Abstract
Two halo-tolerant and alkaliphilic actinomycetes, Nocardiopsis alba OM-4 (GeneBank Number, KC119568) and Nocardiopsis alba TATA-13 (GeneBank Number, KC119569) isolated from the salt-enriched soil of the Coastal Gujarat (India) were studied for their proteases. The low molecular weight (19–20 kDa) alkaline proteases were purified by the hydrophobic interaction chromatography on Phenyl Sepharose 6 FF column. The enzymes were optimally active at 60–70°C and pH 10.0. NaCl enhanced the catalysis and enzyme stability at different temperatures and in the presence of up to 50% concentrations of various solvents. The purified enzymes were resistant against various surfactants and inhibitors, suggesting their potential applications in the detergent industry. The changes in the secondary structures were probed by the circular dichroism spectroscopy at various temperatures and solvents, followed by the K2D analysis. With increasing temperatures, the contents of the α- helices and β-sheets increased in the N. alba OM-4 protease, while a reverse trend was evident for the N. alba TATA-13 protease. On the other hand, the α-helix contents increased accompanied with decreased β-sheets in both proteases in the presence of different solvents.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Enzymes are catalysts produced by living cells to bring about specific biochemical reactions. More than 3000 different enzymes have been identified and some are being used in various applications [1–3]. The use of proteases for industrial applications started in 1960s and since then these enzymes have attracted attention and found applications in various industries, such as pharmaceuticals, leather, food and agriculture [4].
Search for protease is a dynamic process and in this context microbes have continuously served as one of the largest and useful sources [5].
The present study is focused on the characterization of the alkaline serine proteases of the salt-tolerant alkaliphilic actinomycetes, Nocardiopsis alba OM-4 and Nocardiopsis alba TATA-13. The stability of the proteases in various organic solvents was studied. The effect of NaCl on the activity and stability of the enzymes was also assessed. Further, the changes in the protease secondary structure in various solvents and at different temperatures were studied using the CD spectroscopy.
MATERIALS AND METHODS
Materials. The Phenyl Sepharose 6 FF was purchased from Sigma (USA). SDS was obtained from Merck (Germany). Casein was purchased from SISCO Research Laboratories (India). Other chemicals and medium components were obtained from HiMedia Laboratories (India).
Isolation and identification of protease-producing actinobacteria. Saline soil from the coastal region of Gujarat (India) was collected in a sterile container and screened for protease-producing actinobacteria using yeast extract-malt extract (YEME) medium containing (g/L): yeast extract—3.0, malt extract—3.0, peptone—5.0, dextrose—10.0, NaCl—50.0, agar—30.0 (pH 9.0) incorporated with 10 g/L casein. After 7 days of incubation at 28°C, two strains designated as OM-4 and TATA-13 showed the highest zones (diameter of 2.3 and 2.9 cm, respectively) of hydrolysis. These strains were selected for the present study.
OM-4 and TATA-13 were identified based on 16S rRNA sequencing. Extraction of the genomic DNA by soft lysis method and PCR amplification of the 16S rRNA gene were performed as described earlier by Li et al. [6]. The sequences were compared with the available 16S rRNA gene sequences at EzTaxon-e server (http://eztaxon-e.ezbiocloud.net/) [7]. Phylogenetic tree was constructed using the neighbour-joining (NJ) method [8] by MEGA 7.0 software [9] after multiple alignments of the sequences using CLUSTAL_X program [10] using Kimura’s two parameter model [11] with 1000 boot strap replicates [12].
Enzyme purification. The activated cultures were inoculated at 5% (vol/vol) concentration into the gelatin broth containing (g/L): gelatin—10.0, peptone—5.0, yeast extract—5.0 and NaCl—50.0 (pH 9.0), followed by the incubation at 37°C at 120 rpm. The cultures were harvested after 7 days of the incubation and the crude enzyme or cell-free extracts were obtained as supernatants after centrifugation at 13 000 g for 10 min at 4°C. The extracellular alkaline proteases from OM-4 and TATA-13 strains were purified by a two-step hydrophobic interaction chromatography using Phenyl Sepharose 6FF, as described earlier by Gohel and Singh [13, 14]. The purity of the enzymes was judged on SDS-PAGE according to the Laemmli method [15]. The protein bands were visualized by Comassie brilliant blue R-250 staining [16].
Protease assay. The protein content was measured according to the Bradford method using BSA (0–100 µg/mL) as a standard [17]. The Anson-Hagihara’s method [18] was used to measure the alkaline protease activity using casein as a substrate. The protocol was based on the method described earlier [19]. One unit of the alkaline protease was defined as the amount of the enzyme liberating 1 μg of Tyr per min under the assay conditions. Enzyme units were calculated using Tyr (0–100 μg) as a standard.
Temperature and pH profiles. The optimum temperatures for the catalysis of the proteases were obtained by incubating the reaction mixtures at different temperatures within the range of 30–90°C. The protease activity was measured, followed by the calculation of the relative activity for the enzyme. Similarly, the optimum pH for the enzyme activity was calculated after dissolving the substrate in the buffers with different pH values. The enzyme-substrate reaction mixture was incubated at the optimum temperature. In the experiments, 20 mM phosphate (pH 5.8–8.0), 20 mM glysine–NaOH (pH 9.0), 20 mM borax-NaOH (pH 10.0–11.0) and 20 mM KCl–NaOH (pH 12.0–13.0) buffers were used. The protease activities were measured, followed by calculation of the relative activity of the enzyme.
Effect of NaCl on the protease activity and stability. The reaction mixture was supplemented with 0–3 M NaCl in the optimum buffer to monitor the effect of NaCl on the enzyme catalysis. The reaction mixture was then incubated at the optimum temperature and the protease activity was measured. Correspondingly, the enzymes without NaCl and enzymes supplemented with optimum NaCl were incubated at different temperatures between 30 and 80°C to determine the temperature dependence of NaCl treatment on the protease activity. The protease activity was measured at the regular time intervals (0–180 min). The protease activity was measured, followed by the calculation of the relative activity of the enzyme.
Effect of inhibitors and surfactants on the enzyme activity. The influence of various effectors on the protease activity was studied under standard assay conditions, where the reaction mixture was supplemented with 10 mM of inhibitors: phenyl methyl sulfonyl fluoride (PMSF), EDTA, dithiothreitol (DTT) and β-mercaptoethanol. The surfactants, triton X-100 and tween-80 at 0.5% (vol/vol) concentration and 0.5% (wt/vol) SDS, were used. The effect was assessed by considering the control (without inhibitors and surfactants) as 100%.
Catalysis and stability of the alkaline proteases in the presence of various organic solvents. The catalysis of the purified alkaline proteases was performed in the presence of various organic solvents, such as n-decane (5.6), iso-octane (4.5), n-haxane (3.2), benzene (2.1), butanol (0.8), acetone (–0.24) and methanol (–0.75). The solvents were selected based on the LogPow values, as indicated with the solvents in the brackets. The reaction mixture was supplemented with varying concentrations of the solvents from 0 and 50% (vol/vol). The relative activities were then calculated.
The enzyme stability was investigated by incubating the enzymes in the above stated solvents with and without NaCl at 37°C for 24 h. The effect of salt on the OM-4 and TATA-13 protease stability in the presence of different organic solvents was assessed. The enzymes were incubated with NaCl (1–2 M) along with solvent. The relative enzyme activities were calculated and compared with the control (without any solvent).
CD spectroscopic analysis of the purified enzymes. The purified enzymes in 1 M NaCl were subjected to different temperatures (30, 50 and 80°C for 1 h). Similarly, the enzyme preparations were subjected to different solvents, such as benzene and n-decane at 10% (vol/vol) at 30°C for 2 h. The denatured enzymes were then analyzed using J-815 CD spectropolarimeter (JASCO, Germany) within the range of the wavelengths from 180 to 260 nm. The smoothed resultant spectra were average of the 5 consecutive runs. The control spectra were then subtracted from the spectra of the treated samples. The mean residual ellipticity (MRE) was calculated from the Θobs values at the above stated wave length range. MRW was taken as 115 [20].
where MRW is the mean residual weight, L is the path length (0.1 cm), C is the enzyme concentration.
The MRE values, between 190–240 nm, were subjected to online CD software, K2D2 to find out the changes in its secondary protease structures in terms of the percent changes in α-helices and β-sheets.
RESULTS
Isolation and identification of protease-producing actinobacteria. The potential protease-producing strains OM-4 and TATA-13 have been isolated from the saline soil of coastal region of Gujarat (India) by Dr. S.D. Gohel in the laboratory of Prof. S.P. Singh, Department of biosciences, Saurashtra University, Rajkot, India. Based on 16S rRNA gene sequence homology and phylogenetic analysis, OM-4 and TATA-13 were identified as Nocardiopsis alba (Gene Bank Accession Number, KC119568) and Nocardiopsis alba (Gene Bank Accession Number, KC119569), respectively. Further the NJ- tree (Fig. 1) confirmed the strains are different from each other but both strains OM-4 and TATA-13 belong to N. alba.
Purification of N. alba OM-4 and N. alba TATA-13 alkaline proteases. The optimum growth of both the actinomycete isolates, N. alba OM-4 and N. alba TATA-13, was achieved after 7 days at 37°C at 120 rpm of the incubation in the gelatin- containing broth. Under these conditions, the alkaline protease production was 198.2 and 215.3 U/mL for N. alba OM-4 and N. alba TATA-13, respectively. Active protease was isolated from the culture supernatant by ammonium sulfate precipitation and hydrophobic interaction chromatography using Phenyl Sepharose 6 FF column. In case alkaline protease of N. alba OM-4, the two-step purification method yielded 2.24-fold purification, 3.6% yield and 4266 U/mg specific activity. For the protease of N. alba TATA-13, the purification was 2.64-fold leading to 3.8% yield and 4895 U/mg specific activity (Table 1). The purified proteases were observed as single bands on SDS-PAGE with apparent molecular weights as 19 kDa and 20 kDa for N. alba OM-4 and N. alba TATA-13, respectively.
Effects of temperature, pH and NaCl on the enzyme catalysis. The N. alba OM-4 protease exhibited optimum activity of 234.7 U/mL at 60°C, while the N. alba TATA-13 protease had optimum activity of 270.6 U/mL at 70°C (Fig. 2a). The optimum pH for N. alba OM-4 and N. alba TATA-13 proteases was 10.0 in 20 mM borax-NaOH buffer. However, both enzymes were active within the pH range of 8.0–11.0 (Fig. 2b). With respect to salt, 1% (wt/vol) NaCl was optimum for the N. alba OM-4 protease for catalysis; while for the N. alba TATA-13 protease, it was 0.5% (w/v) NaCl (Fig. 2c).
Effect of NaCl on thermostability of alkaline proteases. The effect of NaCl on thermostability of alkaliphilic actinobacteria proteases was assessed. After treatment with 1 M NaCl for 1 h, the enzyme retained 50% of the residual activity at 50°C (Fig. 3). However, in the absence of salt at 60, 70 and 80°C, the protease denatured within 10–30 min. While in the presence of salt, the enzymes retained more than 50% of the residual activity even after 2 h. At 30 and 50°C, the N. alba OM-4 protease was quite stable with 1 M salt but as the temperature increased from 60 to 80°C, the protease required higher salt concentrations for the stabilization. On the other hand, the N. alba TATA-13 protease was relatively more stable with 1 M salt.
Recently in 2017 [1], a solvent tolerant protease from haloalkalophilic bacteria, Paracoccus saliphilus was isolated from shrimp shell waste. This enzyme was stable at 60°C, pH 9.0, and 3.0 M NaCl concentration.
Effects of inhibitors and surfactants on activity of alkaline proteases. The activities purified enzymes were completely inhibited by PMSF and partially inhibited by EDTA, DTT and β-mercaptoethanol (Table 2). The N. alba OM-4 and N. alba TATA-13 proteases were stable in Triton-100 and retained 80% of the original activities after 24 h of the incubation. The enzymes retained 65 and 70% activities in SDS and Tween-80, respectively (Table 2).
Effect of solvents on the enzyme catalysis and stability. Rathinam et al. [33] have described enzymatic dehairing in aqueous medium, a method which appears to be superior than the lime-sulphide based conventional method of leather treatment. It has also been demonstrated that the solvent-enzyme mixture removed hair with better efficiently without any sulphide and lime than aqueous enzyme-lime-sulphide system.
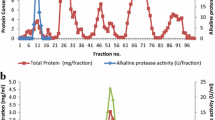
The N. alba OM-4 and N. alba TATA-13 proteases displayed considerable stability in various organic solvents. For the catalysis, both enzymes retained nearly 100% of the activities in the presence of up to 10% (vol/vol) solvents of comparatively higher LogPow values such as benzene, iso-octane and n-decane (Fig. 4).
The effect of solvents on the enzyme catalysis was studied at various salt concentrations. It is evident that 1 and 2 M NaCl concentrations were optimal for the protease activities of N. alba TATA-13 and N. alba OM-4, respectively (Fig. 2c). The presence of salt significantly reduced the detrimental effect of the solvents (Figs. 5 and 6). The enhanced stability of the proteases in the presence of salt might be due to the electrostatic interactions amongst the contrary charges and salt bridges stabilizing the native structure [30]. Salt–bridge interactions play significant role in alleviating many protein structures and can be altered for desirable features of the protein [31].
The time-dependent curves after 30 min—24 h treatment with n-decane, iso-octane, n-hexane, benzene, butanol, acetone and methanol for activities of alkaliphilic actinobacteria proteases are presented in Figs. 5 and 6.
In the presence of 2 M NaCl, N. alba OM-4 and 1 M NaCl, N. alba TATA-13 proteases retained 58% (Fig. 5j) and 65% (Fig. 6j) residual activities, respectively, after 2 h of incubation with 50% benzene. Both proteases lost more than 50% activities when NaCl was excluded.
In the presence of 2 M NaCl, N. alba OM-4 and 1 M NaCl, N. alba TATA-13 proteases displayed 90% (Fig. 5h) and 129% (Fig. 6h) residual activities, respectively, after 2 h incubation with 50% iso-octane. However, without NaCl, the respective activities decreased to 42% (Fig. 5g) and 62% (Fig. 6g). With 50% n-decane in the presence of 2 M NaCl, the N. alba OM-4 and 1 M NaCl N. alba TATA-13 proteases retained 91% (Fig. 5f) and 76% (Fig. 6f) of the residual activities, respectively. However, in the absence of salt, these proteases under the same conditions retained 73 and 71% of the original activities (Figs. 5e and 6e). The enzyme activities decreased after adding n-hexane, methanol and butanol at increased concentrations (Fig. 4). After 1 h of incubation with 20% (vol/vol) n-hexane in the presence of 2 M NaCl, the N. alba OM-4 and 1 M NaCl N. alba TATA -13 proteases retained 50 and 58% of the residual activities, respectively (Fig. 5b and 6b). However, in the absence of salt, under the same conditions, only 19 and 47% of the residual activities were determined (Figs. 5a and 6a).
After 4 h of incubation in the presence of NaCl with 20% n-butanol, N. alba OM-4 and N. alba TATA-13 proteases retained 68% (Fig. 5n) and 55% (Fig. 6n) residual activities, respectively, which is quite higher than those without NaCl (Figs. 5m and 6m). Similar, with 20% (vol/vol) methanol in the presence of NaCl, 55% (Fig. 5d) and 60% (Fig. 6d) of the residual activities were correspondingly detected for N. alba OM-4 and N. alba TATA-13 proteases.
The enzymes retained 95% (Figs. 5j and 6j) of residual activity after 2 h of incubation in 20% benzene with NaCl, while without NaCl, both enzymes retained only 85 and 80% of residual activities, respectively (Figs. 5i, 6i), suggesting the role of salt in the stability of the enzymes.
Both alkaline proteases were highly stable in 20% acetone for 6 h, with 74% residual activities in the absence of salt (Figs. 5k and 6k). However, the addition of salt enhanced the residual activities proteases to 91% (Fig. 5l) and 92% (Fig. 6l).
Both these enzymes exhibited enhanced activities in the presence of NaCl and solvents. In 20% solvent, both proteases were highly stable for 1 h without salt and for 2 h with salt (Figs. 5, 6). Interestingly, even at 50% solvent concentration, the alkaline proteases were found stable for 30 min when salt was included.
On the whole, the results of the catalysis and stability establish the solvent stability of these two alkaline proteases. The kinetic network models to reveal the effect of non-native salt-bridges on α-helix folding have been described [31]. The secondary structures of the proteins determine the functional state of the protein. Therefore, structural studies of the proteases by CD analysis have been undertaken.
CD spectroscopic analysis of the purified proteases. The CD spectroscopy has emerged as an excellent method of determining the secondary structures of the proteins. The spectra were generated after treating the N. alba proteases at different temperatures with different solvents, such as 10% benzene and n-decane. The enzymes exhibited major changes in α-helices and β-sheets contents at 80°C (Fig. 7, Table 2) in contrast to the treatment at 30 and 50°C at which the effect was negligible.
CD spectra analysis of N. alba OM-4 (a) and N. alba TATA-13 (b) proteases treated for 1 h at different temperatures, such as 30 (1), 50 (2) and 80°C (3) and CD spectra analysis of N. albaalba OM-4 (dark black line) and N. alba alba TATA-13 (grey line) proteases treated for 2 h with 10% (vol/vol) n-decane (c) and benzene (d).
In N. alba OM-4 and N. alba TATA-13 proteases, the α-helices accounted for 1.86 and 3.25%, respectively. The β-sheets were 49.03 and 49.36%, respectively at 30°C, showing the native structure of the proteases. At 50°C, the same values for the α-helices and β-sheets were observed. It was revealed that at 50°C after 1 h, the protease retained its native structure. While at 80°C, the content of the α-helices were computed as 2.11 and 1.83% for N. alba OM-4 and N. alba proteases, respectively, while β-sheets were at 51.77 and 47.70%, indicating changes in the native structures of the proteases. It can, therefore, be predicted from the CD analysis that the enzymes are highly stable at 50°C and get denatured at 80°C.
With 10% n-Decane, the contents of α-helices were computed as 8.40 and 1.86% in N. alba OM-4 and N. alba TATA-13 proteases, respectively, while the β-sheets were 1.86 and 49.03%, respectively. The trends reflected significant differences in the native structures of these proteases. However, in the presence of 10% benzene, 3.16 and 8.16% α-helices were revealed in N. alba OM-4 and N. alba TATA-13 proteases, respectively, while β-sheets were detected as 8.16 and 24.05%, respectively (Table 2).
The trends reflected in Table 2 are correlated with the thermostability patterns indicated in Fig. 2. At 80°C, the contents of α-helices increased while those for β-sheets decreased in N. alba OM-4 protease. On the other hand, the contents of two structure types decreased at 80°C in N. alba TATA-13 protease.
While considering the stability in solvents, the changes in the α-helices and β-sheets in both proteases were distinct. The trends reflected in the CD analysis correspond with those highlighted in the protease activities shown in Figs. 5 and 6. Both proteases displayed structural changes in the presence of n-decane and benzene.
To the best of our knowledge, the analysis of the secondary structures has not been reported for the proteases of haloalkaliphilic actinomycetes and archaea of the saline habitats. However, a report on the recombinant leucine aminopeptidase II of Bacillus stereothermophilus suggested changes in the contents of the α-helical structures in response to varying concentrations of Zn2+ [27]. Based on the CD spectroscopic studies, an active mutant of luciferase (rLuc (Arg)) expressed under the same conditions as the native form (rLuc) was confirmed to refold without aggregation in Lampyris turkestanicus [28].
DISCUSSION
Enzymes are obtained from all living organisms. However, microorganisms are usually the first choice due to their broad biochemical diversity, feasibility of cultivation and ease of genetic manipulation [21]. The main obstacle for the industrial applications of enzyme is its relatively lower activity under extreme physical and chemical conditions. Therefore, we focused on the proteases displaying high activity and stability under extreme conditions.
The catalysis and stability of the enzymes at high temperatures is desirable for the applications of proteases [22]. Production and biochemical characterization of an alkaline protease from Aspergillus oryzae CH93 has recently been reported [23]. While comparing our results with that reported in literature, N. alba OM-4 and N. alba TATA-13 enzymes show high stability at 60–80oC after treatment with 20–50% solvents and other denaturing agents. A detergent-stable solvent-tolerant serine thiol alkaline protease was purified from Streptomyces koyangensis [24]. Anorganic solvent-tolerant protease from haloalkalophilic bacterium Paracoccus saliphilus isolated from shrimp shell waste has recently been described for chitin extraction [1]. Earlier, an acid stable protease isolated from N. alba was reported by Kelch et al. [25]. Alkaline protease from N. alba was described by Gohel and Singh [14]. However, none of these studies related to the structural elucidation and solvent resistance of the enzyme.
The CD spectra of the enzyme at higher temperature and in the presence of organic solvents help us to predict the denaturing patterns of the enzyme. The patterns reflected that the α-helix and β-sheet contents remained unchanged for both N. alba proteases, suggesting their thermostable nature. The structural investigations using NMR and X-ray crystallographic studies further add to the structure-function relationships of these proteases.
It was of significant interest to purify proteases from the rarely explored group of the halotolerant alkaliphilic actinomycetes. The enzymes displayed significant activity and stability at higher salt concentration, higher temperatures, alkaline pH and in the presence of different orgainic solvents. The enzymes were also resistant against various surfactants and inhibitors, an important feature for the potential application in the detergent industry. The solvent-tolerant alkaline proteases can be useful in wastewater treatment systems, where toxic organic solvents and/or surfactants are accumulated [29]. The structural elucidation of the enzymes by CD spectroscopy provided clues for the thermal and solvent stability of the enzymes.
ACKNOWLEDGMENT
The haloalkaliphilic actinomycetes employed in this study were originally isolated by Dr. S.D. Gohel in the Laboratory of Prof. S.P. Singh at the Saurashtra University, Rajkot, India. The authors are highly thankful to UGC, India and Saurashtra University, Gujarat for the infrastructural and financial support. BAK acknowledges the award of Senior Research Fellowship by the CSIR, New Delhi, India. FJT and AKS acknowledge the award of UGC-BSR Meritorious Research Fellowship. We are also thankful to CSIR-CSMCRI, Bhavnagar, India for providing the CD spectroscopic facilities.
COMPLIANCE WITH ETHICAL STANDARDS
Conflict of interests. The authors declare that they have no conflict of interest.
Statement on the welfare of animals. This article does not contain any studies involving animals performed by any of the authors.
REFERENCES
Thirumalai, M. and Arunachalam, P., Int. J. Biol. Macromol., 2017, vol. 97, pp. 552–560.
Sarmiento, F., Peralta, R., and Blamey, J.M., Front. Bioeng. Biotechnol., 2015, vol. 3, pp. 148–162.
Mhamdi, S., Ktari, N., Hajji, S., Nasri, M., and Kamoun, A.S., Int. J. Biol. Macromol., 2017, vol. 94, pp. 415–422.
Pant, G., Prakash, A., Pavani, J.V.P., Bera, S., Devirama, G.V.N.S., Kumar, A., et al., J. Taibah Univ. Sci., 2015, vol. 9, pp. 50–55.
Adrio, J.L. and Demain, A.L., Biomolecules, 2014, vol. 4, no. 1, pp. 117–139.
Li, W.J., Xu, P., Schumann, P., Zhang, Y.Q., Pukall, R., Xu, L.H., et al., Int. J. Syst. Evol. Microbiol., 2007, vol. 57, no. 7, pp. 1424–1428.
Kim, O.S., Cho, Y.J., Lee, K., Yoon, S.H., Kim, M., Na, H., et al., Int. J. Syst. Evol. Microbiol., 2012, vol. 62, no. 3, pp. 716–721.
Saitou, N. and Nei, M., Mol. Biol. Evol., 1987, vol. 4, pp. 406–425.
Kumar, S., Stecher, G., and Tamura, K., Mol. Biol. Evol., 2016, vol. 33, no. 7, pp. 1870–1874.
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D. G., Nucleic Acids Res., 1997, vol. 25, no. 24, pp. 4876–4882.
Kimura, M.A., J. Mol. Evol., 1980, vol. 16, pp. 111–120.
Felsenstein, J., Evolution, 1985, vol. 39, pp. 783–791.
Gohel, S.D. and Singh, S.P., Int. J. Biol. Macromol., 2013, vol. 56, pp. 20–27.
Gohel, S. D. and Singh, S. P., J. Chromatogr. B, 2012, vol. 889, pp. 61–68.
Laemmli, U.K. Nature, 1970, vol. 227, p. 680.
Meyer, T.S. and Lamberts, B.L., Biochim. Biophys. Acta, 1965, vol. 107, no. 1, p. 144.
Bradford, M.M., Anal. Biochem., 1976, 72, no. 1–2, pp. 248–254.
Hagihara, B., Enzymes, 1960, vol. 4, pp. 193–213.
Patel, R.K., Dodia, M.S. and Singh, S.P., Proc. Biochem., 2005, vol. 40, pp. 3569–3575.
Greenfield, N.J., Nat. Protoc., 2006, vol. 1, pp. 2876–2890.
Souza, P. M., Aliakbarian, B., Ferreira-Filho, E. X., Magalhães, P. O., Junior, A. P., Converti, A. and Perego, P., Int. J. Biol. Macromol., 2015, vol. 81, pp. 17–21.
Bennur, T., Kumar, A. R., Zinjarde, S., and Javdekar, V., Appl. Microbiol. Biotechnol., 2014, vol. 98, no. 22, pp. 9173–9185.
Salihi, A., Asoodeh, A. and Aliabadian, M., Int. J. Biol. Macromol., 2017, vol. 94, pp. 827–835.
Elhoul, M.B., Jaouadi, N.Z., Rekik, H., Bejar, W., Touioui, S.B., Hmidi, M., et al., Int. J. Biol. Macromol., 2015, vol. 79, pp.871–882.
Kelch, B.A., Eagen, K.P., Erciyas, F.P., Humphris, E.L., Thomason, A.R., Mitsuiki, S. and Agard, D.A., J. Mol. Biol., 2007, vol. 368, no. 3, pp. 870–883.
Rohamare, S.B., Dixit, V., Nareddy, P.K., Sivaramakrishna, D., Swamy, M.J., and Gaikwad, S.M., Biochim. Biophys. Acta—Proteins Proteomics, 2013, vol. 1834, no. 3, pp. 708–716.
Wang, F., Ning, Z., Lan, D., Liu, Y. Yang, B., and Wang, Y., Agric. Food Chem., 2012, vol. 60, no. 1, pp. 165–172.
Tafreshi, N. K., Hosseinkhani, S., Sadeghizadeh, M., Sadeghi, M., Ranjbar, B., and Naderi-Manesh, H., J. Biol. Chem., 2007, vol. 282, no. 12, pp. 8641–8647.
Dodia, M.S., Rawal, C.M., Bhimani, H.G., Joshi, R.H., et al., J. Ind. Microbiol. Biotechnol., 2008, vol. 35, pp. 121–131.
Protein Structure, Stability, and Interactions, Shriver J., Ed., Huntsville: Humana Press, 2009, pp. 227–260. https://doi.org/10.1007/978-1-59745-367-7.
Zhou, G. and Voelz, V.A., J. Phys. Chem. B, 2016, vol. 120, no. 5, pp. 926–935.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Thakrar, F.J., Kikani, B.A., Sharma, A.K. et al. Stability of Alkaline Proteases from Haloalkaliphilic Actinobacteria Probed by Circular Dichroism Spectroscopy. Appl Biochem Microbiol 54, 591–602 (2018). https://doi.org/10.1134/S0003683818100022
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683818100022









 0%,
0%,  5%,
5%,  10%,
10%,  15%,
15%,  20%,
20%,  50%) in the presence of 2 M NaCl and without salt.
50%) in the presence of 2 M NaCl and without salt.
 0%,
0%,  5%,
5%,  10%,
10%,  15%,
15%,  20%,
20%,  50%) in the presence of 1 M NaCl and without salt.
50%) in the presence of 1 M NaCl and without salt.