Abstract
Marker-free transgenic Camelina sativa (L.) plants carrying a synthetic gene for cecropin P1, an antimicrobial peptide, under the control of the cauliflower mosaic virus 35S RNA promoter have been obtained and analyzed. The plants were transformed with an agrobacterial binary vector free of selective genes of antibiotic and herbicide resistance. The marker-free transformants were screened via measurement of the antibacterial activity of cecropin P1 and enzyme immunoassay. The obtained plants exhibited an increased resistance to infection with the bacteria Erwinia carotovora, the fungi Fusarium graminearum, and oxidative stress during infection. Analysis of the fatty acid composition of seed oil showed an increased amount of α-linolenic acid in the transgenic Camelina lines as compared to unmodified plants. The results indicate that the cecropin P1 gene can be included in an integral antistress plant-protective system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Camelina (Camelina sativa (L.)) is an oil plant of the Brassicaceae family [1]; it is also known as German sesame, ginger, and false flax. It is an annual herb, the seeds of which contain a unique composition of fatty acids. The value of camelina oil in its high content of polyunsaturated ω3 fatty acids, the proportion of which is equal to 35‒40% [2]. This plant oil also contains large amounts of carotenoids that exceed their contents in sunflower, soybean, and other oils. It is also rich in vitamin E, a powerful antioxidant: the content of this compound is 105 mg per 100 mL of oil, which is 2.5 times higher than that in rapeseed and 7 times higher than that in linseed oil [3]. False flax is mainly cultivated in North America, Australia, and the northern regions of Europe. In Russia, the plant is grown to maintain crop rotation with sunflower and cereals; seed cake is used as a farm animal feed.
The crop is grown in limited quantities for oil production in the Volgograd, Orlov, and Saratov regions. Studies for the production of transgenic camelina plants as producers of valuable compounds for biotechnology are in progress [4]. Despite their generall high resistance, camelina plants are sensitive to bacterial and fungal pathogens, which cause wilting and root rot. In order to increase resistance to phytopathogens, a method of plant transformation with genes for antimicrobial peptides (AMPs) with a wide spectrum of antibiotic and fungicidal activities is used [5]. The gene for the AMP cecropin P1 isolated from mammalian nematodes was shown to be useful as a target gene. The corresponding protein belongs to a group of linear cysteine-free α-helical peptides [7]. The aforementioned gene consists 31-member amino acid sequence and is characterized by high activity against gram-positive and gram-negative bacteria. The activity of cecP1 against pathogenic fungi has also been reported [8, 9].
Selective genes of antibiotic and herbicide resistance [10] and reporter genes [11] have traditionally been used to select transgenic plants. However, plants obtained with the use of constructs free of selective genes are more attractive due to their biosafety in terms of the dangerous transfer of primarily herbicide resistance genes to wild relatives of transgenic plants and antibiotic resistance genes to soil bacteria or bacteria occurring in the human gastrointestinal tract. The obtainment of marker-free plants that do not contain selective genes for antibiotic or herbicide resistance in their genomes is a promising direction in the creation of safer transgenic plants.
The creation of marker-free transgenic plants via the transfer of T-DNA without selective gene sequences has a number of advantages: (1) it eliminates the need for the use of additional, often labor-consuming, methods to remove the selective gene from the transgenic plant genome; (2) transformants are not subjected to selective stress, which can lead to DNA hypermethylation and, as a consequence, the silencing of target genes [12, 13]; (3) the genome of transformants contains less genetic junk, and the shorter is the genetic construct transferred to plants, the higher is the probability of target-gene expression [14].
We previously obtained camelina plants containing the cecP1 gene and a selective gene for neomycin phosphotransferase II (npt II) that confers kanamycin resistance to transgenic plants [9].
The goal of the present work was to obtain and conduct a physiological and biochemical analysis of marker-free Camelina sativa (L.) plants capable of expressing the gene for the cecropin P1 peptide.
MATERIALS AND METHODS
Plant Material
Camelina plants (Camelina sativa (L). Crantz) of the domestic Omskii variety supplied by the Siberian Pilot Station of the All-Russia Research Institute of Oil Crops by V.S. Pustovoit (Krasnodar, Russia) were objects of study. The seeds were sterilized for 1.5 min in 70% ethanol (Labtekh, Russia) and then for 2 min in 20% sodium hypochlorite. They were washed three times for 10 min in sterile distilled water. The seeds were then germinated on MS medium [15] containing agar (7 g/L), sucrose (30 g/L), pH 5.8, and an ordinary salt set (Labtekh). The plants were cultivated at 22–24°C with a 16-h light day and an illumination of 2 klx. The rooted plants were transferred to a greenhouse.
Bacterial Strains and Plasmids
The bacteria Agrobacterium tumafaciens LBA 4404(pAL4404) [16], which contain the pВМ::сесР1 vector with a synthetic gene for the antibacterial peptide of cecropin P1 were used for genetic transformation of the plants. The bacterial culture was grown overnight in LB medium [18] and diluted to OD600 = 1.0; Tween 20 (USB, United States), a surfactant, was added to a concentration of 0.075% [19].
The phytopathogenic bacterial strain Erwinia carotovora ssp. carotovora В15 from the Horticulture Сentre (Canada) and the phytopathogenic fungal strain Fusarium graminearum Schwabe 1839 (VKM F-2381) from the All-Russia Collection of Microorganisms (Pushchino, Russia; VKM@ibpm.pushchino.ru) were also used. The bacterial strain was grown on LB medium, whereas the fungal strain was cultured on PGA containing extract of thermostable proteins from potato tubers (200 g/L), glucose (15 g/L), and agar (25 g/L) (Difco, United States) [20].
Camelina Transformation
The genetic transformation was performed via agroinfiltration of immature flower buds [21]. The plants were placed in a sealed chamber 5–7 days before flowering, and the flower buds were immersed in the Agrobacterium suspension (OD600 = 1.0) and held in a vacuum (104 Ра) for 2–10 min. Three plants were used for each of the three experiments. The plants were then placed on a horizontal surface of wet filter paper, covered with the same paper, and kept for 24 h at room temperature. The plants were then transferred to a greenhouse and grown in the conditions of long daylight hours (16 h) until the end of vegetation. The seeds were collected and stored at 4°C. The seeds were sterilized and germinated on MS medium for further study.
Western Blot Analysis
Cell-free extracts were obtained [22] to assess the сесР1 gene expression in the transgenic plants. The electrophoresis was carried out in the tricine system of SDS-PAG; the proteins were transferred onto a PDVF nylon membrane (Amersham Pharmacia Biotech, GB) [23]. The Western blot of cecropin P1 was performed with the use of rabbit polyclonal antibodies to this synthetic peptide and antirabbit immunoglobulins conjugated with horse radish peroxidase. The artificial cecP1 was obtained via solid-phase synthesis [24]. The membranes were developed with the ECL chemiluminescent system (Pierce, United States).
DNA Isolation
The DNA template for PCR analysis was isolated from 3-week-old transgenic plants [25]. For this purpose, primers for the cecP1 gene, 5′-CGGGATCCATGGGCTCTTG-3′ and 5′-CGAGATCTCTACTTAGCGCGGC-3′ (Evrogen, Russia), were also used. Therefore, the reaction mixture contained 0.1 μg of the camelina DNA template, 10 mM Tris-HCl (Sigma), pH 8.8 at 25°C, 50 mM KCl (Labtekh), 0.1% Triton X-100 (USB), 1.5 mM MgCl2 (Labtekh), a mixture of 0.2 mmol dNTP, primers (50 pmol of each), and 2.5 U of Taq-polymerase (Thermo, Lithuania). The reaction occurred in an MJ-Mini Personal Thermal Cycler amplifier (Bio-Rad, United States) in a volume of 25 μL according to the following protocol: 5 min, 94°C; 30 cycles—1 min, 94°C; 30 s, 55°C; 30 s, 72°C; and then 7 min at 72°C. The possible contamination of plant DNA preparations by trace amounts of agrobacterial DNA was detected via the amplification of a 670-bp fragment of the VirB1 gene with the following primers: F-GGCTACATCGAAGATCGTATGAATG and R-GACTATAGCGATGGTTACGATGTTGAC. The amplification products were analyzed via PAGE in 6% gel in a Tris-borate buffer in a VF-10 chamber for vertical electrophoresis, 10 × 10 cm (Helicon, Russia). A preparation of 100 bp of PLUS (MEDIGEN Laboratory, Russia) served as the molecular mass markers.
Antimicrobial Activity of Plant Extracts
The effect of transgenic plant extracts on the cell growth of the phytopathogenic bacteria Erwinia carotovora was assessed by the agar-diffusion method [26]. To this end, the plant leaves were ground in a porcelain mortar with liquid nitrogen, and the extraction buffer (10% glycerol (AppliChem, Germany), 40 mM EDTA (Dia-M, Russia), 150 mM NaCl (Labtekh), 100 mM NH4Cl (Labtekh), 4 mM phenylmethylsulfonyl fluoride (Serva, United States), 10 mM Tris-HCl, pH 7.5 (Sigma), 3.0 mg/mL dithiotreitol (Sigma), 0.2 mg/mL leupeptin (Serva), 0.2 mg/mL trypsin inhibitor (USB), and 2 mg/mL bovine serum albumin (Sigma)) was then added. The grinding was continued until a homogenous suspension was obtained.
The extracts were centrifuged for 20 min at 10 000 g, and the antibiotic activity in the supernatant was measured. Petri dishes with a diameter of 9 cm contained 25 mL of 1.5% agar with the bacterial suspension (108 cells/mL). Wells 5 mm in diameter were made in the agar, and cell-free extracts of transgenic and nontransgenic plants were placed in them with preliminary adjustment of the total protein concentration to 1 mg/mL [28]. The agar blocks were incubated for 8 h at 4°C to allow agar diffusion of the extracts and then kept at 25°C for 2 days. The sterile zones around the wells were then localized.
Biotests with Isolated Leaves and Plants
In order to assess the resistance of transgenic plants to phytopathogens, young leaves were contaminated with an E. carotovorа suspension (103–105 cells/mL) or with fragments of the mycelium of Fusarium graminearum fungi. Young leaves of untransformed plants served as control. The leaf stalks of transgenic and nontransgenic plants were contaminated with phytopathogenic bacteria or fungi, placed on the agar MS medium in Petri dishes, and kept closed at 24°C with 16 h of daylight; the damage rate was assessed after 1–14 days of incubation, depending on the pathogen species. The entire plants were infected via injection with a needle moistened in a suspension of the pathogenic bacteria. The fungal pathogen was introduced by the placement of agar fragments with the mycelium in the leaf internodes. Four leaves were contaminated in each experimental variant.
The rate of the pathogen-caused damage was assessed by the disease index [29]. According to the method, there were five categories of symptom severity in individual seedlings: 0, no visible symptoms; 1, symptoms are only manifested at the top of a seedling; 2, the whole seedling body is affected, but the seedling still stands straight; 3, the seedling fell but is still green and juicy; and 4, the seedling wilted and withered. With consideration of the symptom severity in individual seedlings, the disease index was determined from the formula
where A, B, C, D, and Е are the numbers of seedlings with symptoms 0, 1, 2, 3, and 4, respectively. If all of the seedlings died, the index was equal to 100. If none of the tested seedlings had any visible symptoms, the index was 0. The lower the index was, the higher was the degree of plant resistance to disease.
Resistance to Oxidative Stress of Plants Infected with Phytopathogens
To estimate this type of resistance in experimental and control plants, the leaves were inoculated with a suspension of E. carotovora bacteria with a density of 103–105 cells/mL. As a control, pathogen-untreated leaves of both plant groups were immersed in distilled water. The excess water was shaken off, and the leaves were placed in Petri dishes on a wet filter paper. Three leaves from three plants of each of five tested lines were analyzed. Leaf cuttings were made and combined, and the parameters of the oxidative stress were measured after 2 days. The SOD activity was determined as follows: the plant material (100 mg) was homogenized in 100 mM phosphate buffer, pH 6.8 (1 mL), and the reaction with nitroblue tetrazolium was then carried out [30]. The amount of enzyme causing the 50% inhibition of the photochemical reduction of nitroblue tetrazolium was taken as the unit (U) of SOD activity. The specific activity (U/(min μg total soluble protein)) was also calculated; the SOD specific activity was represented in U/μg protein.
Isolation of Fatty Acids and Analysis of their Content and Composition in Camelina Seeds
Sample preparation. The conversion of triglycerides and phospholipids of the camelina seeds to methyl ethers of the corresponding fatty acids was performed via nonextraction methylation with sodium methoxide and boron trifluoride in methanol [31]. Each sample was disintegrated in a coffee grinder and sieved through 0.4-mm holes. Margaric acid (2 mg) in toluene (0.2 mL), an internal standard, and 1 M solution of sodium methoxide in methanol were added to an aliquot (16–20 mg) of the powdery sample, and the mixture was heated at 70°C for 20 min. A 15% boron trifluoride solution in methanol (500 μL) was added, and the heating was performed for another 20 min. Water (1 mL) and distilled heptane (2 mL) were introduced, and the fatty acid methyl ethers were extracted by the organic phase and subjected to chromatography.
Chromatography. A VARIAN 3900 analytical gas chromatograph was used with the following process parameters: column 15 m × 0.2 mm × 0.2 μm with the polar phase of Supelkovaks-10; helium as a carrier gas (1.8 mL/min); sample input via division of the gas flow in a ratio of 1 : 65; volume of the introduced liquid sample of 2.0 μL; temperature of the analytical program from 100°C for 30 s to 245°C for 5 min with a step of 10°C/min; temperatures of the evaporator and flame ionization detector of 260 and 255°C, respectively.
The signal was registered with the Multichrom-1.5 software.
The chromatographic peaks of the methyl ethers of the individual fatty acids were identified with standard calibration mixtures of saturated and unsaturated fatty esters of ME-14 and ME-20 (Supelko).
The quantification of fatty acid components in camelina oil was performed with an internal standard, margaric acid, which was added to the studied material immediately before its chemical derivatization. The calibration coefficients of С16:0, С18:0, С18:1, С18:2, and С18:3 acids for margaric acid С17:0 were preliminarily determined via chromatography of a solution of known concentrations of the above compounds with reactive purity (no less than 98%); for the minor components, the coefficients were taken to be equal to that of С18:1 for С17:0.
Statistical Analysis
Statistical processing of the data was performed with the programs Statistica 6.0 and Microsoft Excel 2007. The measurements were carried out in three analytical and biological replications. The graphs and diagrams contain the mean values and their standard deviations. The significance of differences between the values was assessed with the nonparametric Mann–Whitney U test.
RESULTS AND DISCUSSION
Transformation of Camelina
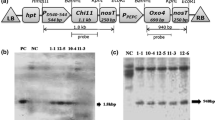
The genetic construct [17] used for the transformation did not contain the genes for antibiotic and herbicide resistance in the T-DNA area, which was integrated in the plant genome (Fig. 1).
Scheme of pBM::сесP1 binary vector: CaMV 35S—see Abbreviations; pACaMV—signal of polyadenylation of mosaic cauliflower virus; сесP1—see Abbreviations; KmR—kanamycin-resistance gene for bacteria selection; oriV—replication origin; TL and TR—left and right borders of DNA, respectively; ColE1—replication origin of ColE1 plasmid; R, K, Sm, B, SI, and SphI—restriction sites for EcoR1, KpnI, SmaI, BamHI, SalI, and SphI, respectively.
The false flax plants were transformed via the agroinfiltration of immature buds [19]. Different times (2–10 min) were used for this purpose (Table 1). The seeds (50 pieces in each experiment) were sterilized and germinated on Petri dishes on MS medium after transformation for the selection of transformed plants (Fig. 2).
Screening and Analysis of Transgenic Plants
Plants synthesizing the cecP1 AMP were screened by Western blotting and the detection of antibiotic activity in the extracts. In the first case, Western blotting of leaf extracts containing about 100 μg of total protein was carried out. Some plant extracts contained the desired peptide with a molecular mass of 3.3 kDa, which corresponded to the mature cecP1 form (Fig. 3).
Figure 3 shows that the detection of the target product in the transgenic plants allows the identification of lines with various levels of positive signal, i.e., it was possible to assess simultaneously the occurrence and intensity of cecP1 production. We obtained 44 lines of сесР1-containing plants. The presence of the cecP1 gene in transgenic plants of the T0 generation, which emitted a positive signal in Western blot, was confirmed by PCR. When PCR products with specific primers were separated by electrophoresis, the expected 102-bp fragment corresponding to the full-length cecP1 gene, which encodes the mature form of the peptide, was revealed (Fig. 4a). Samples of the plant tissues were preliminarily tested for the absence of agrobacterial contamination; to this end, PCR with primers to the VirB1 gene was performed. All of the analyzed samples were free of contamination (data not shown).
PCR analysis of DNA of transgenic camelina plants of T0 (a) and T1 (b) generations containing the cecP1 gene with primers for this gene: M—DNA MM marker (100 bp PLUS, MEDIGEN Laboratory); (1)–(10) DNA products of independent transformed lines 1–10, respectively; C(–)—untransformed plant (negative control); C(+)—plasmid containing the cecP1 gene (positive control).
To obtain plants of the Т1 generation, ten randomly selected lines of the transgenic Т0 samples were planted in a greenhouse before flowering and at the end of vegetation. The plants had normal phenotypes and retained the capacity of viable seed formation as a result of self-pollination. The obtained seeds were collected from camelina of the Т0 generation and planted on MS medium. The seedlings of each line were analyzed via PCR for the presence of the сесР1 gene. Analysis of the Т1 generation showed the presence of the desired cecP1 gene in most transgenic plants (Fig. 4b). In five of the ten tested lines, the ratio of the number of plants with and without the сесР1 gene was 3 : 1, which suggests that one copy of the transgene was integrated into their nuclear genome.
The second method of plant screening is associated with analysis of the antibacterial activity of the studied extracts. The activity level was in accordance with the size of the zone of inhibited Erwinia carotovora growth on agar medium around the wells with added leaf extract from transgenic or nontransgenic plants containing the equal protein amounts. The extracts of control plants were almost completely devoid of antibacterial activity. Figure 5 shows the results of analysis of the antibiotic activity of various amounts of the synthetic cecP1 peptide (34 ng, well (7); and 3.4 ng, well (6)) and five lines of the transformed сесР1-containing plants (wells (1)–(5)).
Effect of extracts of transgenic false flax plants on the growth of the E. carotovora bacterial pathogen. Wells (1)–(5) leaf extracts from five independent transgenic lines, 1–5, respectively; (6) synthetic cecropin P1 (3.4 ng); (7) synthetic cecropin P1 (34 ng); (8) leaf extract from untransformed plant.
The radius of the area of inhibited bacterial growth around wells with transgenic plant extracts was equal to 2–10 mm. The addition of fixed amounts of the synthetic cecP1 to the control-plant extracts established that the content of this peptide in various transgenic lines was 0.003–0.03% of the total soluble protein.
When marker-free, сесР1-containing plants were obtained, the frequency of their genetic transformation was determined as the ratio of the number of seedlings that germinated from the seeds on a nonselective medium and generated positive signals of the cecP1 peptide occurrence to the total number of cultivated extracts. The dependence of the frequency of camelina transformation on the time of agroinfiltration was studied. Five minutes proved to be the optimal interval for the exposure of immature buds to infiltration with A. tumafaciens; the frequency of the transformation in this case reached 25.4% (Table 1).
Plant Resistance to Phytopathogens
The obtained cecP1-bearing plants manifested an increased resistance to the phytopathogenic microorganisms Erwinia carotovora and Fusarium graminearum (Figs. 6a and 6b). Contamination traces were already visible on the leaves of the control plants after the first day. The leaves completely died on the second day.
Enhanced resistance of individual leaves of transgenic camelina plants to phytopathogenic microorganisms E. carotovora (a) and P. graminearum (b) a week after infection: (1) leaves of transformed plants (complete preservation of initial density and color); (2) leaves of control plants (necrosis and tissue death).
The degree of plant damage by the fungal pathogen was also assessed 8–14 days after contamination. By the end of the second week, tissue necrosis and death were observed in the control-plant leaves, whereas the leaves of transgenic plants remained virtually without any signs of damage. The degree of disease severity of whole seedlings was assessed based on the disease index (formula (1)). This index, in terms of infection with E. carotovora, did not exceed 30% in four transgenic false flax lines. At the same time, the disease index in the control plants already reached 100% on the third day (Fig. 7).
The data in Fig. 7 indicated that transgenic plants showed enhanced resistance to infection as compared to control plants due to the expression of the cecP1 AMP.
Fatty Acid Composition in Camelina Seeds
Table 2 presents the fatty acid composition of total lipids from camelina seeds (in % of total fatty acids). It was established that the increase in the proportion of the unsaturated α-linolenic fatty acid from 5 to 27% in transgenic plants (depending on the line) is higher than that in the control (untransformed plants). The increased content of the aforementioned fatty acid makes the oil from the transgenic plants more nutritionally valuable.
Plant Resistance to Oxidative Stress
The resistance of transgenic plants to oxidative stress caused by contamination with the pathogenic bacteria E. carotovora was studied. The SOD activity was used as a stress response marker and was measured in both uninfected and contaminated plants.
It is known that stress-resistant plants are characterized by a lower SOD activity and less oxidative damage [32, 33]. In this work, an increase in SOD activity was observed in both control and transgenic plant groups under stressful conditions. However, the enzyme activity in the leaves of control contaminated plants increased by two times, whereas this value increased by 1.12–1.49 times in the transgenic plants (Fig. 8).
It is assumed that the relatively low SOD activity in the leaves of infected transgenic plants as compared to that in the leaves of control infected plants may result from the synthesis of the antimicrobial cecP1 peptide with its protective effect.
The involvement of the cecP1 gene in the antistress system in terms of the intensification of various adaptive reactions in plants was established previously [34]. It was also found that the rate of the photosynthesis in cecP1-expressing plants decreased with contamination, and the resistance to the oxidative paraquat-caused stress was enhanced during contamination [34]. An increased resistance of cecP1-expressing false flax plants (Camelina sativa (L.)) to salt stress was also observed [9]. An extract of transgenic сесР1-containing kalanchoe plants (Kalanchoe pinnata) added to a culture medium intensified growth and stimulated rhizogenesis in calluses of crystal grass (Mesebryanthemum crystallinum) [35]. The enhanced biological activity of the cecP1 AMP was demonstrated in extracts of сесР1-containing K. pinnata plants with the use of not only plants but also animals (wound-healing and immunomodulatory effects) [36–38].
The results indicated the enhanced biological activity of plant extracts producing the antimicrobial P1 peptide. Our results are in agreement with data showing that the constitutive expression of cecP1 genes in rice [39] and msrA3 gene in potato [40] increased the expression of a number of the plant’s own genes and intensified the systems of antimicrobial and antioxidant protection. In plants producing cecropin A, both genes for protection from phytopathogens and oxidative stress and genes that increase photosynthetic activity in plants during bacterial contamination (the pet gene, which is responsible for the synthesis of a copper-containing protein of plastocyanin during electron transfer, the atp gene for ATP-synthetase, the cab gene, which is responsible for the light-harvesting complexes I and II) were characterized by increased expression. Transgenic plants bearing the msrA3 gene for AMP manifested an increased resistance to the phytopathogen Fusarium solani and to abiotic stress caused by darkness, injury, and heat.
The study of the fatty acid composition of false flax seeds in this work showed an increased content of linolenic unsaturated fatty acid in transgenic lines as compared to that in control plants. According to the reported data, the increase in the degree of unsaturation in fatty acids during leaf development is associated with the biogenesis of chloroplasts in which thylakoid membranes are characterized by a high (up to 85–90%) degree of polyunsaturation [41]. The increased content of linolenic acid allowed tomato transgenic plants with increased ω3 gene expression (responsible for fatty acid denaturation) to preserve a high photosynthesis rate in comparison with the wild-type photochemical activity and prevented the low-temperature photoinhibition of photosystems I and II [42]. It is known that linolenic acid, which contains three double bonds, is an intermediate product of the degradation of plant lipids. An intensification of this process and, as a result, an increase in the content of catabolic intermediates leads to activation of the synthesis of new polymeric molecules, instead of degradation. Some linolenic acid derivatives have the properties of hormones, metabolic activators, or inhibitors, which allows them to accelerate growth and modify morphogenesis in plants. There is also data on their fungicidal, repellent, and antitumor activities. Therefore, a change in the ratio of these compounds during stress can influence adaptation processes in plant cells [43]. In the experiments described here, the degree of unsaturation of the fatty acids was a consequence of the integration of the cecP1 gene in the camelina genome.
CONCLUSIONS
Thus, the work produced marker-free, transgenic, false flax plants with increased safety due to the absence in the genome of a large portion of heterologous DNA, in particular, the nptII gene for kanamycin resistance. At present, there is increased interest in camelina due to its potential for biotechnological use. It was shown that the addition of false flax to food for animals, fish, and poultry improves the quality of meat, fish, and eggs [44]. This plant releases some physiologically active compounds that have a beneficial effect on neighboring crops and show allelopathic activities against weeds that inhibit their growth [45]. The camelina growing areas and global demand for its seeds continue to increase, with use of the plant oil not only as a valuable nutritional product but also as a medicine for wounds and burns; this oil also has a high potential in biofuels, industrial oils, and lubricants.
The obtainment and physiological and biochemical study of various false flax domestic varieties is an important task of current biotechnology. The cecP1-expressing camelina plants obtained in this work are highly safe and greatly resistant to phytopathogens, and their seeds contain enhanced amounts of a valuable α-linolenic fatty acid. It seems probable that the expression of the genes for antimicrobial peptides can activate the integral antistress protective system in plants and induce changes in the synthesis of new, biologically active compounds including fatty acids. This observation will be the subject of ongoing research.
REFERENCES
Vasil’chenko, I.T., Genus 687. Camelina Crantz, in Flora SSSR. V 30 tomakh (Flora of the USSR, in 20 vols), Komarov, V.L., Ed.-in-Chief, Bush, N.A, Ed., Leningrad: Akad. Nauk SSSR, 1939, vol. VIII, pp. 596–602.
Putnam, D.H., Budin, J.T., Field, L.A., and Breene, W.M., Camelina: a promising low-input oilseed, in New Crops, Janick, J. and Simon, J.E., New York: Wiley, 1993, pp. 314–322. http://www.hort. purdue.edu/newcrop/proceedings1993/v2-314.html
Zubr, J., Oil-seed crop: Camelina sativa, Ind. Crop Prod., 1997, vol. 6, pp. 113–119. https://doi.org/10.1016/S0926-6690
Liu, Z., Brost, J., Hutcheon, C., et al., Transformation of oilseed crop Camelina sativa by Agrobacterium-mediated floral dip and simple large-scale screening of transformants, In Vitro Cell. Dev. Biol. Plant., 2012, vol. 48, pp. 462–468. https://doi.org/10.1007/s11627-012-9459-7
Motesinos, E., Antimicrobial peptides and plant disease control, FEMS Microbiol. Lett., 2007, vol. 210, pp. 1–11. https://doi.org/10.1111/j.1574-6968.2007.00683.x
Andersson, M., Boman, A., and Boman, H.G., Ascaris nematodes from pig and human make three antibacterial peptides: isolation of cecropin P1 and two ASABF peptides, Cell. Mol. Life Sci., 2003, vol. 60, pp. 599–606. https://doi.org/10.1007/s000180300051
Martemyanov, K.A., Spirin, A.S., and Gudkov, A.T., Synthesis, cloning and expression of genes for antibacterial peptides: cecropin, magainin, and bombinin, Biotechnol. Lett., 1996, vol. 18, pp. 1357–1362. https://doi.org/10.1007/BF00129335
Zakharchenko, N.S., Rukavtsova, E.B., Gudkov, A.T., et al., Expression of the artificial gene encoding anti-microbial peptide cecropin P1 increases the resistance of transgenic potato plants to potato blight and white rot, Dokl. Biol. Sci., 2007, vol. 415, pp. 267–269. https://doi.org/10.1134/S0012496607040059
Zakharchenko, N.S., Kalyaeva, M.A., and Bur’yanov, Ya.I., Expression of cecropin P1 gene increases resistance of Camelina sativa (L.) plants to microbial phytopathogens, Russ. J. Genet., 2013, vol. 49, no. 5, pp. 523–529. https://doi.org/10.7868/S0016675813050147
Angenon, G., Dillen, W., and van Montagu, M., Antibiotic-resistance markers for plant transformation, in Plant Molecular Biology Manual, Gelvin, S.B. and Schilperoort, R.A., Eds., Dordrecht: Kluwer, 1994, pp. 1–13.
Herrera-Estrella, L., Leon, P., Olsson, O., and Teeri, T.H., Reporter genes for plants, in Plant Molecular Biology Manual, Gelvin, S.B. and Schilperoort, R.A., Eds., Dordrecht: Kluwer, 1994, pp. 1–32.
Schmit, F., Oakeley, E.J., and Jost, J.P., Antibiotics induce genome-wide hypermethylation in cultured Nicotiana tabacum plants, J. Biol. Chem., 1997, vol. 272, pp. 1534–1540.
Yisraeli, J. and Szyf, M., Gene methylation patterns and expression, in DNA Methylation: Biochemistry and Biological Significance, Razin, A., Eds., Berlin: Springer Verlag, 1984.
Etchberger, J.F. and Hobert, O., Vector-free DNA constructs improve transgene expression in C. elegans,Nat. Methods, 2008, vol. 5, p. 3.
Murashige, T. and Skoog, F., A revised medium for rapid growth and bioassays with tobacco cultures, Physiol. Plant., 1962, vol. 15, pp. 473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Ooms, G., Hooykaas, J.J., et al., Octopine Ti-plasmid deletion mutants of Agrobacterium tumefaciens with emphasis on the right side of the T-region, Plasmid, 1982, vol. 7, pp. 15–29.
Zakharchenko, N.S., Pigoleva, S.V., Yukhmanova, A.A., and Bur’yanov, Ya.I., Use of the gene of antimicrobial peptide cecropin P1 for producing marker-free transgenic plants, Russ. J. Genet., 2009, vol. 45, no. 8, pp. 929–933. https://doi.org/10.1134/S1022795409080067
Sambrook, J., Fritsch, E.E., and Maniatis, T., Molecular Cloning: A Laboratory Manual, New York: Cold Spring Harbor Lab. Press, 1989.
Das, P. and Joshi, N.C., Minor modifications in obtainable Arabidopsis floral dip method enhances transformation efficiency and production of homozygous transgenic lines harboring a single copy of transgene, Adv. Biosci. Biotechnol., 2011, vol. 2, pp. 59–67.
Naumov, N.A., Metody mikologicheskikh i fitopatologicheskikh issledovanii (Methods of Mycological and Phytopathological Studies), Leningrad: Sel’khozizdat, 1937.
Cljugh, S.J. and Bent, A.F., Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thalianta,Plant J., 1998, vol. 16, pp. 735–743.
Jan, P.-S., Huang, H.-Y., and Chen, H.-M., Expression of a synthesized gene encoding cationic peptide cecropin B in transgenic tomato plants protects against bacterial diseases, Appl. Environ. Microbiol., 2010, vol. 4, pp. 769–775.
Promega Protocols and Application Guide, Madison, USA, 1991, pp. 262–265.
Rodionova, L.N., Zagranichny, V.E., Rodionov, I.L., et al., The total solid phase synthesis of the subunit of cGMP phosphodiesterase from bovine retina and some physicochemical properties of the synthetic protein, Russ. J. Bioorg. Chem., 1997, vol. 23, no. 12, pp. 823–838.
Edwards, K., Johnstone, C., and Thompson, C., A simple and rapid method for the preparation of plant genomic DNA for PCR analysis, Nucleic Acids Res., 1991, vol. 19, p. 1349. https://doi.org/10.1093/nar/19.6.1349
Ohshima, M., Mitruhara, I., Okamoto, M., et al., Enhanced resistance to bacterial diseases of transgenic tobacco plants overexpressing sarcotoxin IA, a bactericidal peptide of insect, J. Biochem., 1999, vol. 125, pp. 431–435.
Draper, J., Scott, R., and Hamil, J., Transformation of dicotyledonous plant cells using the Ti plasmid of Agrobacterium tumefaciens and the Ri plasmid of A. rhizogenes, in Plant Genetic Transformation and Gene Expression. A Laboratory Manual, Draper, J., Scott, R., Armitage, P., and Walden, R., Eds., Oxford: Blackwell Sci. Publ., 1988, pp. 69–160.
Bradford, M.M., A rapid and sensitive method of the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding, Anal. Biochem., 1976, vol. 72, pp. 248–254. https://doi.org/10.1016/0003-2697
Nariyoshi, K., Hitoshi, K., and Takayuki, A.B.E., Control of tomato bacterial wilt without disinfections using a new functional polymer that captures microbial cells alive on the surface and is highly biodegradable, BioSci. Biotechnol. Biochem., 2005, vol. 69, pp. 326–333. https://doi.org/10.1271/bbb.69.326
Beauchamp, C.O. and Fridovich, I., Superoxide dismutase: improved assays and an assay applicable to acrylamide gels, Anal. Biochem., 1971, vol. 44, pp. 276–287.
Griffiths, M.J., van Hille, R.P., and Harrison, S.T., Selection of direct transesterification as the preferred method for essay of fatty acid content of microalgae, Lipids, 2010, vol. 45, no. 11, pp. 1053–1060. . PMID: https://doi.org/10.1007/s11745-010-3468-220820931
Mittova, V., Tal, M., Volokita, M., and Guy, M., Up-regulation of the leaf mitochondrial and peroxisomal antioxidative systems in response to salt-induced oxidative stress in the wild salt-tolerant tomato species Lycopersicon pennellii,Plant Cell Environ., 2003, vol. 26, pp. 845–856. PubMed: 12803612
Kreslavski, V.D., Los, D.A., Allakhverdiev, S.I., and Kuznetsov, Vl.V., Signaling role of reactive oxygen species in plants under stress, Russ. J. Plant Physiol., 2012, vol. 59, no. 2, pp. 141–154. https://doi.org/10.1134/S1021443712020057
Zakharchenko, N.S., Buryanov, Ya.I., Lebedeva, A.A., et al., Physiological features of rapeseed plants expressing the gene for an antimicrobial peptide cecropin P1, Russ. J. Plant Physiol, 2013, vol. 60, no.,3 pp. 411–419. https://doi.org/10.1134/S1021443713030163
Zakharchenko, N.S., Furs, O.V., Pigoleva, S.V., et al., Biological activity of leaf extracts from cecropin P1-synthesizing Kalanchoe plants: pharmacological prospects, Russ. J. Plant Physiol, 2018, vol. 65, no. 1, pp. 136–142. https://doi.org/10.7868/S0015330318010074
Lebedeva, A.A., Zacharchenko, N.S., Trubnikova, E.V., et al., Bactericide, immunomodulating, and wound healing properties of transgenic Kalanchoe pinnata synergize with antimicrobial peptide cecropin P1 in vivo, J. Immunol. Res., 2017. https://doi.org/10.1155/2017/4645701
Zakharchenko, N.S., Belous, A.S., Biryukova, Y.K., et al., Immunomodulating and re-vascularizing activity of Kalanchoe pinnata synergize with fungicide activity of biogenic peptide cecropin P1, J. Immunol. Res., 2017. https://doi.org/10.1155/2017/3940743
Belous, A.S., Shevelev, A.V., Trubnikova, E.V., et al., Wound treatment with transgenic Kalanchoe leaf extract with cecropin P1 (histological examination), Vestn. Ross. Gos. Med. Univ., 2017, vol. 1, pp. 70–78.
Campo, S., Manrique, S., Garcia-Martinez, J., and San, SegundoS., Production of cecropin A in transgenic rice plants has an impact on host gene expression, Plant Biotechnol. J., 2008, vol. 6, pp. 585–608. https://doi.org/10.1111/j.1467-7652.2008.00339.x
Goyal, R.K., Hancock, R.E.W., Mattoo, A.K., and Misra, S., Expression of an engineered heterologous antimicrobial peptide in potato alters plant development and mitigates normal abiotic and biotic responses, PLoS One, 2013, vol. 8, no. 10. e77505. PubMed: PMC3797780.
Murphy, D.J., The molecular organisation of the photosynthetic membranes of higher plants, Biochim. Biophys. Acta, 1986, vol. 864, pp. 33–94.
Liu, X-Y., Li, B., Yang, J-H., et al., Overexpression of tomato chloroplast omega-3 fatty acid desaturase gene alleviates the photoinhibition of photosystems 2 and 1 under chilling stress, Photosynthetica, 2008, vol. 46, no. 2, pp. 185–192.
Tarchevsky, I.A., The regulatory role of the degradation of biopolymers and lipids, Russ. J. Plant Physiol., 1992, vol. 39, pp. 1215–1223.
Cherian, G., Campbell, A., and Parker, T., Egg quality and lipid composition of eggs from hens fed Camelina sativa,J. Appl. Poult. Res., 2009, vol. 18, pp. 143–150. https://doi.org/10.3382/japr.2008-00070
Davis, P.B., The invasion potential and competitive ability of Camelina sativa (L.) Crantz (Camelina) in rangeland ecosystems, MSc Thesis, Bozeman, MT, USA: Montana State University, 2010. http://etd.lid.montana.edu/etd/2010/davis/DavisP0510.pdf
ACKNOWLEDGMENTS
The authors are grateful to I.L. Rodionov for the synthesis of the cecropin P1 peptide and to A.G. Laman for the production of antibodies to cecropin P1 and KLH-CEC conjugates.
Funding
The work was financially supported by the State Assignments no. 0101-2014-0046 and RK 01201352439 with partial financing from the Russian Foundation for Basic Research (projects 18-08-00752 and 16-04-00623).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest.
This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by I. Gordon
Abbreviations: AMPs—antimicrobial peptides; сесР1—cecropin P1; cecP1—cecP1 gene; CaMV 35S—35S RNA promoter of cauliflower mosaic virus; dNTP—deoxynucleoside triphosphate(s); LB medium—nutrient medium of Luriya Bertani; MS medium—hormone-free Murashige—Skoog medium; OD600, optical density at wavelength of 600 nm; PAGE—polyacrylamide gel electrophoresis; PGA—potato-glucose agar; SOD—superoxide dismutase.
Rights and permissions
About this article
Cite this article
Zakharchenko, N.S., Furs, O.V., Pigoleva, S.V. et al. Obtainment and Analysis of Marker-Free Oil Plants Camelina sativa (L.) Expressing of Antimicrobial Peptide Cecropin P1 Gene. Appl Biochem Microbiol 55, 888–898 (2019). https://doi.org/10.1134/S0003683819090096
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683819090096