular cyanobacteria.
Comparison of the composition of Hli proteins in the unicellular cyanobacteria Synechocystis and the multicellular extremophilic cyanobacteria A. platensis, which contain long-wavelength forms of chlorophyll [8], showed differences in the content of Hli proteins in the composition of pigment–protein complexes of the photosystems. It is possible that the differences in the composition of Hli proteins in unicellular and multicellular cyanobacteria reflect characteristics of the photoprotection mechanism of these cyanobacteria.
CONCLUSIONS
The study of the mechanisms of photoprotection in different types of cyanobacteria makes it possible to expand our knowledge of the photoprotective functions of single-stranded, light-induced Hli proteins. These studies open up new possibilities for the selection of varieties and hybrids that are resistant to excessive light and high-yielding varieties and hybrids, the photosynthetic apparatus of which will make it possible to use absorbed light energy more efficiently. The mechanism of the functioning of stress proteins can also be used to create systems for artificial photosynthesis.
REFERENCES
Khetkorn, W., Rastogi, R.P., Incharoensakdi, A., Lindblad, P., Madamwar, D., and Pandey, A., Bioresour. Technol., 2017, vol. 243, pp. 1194–1206.
Babele, P.K., Kumar, J., and Chaturvedi, V., Front. Microbiol., 2019, vol. 10. Art. 1315. https://doi.org/10.3389/fmicb.2019.01315
Jensen, P.E. and Leister, D., F1000Prime Rep., 2014, vol. 6, Art. 40. https://doi.org/10.12703/P6-40
Tibiletti, T., Rehman, A.U., and Vass, I., Photosynth. Res., 2018, vol. 135, pp. 103–114.
Komenda, J. and Sobotka, R., Biochim. Biophys. Acta, 2016, vol. 1857, pp. 288–295.
Akulinkina, D.V., Bolychevtseva, Yu.V., Elanskaya, I.V., Karapetyan, N.V., and Yurina, N.P., Biochemistry (Moscow), 2015, vol. 80, no. 10, pp. 1254–1261.
Sharapova, L.S., Akulinkina, D.V., Bolychevtseva, Yu.V., Elanskaya, I.V., and Yurina, N.P., Appl. Biochem. Microbiol., 2019, vol. 55, no. 1, pp. 52–58.
Karapetyan, N.V., Bolychevtseva, Yu.V., Yurina, N.P., Terekhova, I.V., and Shubin, V.V., Biochemistry (Moscow), 2014, vol. 79, no. 3, pp. 213–220.
Lichtenthaler, H.K., Methods Enzymol., 1987, vol. 148, pp. 350–382.
Schagger, H. and von Jagow, G., Anal. Biochem., 1991, vol. 199, pp. 223–231.
Fujisawa, T., Narikawa, R., Okamoto, S., Ehira, S., Yoshimura, H., Suzuki, I., et al., DNA Res., 2010, vol. 17, pp. 85–103.
Nowicka-Krawczyk, P., Muhlsteinova, R., and Hauer, T., Sci. Rep., 2019, vol. 9. Art. 694. https://doi.org/10.1038/s41598-018-36831-0
Shukla, M.K., Llansola‑Portoles, M.J., Tichy, M., Pascal, A.A., Bruno, R., and Sobotka, R., Photosynth. Res., 2017, vol. 137, pp. 29–39.
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., and Zhang, Y., Nat. Methods, 2015, vol. 12, pp. 7–8.
Roy, A., Kucukural, A., and Zhang, Y., Nat. Protoc., 2010, vol. 5, pp. 725–738.
Zhang, Y., BMC Bioinformatics, 2008, vol. 9, p. 40. https://doi.org/10.1186/1471-2105-9-40
Ruban, A.V., Plant Physiol., 2016, vol. 170, pp. 1903–1916.
Fan, M., Li, M., Liu, Z., Cao, P., Pan, X., Zhang, H., Zhao, X., Zhang, J., and Chang, W., Nat. Struct. Mol. Biol., 2015, vol. 22, pp. 729–735.
Kopf, M., Klahn, S., Scholz, I., Matthiessen, J.K., Hess, W.R., and Voss, B., DNA Res., 2014, vol. 21, pp. 527–539.
Staleva, H., Komenda, J., Shukla, M.K., Slouf, V., Kana, R., Polivka, T., and Sobotka, R., Nat. Chem. Biol., 2015, vol. 11, pp. 287–291.
Kaneko, T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., et al., DNA Res, 1996, vol. 3, pp. 109–136.
Funk, C. and Vermaas, W., Biochemistry, 1999, vol. 38, pp. 9397–9404.
Funding
This work was partially supported by the Russian Foundation for Basic Research (project no. 190400798).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Rights and permissions
About this article
Cite this article
Sharapova, L.S., Yurina, N.P. Identification of the Stress Hli Protein in the Pigment–Protein Complexes of Arthrospira platensis. Appl Biochem Microbiol 57, 706–711 (2021). https://doi.org/10.1134/S0003683821060119
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683821060119