Abstract
KT2 is a cationic antimicrobial peptide belonging to Crocodylus siamensis leucrocin I analogs. The mode of action of this compound taken at lethal concentration includes translocation into bacterial cells where binding to DNA is presumed to occur. However, the effects of KT2 on bacterial membrane have not been completely elucidated to date. In this study, a LIVE/DEAD staining technique was used to estimate the appropriate time of peptide-bacteria interaction. The results indicated more than 90% of Escherichia coli population was killed at density of ∼5 × 108 CFU/mL within 30 min after treatment with KT2 at MIC and 10 × MIC. The effects of KT2 on bacterial cells were investigated by the atomic force microscopy (AFM). At near MICs, the peptide induced heavy indentation of the bacterial surface as well as cellular collapse. Conversely, at concentrations of several times the MIC the potential to kill bacteria was greatly increased as judged by the induction of multiple membrane buds on the cell surface. Therefore, the collected results indicate that KT2 can cause different effects on bacterial surface which are positively correlated in magnitude and severity with peptide concentration via membranolytic effects.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Increasing bacterial resistance to multiple antibiotics represents a global problem of significant impact on hospital health care. Reduced effectiveness of conventional drugs in patients who are infected by some antibiotic resistant bacteria often leads to failure of therapy with potentially lethal consequences [1]. Thus, the discovery and development of novel classes of antibiotic agents constitutes an important requisite to tackle this urgent public problem. Recently, antimicrobial peptides (AMPs) have been proposed as a novel alternative to traditional antibiotics as these peptides reveal a great potential to kill a broad spectrum of pathogenic bacteria both at low concentration within a few minutes and with less toxicity to human cells than established antibiotic agents [2].
In nature, a variety of AMPs have been found to occur in diverse natural sources, including plant and animal species [3], while some have also been designed and chemically synthesized in order to improve their potency, specificity and activity [4, 5]. With respect to the antimicrobial mechanisms of AMPs, the bacterial membrane and numerous intracellular components have been identified as the two most important targets. In the first case, membrane targeting of peptides involves interaction with the bacterial cell surface, insertion into the cell membrane, and disruption of the membrane via a number of specific pathways. They have been categorized into the barrel stave model (e.g. alamethicin) [6, 7], torroidal pore model (e.g. LL-37, protegrin-1, and magainin-2) [8–10], and different carpet models (e.g. dermaseptin S, cecropin P1 and caerin 1.1) [11–13]. On the other hand, many AMPs, for example buforin II, indolicidin, and PR-39, have the potential to translocate across the cell membrane into the cytoplasm, which contains several types of intracellular molecules with specific biological function, such as nucleic acids (DNA/RNA) and proteins/enzymes [14–16]. Furthermore, some AMPs were shown to employ a dual mode of action on bacterial cells, combining membrane targeting and interaction with intracellular components to varying extent. For example, Bac7(1-35) and P-Der are cationic AMPs, which correspond to the N-terminus of Bac7 and a C-terminally amidated hybrid of flounder pleurocidin and frog dermaseptin, respectively. At their lowest inhibitory concentrations, Bac7(1-35) and P-Der are less capable of damaging bacterial membranes but possess the ability to translocate across the cell membrane and possibly inhibit molecular (nucleic acids and/or proteins) synthesis, while rapidly killing bacteria by damaging the cell membrane at multiples of the minimal inhibitory concentration (MIC) [17, 18]. Regarding this evidence, it should always be considered that some cationic AMPs may provide activity against bacterial cells either by a membranolytic or non-membranolytic mechanisms depending on their concentration or the peptide-to-lipid ratio.
KT2 and RT2 are cationic AMPs derived from Crocodylus siamensis leucrocin I, designed specifically based on the amphipathic properties derived from a helical wheel projection [19]. The peptides are comparably Trp-rich and consist of 17 amino acid residues. Cationic KT2 and RT2 were shown to display a strong potential to kill both prokaryotic bacteria and eukaryotic cancer cells, but proved remarkably non-toxic to eukaryotic Vero cells, macrophage RAW 264.7 and human red blood cells [19–21]. In addition, at concentrations below the respective MICs, both peptides inhibited the initiation of biofilm formation of E. coli O157:H7 and were found capable of killing the bacteria after biofilm progression. The use of confocal microscopy could provide clear evidence that translocation of the peptides into bacterial cells occurs at 2 × MICs. Moreover, in vitro experiments demonstrated the DNA binding ability of KT2 and RT2, for which the strongest activities were observed at a 1 : 1 and 1 : 2 DNA : peptide weight ratio, respectively. Regarding these results, the antibacterial mechanism of both peptides was assumed to originate from their considerable membrane permeation and DNA binding ability. Furthermore, at supra-MICs (10 × MIC) KT2 and RT2 could affect Gram-negative E. coli ATCC 25 922 and Gram-positive Staphylococcus epidermidis ATCC 12 228 bacteria, as demonstrated by scanning electron microscopy (SEM) [19]. However, although SEM can provide valuable insight on the effects of the peptides on bacteria, the cells must be fixed with low concentrations of glutaraldehyde and dehydrated using a concentration gradient of ethanol, which may result in the formation unexpected artifacts on the bacterial surface. Therefore, the effects induced by KT2 and RT2 on bacteria have been further evaluated using complementary microscopic techniques.
Among the variety of available microscopic techniques, atomic force microscopy (AFM) provides high performance and extremely high resolution of images, facilitating the acquisition of topographic magnification in three dimensions. This technique has been applied in the measurement of surface properties of diverse samples and serves nowadays as a powerful tool to study the surface features of inorganic and biological materials [22]. Especially with respect to the observation of bacterial cells, AFM has an enormous potential to visualize living material since the technique does not require any organic fixing or dehydration during bacterial cell preparation. Consequently, many previous studies utilized AFM for the investigation of membranolytic effects in the elucidation of bacterial killing mechanisms of antimicrobial substances, such as chitosans [23, 24], nitric oxide [25], silver ions [26], and antimicrobial peptides [27–30]. Therefore, this study aimed to determine the effects of antimicrobial peptides KT2 and RT2 at different concentrations in terms of morphological and surface changes on E. coli using the AFM method.
MATERIALS AND METHODS
Peptides. KT2 (NH2-NGVQPKYKWWKWWKKWW-NH2) and RT2 (NH2-NGVQPKYRWWRWWRRWW-NH2) were chemically synthesized and purified to >95% by Eurofins Genomics K.K. (Tokyo, Japan).
Bacteria. Two types of bacteria were used in this study, E. coli BL21 and S. epidermidis NBRC 12 993. Bacteria were cultured using nutrient broth (HiMedia Laboratories, India) and grown at 37°C with shaking at 180 rpm.
Determination of antimicrobial activity. Minimum inhibitory concentrations (MICs) were determined by a liquid culture-based growth inhibition assay [31]. The MIC was defined as the lowest peptide concentration that completely inhibited bacterial growth. Peptide solutions were prepared in a serial 2-fold dilution [19]. Then, 10 µL of each peptide sample was mixed thoroughly with 100 µL of bacterial suspension (∼1 × 106 CFU/mL) in culture medium and incubated at 37°C for 16–20 h. Bacterial growth was measured at OD600 using a microliter plate reader. Ampicillin (1 mg/mL) and 20 mM Tris-HCl buffer (pH 7.4) were used as positive and negative controls, respectively.
Moreover, peptide samples at a concentration of 10 × MICs were prepared. Next, 10 µL of each sample was transferred into a sterile micro-centrifuge tube containing 100 µL of bacterial suspension (∼1 × 106 CFU/mL) and incubated at room temperature for 5 min. Antimicrobial effects of peptides on bacteria were observed by a bright-field microscope (100 × magnification), compared with the control (untreated) cells.
Live/dead staining. The viability of immobilized E. coli BL21 on poly-L-lysine coated glass slides (Sigma-Aldrich, USA) was assessed by the BacLight LIVE/DEAD staining system according to the manufacturer’s protocol (Molecular Probes, USA). KT2 solutions were prepared at a concentration equal to the MIC and 10 × MIC towards E. coli BL21. Then, 10 µL of peptide samples was mixed separately to 100 µL of bacterial suspension (∼5 × 108 CFU/mL) and incubated for 60 min at room temperature. During the course of the incubation, 7 µL of each mixture was taken at 5, 10, 15, 30, and 60 min, and compared with an untreated cell solution at equal incubation times. Aliquots were applied individually on glass slides and co-incubated for 10 min with BacLight LIVE/DEAD staining solution. Live cells that are bound to SYTOX-9 showed green fluorescence, whereas dead cells in which nucleic acids were able to interact with propidium iodide fluoresced in red. Microscopic images were acquired using an Eclipse E600 microscope (Nikon, Japan) equipped with a DS-Qi1Mc monochrome quantitative digital camera (Nikon, Japan) and processed with NIS-elements software (Nikon, Japan) and/or Adobe Photoshop CS6 (Adobe, USA).
Preparation of the AFM samples.E. coli BL21 cells were cultured in nutrient broth at 37°C into the log phase. Cells were harvested by centrifugation at 9500 g for 3 min, washed with 0.89% NaCl and then diluted to ∼5 × 108 CFU/mL as an initial population, while varying concentrations of KT2, specifically 4.73 µM (0.5 × MIC), 9.45 µM (MIC), and 94.50 µM (10 × MIC), were used in the reaction tests. Briefly, 10 µL of peptide at each concentration was mixed together with 100 µL of the initial bacterial population and incubated for 5 or 60 min at room temperature. After incubation, 10 µL of the peptide-treated bacterial suspensions was applied onto glass slides coated with 4% 3-aminopropyl triethoxysilane (Sigma-Aldrich, USA) and allowed to dry for 5 min before imaging.
AFM imaging. Imaging was performed in air with a NX10 AFM (Park System, Korea) using a scanner with 7 nm silicon noncontact cantilever with a resonance frequency of approximately 330 Hz and a force constant of approximately 42 N/m. Initially, the area of the bacterial cells was located using a scan speed of 0.5 Hz and a resolution of 128 × 128 pixels in 50 × 50 µm. Single cells were selected randomly and imaged with a scan speed of 0.3 Hz and a resolution of 512 × 512 pixels in 5 × 5 µm. Then, the surface of each single cell was closely observed with the same speed and resolution in 1 × 1 µm. All topographic images were obtained simultaneously in every single scan. Cell height, size, and membrane roughness were quantified by the XEI program software (Park System, Korea).
RESULTS
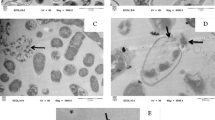
Determination of antimicrobial activity of KT2 and RT2 using AMP. KT2 and RT2 were chemically synthesized and their efficacy evaluated against E. coli and S. epidermidis. Table 1 shows the characteristics of the synthetic peptides and the determined MICs against both bacterial species. As seen from the table, the MICs of KT2 and RT2 were 9.45 and 4.32 µM against E. coli and 18.90 and 0.20 µM against S. epidermidis, respectively. Based on these results, RT2 exhibited the higher antibacterial activity than KT2, especially against S. epidermidis. Moreover, to examine potential effects of the peptides at high concentrations on cell morphology, E. coli and S. epidermidis were incubated with KT2 and RT2 at 10 × MICs for 5 min and then examined by light microscopy (Fig. 1). The cell shape of E. coli was found to be drastically altered by both KT2 and RT2 peptide treatment (Figs. 1b, 1c). The cell suspension contained irregularly rod-shaped cells and tiny dot-like structures that probably correspond to cellular debris, while in the case of S. epidermidis no visual effects (Figs. 1e, 1f) in comparison with untreated cells could be observed (Figs. 1a, 1d). In addition, aggregation of RT2 on the E. coli strain was found after the test, as indicated by arrows (Fig. 1c). Due to being the only combination to display well-resolved visual evidence, only KT2 and E. coli were selected for subsequent investigation using AFM.
Determination of bacterial viability after treatment with of KT2 and RT2 using AMP.E. coli cells were incubated with KT2 at the MIC and 10 × MIC, and the cell viability was analyzed by BacLight LIVE/DEAD staining. As shown in images (a–o) of Fig. 2, green spots designate live E. coli cells, while yellow or red spots originate from dead E. coli cells in which the cell membranes were damaged by KT2. Subsequently, to determine the appropriate incubation time for AFM observation, the total counts of dead and live cells was determined for each image to calculate the percentages of both dead and live cells in the same bar (Fig. 2, bar graph). During 60 min incubation, KT2 at the MIC killed E. coli in a time-dependent manner. About 96% cells died within 30 min after incubation at MIC, while no living cells could be detected after 5 min incubation at 10 × MIC. Accordingly, we concluded that 30 min incubation at the MIC was sufficient for further AFM observation.
Viability of E. coli cells after treatment with KT2 at the MIC and 10 × MIC observed by fluorescence microscopy. Bacteria were tested using the live–dead fluorescent assay, in which living cells fluoresce in green while red fluorescence indicates dead cells (a–o): without treatment with peptides (a–e) and after treatment with KT2 at the MIC (f–j) and 10 × MIC (k–o) for 5, 10, 15, 30, and 60 min, respectively. Bar graph indicates both % dead cells (black bars) and % live cells (gray bars), which correlates to the number of dead and live cells in the fluorescence microscopic images. In bar graph: 1—control; 2—MIC and 3—10 × MIC.
Determination of antimicrobial effects on bacterial surface. To determine the effects of KT2 treatment on the E. coli cell surface, AFM images of the bacteria under various conditions were obtained. Untreated E. coli cells dried in air were imaged and are presented in Fig. 3. To quickly localize bacterial cells, the AFM image was initially taken at low magnification in a scanning area of 50 × 50 µm (Fig. 3a). The individual cells were visualized by scanning at the same size (Figs. 3b, 3c). The images of untreated E. coli cells showed a sharp and discrete shape in conjunction with slight roughness at the middle of the cell. The average measured length, width, and height of the E. coli cells were ∼3.9 µm, ∼1.1 µm, and ∼140 nm, respectively (Fig. 3d). The nanostructure of the surface of E. coli was imaged by scanning the cells at a size of 1000 × 1000 nm (Fig. 3e). The cell surface appeared to be slightly corrugated; however, no pores or ruptures could be detected.
Whole cell and cell surface determination of peptide-untreated E. coli (control) imaged by AFM. (a)—topographic image of bacterial cells in a scale of 50 × 50 µm; (b)—topographic image of a single cell on a scale of 5 × 5 µm; (c)—3D reconstruction of the single cell, based on height data, on a scale of 5 × 5 µm; (d)—section profiles corresponding to the length and the width of the cell visible in the topographic image; (e)—cell surface of the single cell on a scale of 1000 × 1000 nm.
AFM images used for the assessment of dose-dependent effects of KT2 treatment at sub-MIC on the cellular surface structure of E. coli cells are compiled in Fig. 4. Although the overall cell shape was not altered, the cell surface displayed heavy roughness (Figs. 4a, 4b). The length, width, and height of KT2 treated cells were approximated to be ∼4.1 µm, ∼1.12 µm, and ∼360 nm, respectively (Fig. 4c). Figure 4d shows the cell surface morphology at a range of 1000 × 1000 nm, which indicates minor indentation on the E. coli surface due to an increase of membrane permeability as well as concomitant loss of cell integrity induced by KT2 treatment. Figures 5a, 5b show the representative AFM image of the KT2 treated E. coli cells at MIC. Both images of the same cell indicate the collapse of the central part of the cell, albeit no roughness was found on the cell surface. The length, width, and height of the cell were determined as ∼2.7 µm, ∼1.25 µm, and ∼200 nm, respectively (Fig. 5c). Upon cellular collapse, the bacterial cell wall was focused and imaged in high resolution (Fig. 5d). At the size around 1000 × 1000 nm, the cell wall of the bacterium showed unequivocal signs of collapse. In addition, treatment of the E. coli cells with KT2 at a higher concentration (10 × MIC) was investigated and the effects were again visualized by AFM imaging. A topographic image of a single cell was taken (Fig. 6a) and subsequently transformed into a 3D image depicted in Fig. 6b. As it seen in both figures, the bacterial cell suffered substantial damage from interaction with the peptide and many small micelle-like membrane patches were found distributed across the whole surface of the cell. Notably, in this case the measurement of cellular dimensions could not be conducted with sufficient accuracy (Fig. 6c). Figure 6d shows the formation of several blebs or buds on cell surface in nanoscale resolution (1000 × 1000 nm).
Whole cell and cell surface determination of E. coli treated with KT2 at 0.5 × MIC imaged by AFM. (a)—topographic image of a single cell on a scale of 5 × 5 µm; (b)—3D reconstruction of the single cell, based on height data, on a scale of 5 × 5 µm; (c)—cell surface of the single cell on a scale of 1000 × 1000 nm; (d)—section profiles corresponding to the length and the width of the cell visible in the topographic image.
Whole cell and cell surface determination of E. coli treated with KT2 (MIC) imaged by AFM. (a)—topographic of a single cell on a scale of 5 × 5 µm; (b)—3D reconstruction of the single cell, based on height data, on a scale of 5 × 5 µm; (c)—cell surface of the single cell on a scale of 1000 × 1000 nm; (d)—section profiles corresponding to the length and the width of the cell visible in the topographic image.
Whole cell and cell surface determination of E. coli treated with KT2 at 10 × MIC imaged by AFM. (a)—topographic of a single cell on a scale of 5 × 5 µm; (b)—3D reconstruction of the single cell, based on height data, on a scale of 5 × 5 µm; (c)—cell surface of the single cell on a scale of 1000 × 1000 nm; (d)—section profiles corresponding to the length and the width of the cell visible in the topographic image.
In addition, the area of the micelle-like membrane segments was also estimated by the XEI program software. The determined sizes of the micelle-like membrane areas were classified into small (<50 nm2), middle (50–100 nm2), and large (>100–300 nm2), as shown in Table 2. Small-sized area content had a frequency of 53.4% and was of 99–417 nm length. While the middle-sized area of micelle-like membranes had a frequency of 25.2% and 288–518 nm length, the large-sized area displayed a frequency of 21.4% and measured 429–760 nm in length.
DISCUSSION
Within the recent years, AMPs have gained considerable attention as potential alternative agents against drug-resistant bacteria. Such peptides have been obtained from a variety of different sources (i.e. bacteria, plants, and animals) and show several peculiar physical and biological features [32]. Among these unique features, the cationic charge of AMPs has been speculated to be a major prerequisite for conferring efficacy in killing bacteria [3]. In idealized models explaining the killing mechanism of AMPs, most cationic peptides act on the bacterial cell surface as the primary target, and their effects have been elucidated using different microscopic techniques. In this study, AFM was used for the investigation of antimicrobial effects of KT2 and RT2 because it has the potential to measure the physical changes to surface morphology in live bacteria and allows visualization of high-resolution images [33–35].
To determine the antimicrobial efficacy of KT2 and RT2, Gram-negative E. coli BL21 and Gram-positive S. epidermidis NBRC 12993 were treated with peptide solutions in different concentrations. Even at low concentrations, KT2 and RT2 were capable of completely killing both bacterial strains. In addition, E. coli and S. epidermidis were incubated with KT2 and RT2 at high concentrations (10 × MICs) for short duration (5 min) in order to elucidate the effect of peptide treatment by light microscopic observation. These experiments provided evidence for substantial aggregation of RT2, which interfered with the visualization of the bacterial cells. Moreover, after treatment with KT2, several black spots surrounding the E. coli cells were found, which presumably correspond to micelle-like membrane areas formed upon interaction of the peptide with the bacterial cell membrane. However, a previous study by [20] proposed that the antibacterial mechanism of KT2 and RT2 treatment at low concentrations involves translocation of the peptide into bacterial cells and subsequent binding to intracellular DNA. Notably, this conclusion cannot be drawn from our results and we thus assume that KT2 is able to employ different modes of action on bacteria depending on its concentration.
Prior to AFM analysis, the number of live and dead bacterial cells after treatment with KT2 at different concentrations and incubation times was assayed as described previously by using the BacLight LIVE/DEAD staining system, which contains the two specific fluorescent dyes SYTOX green and Propidium iodide [36]. Bacterial cells treated with KT2 at the MIC and 10 × MIC in the present study exhibited spectra of both fluorescent dyes, thus providing direct evidence for the ability of KT2 to disturb bacterial membrane permeability and further suggesting that the peptide may exert some effects on the bacterial cell surface. In addition, the efficacy of KT2 in killing bacteria at both concentrations showed significant dependence on the incubation time. At the highest concentration, KT2 showed strong activity and affected complete killing of bacteria within 5 min or less. In contrast, lower peptide concentration led to inhibition of bacteria at a very slow rate and most bacterial cells were killed within 30 min. Hence, in order to be able to observe the effects of peptide treatment, a 30 min-incubation period between peptide and bacteria was selected in the subsequent AFM imaging experiment.
AFM monitoring of bacterial cells treated with KT2 provided valuable topographic 2D and 3D data for the interpretation of antibacterial effects in correlation with peptide concentration. At sub-lethal concentration, several indentations were found located on the surface of the bacterial cell, suggesting that KT2 molecules might initially be randomly distributed on the bacterial surface before exhibiting antimicrobial effects via insertion into cell membrane. The result is excellent agreement with a report of Anunthawan et al. [20], who reported that KT2 at low concentration is able to translocate into cytoplasm of live bacterial cells and bind to DNA in vitro. In contrast, treatment at the lethal KT2 concentration was accompanied by a reduced number of indentations visible on the surface of the cells. Instead, extended segments of the cell wall were found to be collapsed, suggesting that after peptide interaction on the bacterial membrane, gradual accumulation of neighboring peptide molecules at the same location might induce powerful membranolytic effects, thereby promoting the collapse of adjacent cellular regions. These results correlate well with findings reported previously by Li et al. [28], studying the antimicrobial action of MSI-78 peptide on Gram-negative bacteria by AFM [28]. At supra-lethal concentration, this peptide revealed strong efficacy to cause cellular disintegration in conjunction with the formation of micelle-like membrane patches, blisters and bubbles on the bacterial surface. Regarding these experimental observations, Hartmann with coworkers [37] proposed that at higher peptide concentrations, AMPs promote the disruption of the inner membrane and the release of cytoplasmic cellular content into the periplasmic space, which manifests itself in the formation of blisters or bubbles. Because micelle-like membrane segments were clearly visible upon treatment with KT2 at supra-lethal concentrations, the killing mechanism of the peptide is hypothesized to comply with the carpet model [3]. Regarding the AFM results, KT2 shows great potential to alter the physical morphology of bacterial surfaces and damage the bacterial membrane. Moreover, it is believed that the peptide employs a dual mode of action to kill bacteria via a combination of non-membranolytic and membranolytic modes, depending on its concentration.
Thus, this study represents the first nanoscale demonstration of antimicrobial action exerted by the antimicrobial peptide KT2 on the surface of Gram-negative E. coli. The use of AFM provided high-resolution images capturing the effects of the KT2 treatment and allowed to gain a better understanding how KT2 acts on Gram-negative bacterial cells at different concentrations. At sub-lethal or lethal concentrations, peptide treatment caused specific morphological abnormalities, ranging from the formation of indentations on the cell surface to the complete collapse thereof. At a higher concentration, KT2 induced the formation of blisters or bubbles on the surface and subsequently generated micelle-like membrane patches, which are taken as indicator for severe cellular damage. Therefore, our results clearly demonstrate that KT2 disrupts the bacterial cell surface in different ways depending on its concentration, ultimately causing the death of the bacterial cell.
REFERENCES
Ventola, C.L., Pharm. Ther., 2015, vol. 40, no. 4, pp. 277–283.
Matsuzaki, K., Biochim. Biophys. Acta, 2009, vol. 1788, no. 8, pp. 1687–1692.
Brogden, K., Nat. Rev. Microbiol., 2005, vol. 3, no. 3, pp. 238–250.
Blondelle, S. and Lohner, K., Biopolymers, 2000, vol. 55, no. 1, pp. 74–87.
Hong, S., Park, T., and Lee, K., Peptides, 2001, vol. 22, no. 10, pp. 1669–1674.
Bechinger, B., Biochim. Biophys. Acta, 1999, vol. 1462, no. 1–2, pp. 157–183.
Yang, L., Harroun, T., Weiss, T., Ding, L., and Huang, H., Biophys. J., 2001, vol. 81, no. 3, pp. 1475–1485.
Wildman, K. H., Lee, D., and Ramamoorthy, A., Biochemistry, 2003, vol. 42, no. 21, pp. 6545–6558.
Yamaguchi, S., Hong, T., Waring, A., Lehrer, R., and Hong, M., Biochemistry, 2002, vol. 41, no. 31, pp. 9852–9862.
Yang, L., Harroun, T., Heller, W., Weiss, T., and Huang, H., Biophys. J., 1998, vol. 75, no. 2, pp. 641–645.
Gazit, E., Boman, A., Boman, H., and Shai, Y., Biochemistry, 1995, vol. 34, no. 36, pp. 11 479–11 488.
Pouny, Y., Rapaport, D., Mor, A., Nicolas, P., and Shai, Y., Biochemistry, 1992, vol. 31, no. 49, pp. 12 416–12 423.
Wong, H., Bowie, J., and Carver, J., Eur. J. Biochem., 1997, vol. 247, no. 2, pp. 545–557.
Boman, H., Agerberth, B., and Boman, A., Infect. Immun., 1993, vol. 61, no. 7, pp. 2978–2984.
Park, C., Kim, H., and Kim, S., Biochem. Biophys. Res. Commun., 1998, vol. 244, no. 1, pp. 253–257.
Subbalakshmi, C. and Sitaram, N., FEMS Microbiol. Lett., 1998, vol. 160, no. 1, pp. 91–96.
Patrzykat, A., Friedrich, C., Zhang, L., Mendoza, V., and Hancock, R., Antimicrob. Agents Chemother., 2002, vol. 46, no. 3, pp. 605–614.
Podda, E., Benincasa, M., Pacor, S., Micali, F., Mattiuzzo, M., Gennaro, R., and Scocchi, M., Biochim. Biophys. Acta, 2006, vol. 1760, no. 11, pp. 1732–1740.
Anunthawan, T., Yaraksa, N., Phosri, S., Theansungnoen, T., Daduang, S., Dhiravisit, A., and Thammasirirak, S., Bioorg. Med. Chem. Lett., 2013, vol. 23, no. 16, pp. 4657–4662.
Anunthawan, T., Fuente–Núñez, C.D.L., Hancock, R., and Klaynongsruang, S., Biochim. Biophys. Acta, 2015, vol. 1848, no. 6, pp. 1352–1358.
Theansungnoen, T., Maijaroen, S., Jangpromma, N., Yaraksa, N., Daduang, S., Temsiripong, T., et al., Protein J., 2016, vol. 35, no. 3, pp. 202–211.
Ozkan, A., Topal, A., Dana, A., Guler, M., and Tekinay, A., Micron, 2016, vol. 89, pp. 60–76.
Eaton, P., Fernandes, J., Pereira, E., Pintado, M., and Malcata, F.X., Ultramicroscopy, 2008, vol. 108, no. 10, pp. 1128–1134.
Eaton, P., Fernandes, J., Pereira, E., Pintado, M., and Malcata, F.X., Ultramicroscopy, 2009, vol. 109, no. 8, pp. 854–860.
Deupree, S. and Schoenfisch, M., Acta Biomater., 2009, vol. 5, no. 5, pp. 1405–1415.
Yang, X., Yang, W., Wang, Q., Li, H., Wang, K., Yang, L., and Liu, W., Talanta, 2010, vol. 81, no. 4–5, pp. 1508–1512.
da Silva, A.Jr. and Teschke, O., Biochim. Biophys. Acta, 2003, vol. 1643, no., pp. 95–103.
Li, A., Lee, P., Ho, B., Ding, J., and Lim, C., Biochim. Biophys. Acta, 2007, vol. 1768, no. 3, pp. 411–418.
Mecke, A., Lee, D., Ramamoorthy, A., Orr, B., and Holl, M.B., Biophys. J., 2005, vol. 89, no. 6, pp. 4043–4050.
Rossetto, G., Bergese, P., Colombi, P., Depero, L., Giuliani, A., Nicoletto, S., and Pirri, G., Nanomedicine, 2007, vol. 3, no. 3, pp. 198–207.
Yaraksa, N., Anunthawan, T., Theansungnoen, T., Daduang, S., Araki, T., Dhiravisit, A., and Thammasirirak, S., J. Antibiot., 2014, vol. 67, no. 3, pp. 205–212.
Bahar, A.A. and Ren, D., Pharmaceuticals, 2013, vol. 6, no. 12, pp. 1543–1575.
Mularski, A., Wilksch, J., Wang, H., Hossain, M., Wade, J., Separovic, F., et al., Langmuir, 2015, vol. 31, no. 22, pp. 6164–6171.
Mularski, A., Wilksch, J., Hanssen, E., Strugnell, R., and Separovic, F., Biochim. Biophys. Acta, 2016, vol. 1858, no. 6, pp. 1091–1098.
Shaw, J., Epand, R., Hsu, J., Mo, G., Epand, R., and Yip, C., J. Struct. Biol., 2008, vol. 162, no. 1, pp. 121–138.
Yamashita, H., Taoka, A., Uchihashi, T., Asano, T., Ando, T., and Fukumori, Y., J. Mol. Biol., 2012, vol. 422, no. 2, pp. 300–309.
Hartmann, M., Berditsch, M., Hawecker, J., Ardakani, M. F., Gerthsen, D., and Ulrich, A.S., Antimicrob. Agents Chemother., 2010, vol. 54, no. 8, pp. 3132–3142.
ACKNOWLEDGMENTS
Additionally, we would like to thank the Protein and Proteomics Research Center for Commercial and Industrial Purposes (ProCCI), Department of Biochemistry, Faculty of Science, Khon Kaen University, Thailand as well as the School of Natural System, College of Science and Engineering, Kanazawa University, Kakuma-machi, Japan for providing essential laboratory facilities.
Moreover, we would like to thank Mr. Anuchit Ruangvittayanon and Dr. Worawikunya Kiatponglarp for the AFM assessment, as well as the Synchrotron Light Research Institute (Public Organization), Thailand, for providing access to the AFM equipment.
Funding
This work was financially supported by the Research and Researchers for Industries (RRI), the Thailand Research Fund, and Sriracha Moda Co., Ltd., Chon Buri, Thailand.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Rights and permissions
About this article
Cite this article
Theansungnoen, T., Jangpromma, N., Anwised, P. et al. Membranolytic Effects of KT2 on Gram-Negative Escherichia coli Evaluated by Atomic Force Microscopy. Appl Biochem Microbiol 55, 495–505 (2019). https://doi.org/10.1134/S0003683819050144
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683819050144