Abstract
The bacterial composition of permafrost samples taken during drilling of frozen marine sediments in the area of Barentsburg coal mine on the east coast of Grønfjord Bay of Western Spitsbergen has been studied. The study was based on the analysis of the V4 region of the 16S rRNA gene, carried out using next generation sequencing, as well as using classical microbiological methods (direct luminescence microscopy and aerobic cultivation).The total cell number in permafrost samples ranges from 6.73 ± 0.73 × 106 to 3.37 ± 0.19 107 cells per g. The number of cultivable aerobic bacteria in frozen samples on 1/5 TSA and R2A media ranges from 0 to 6.20 ± 0.45 × 104 CFU/g. Isolates of aerobic bacteria were identified by 16S rRNA gene analysis as representatives of the genera Arthrobacter, Pseudarthrobacter, Psychrobacter, and Rhodoferax. The dominant phyla of the domain Bacteria were Actinobacteria, Proteobacteria, Chloroflexi, Nitrospirae and Firmicutes. As a result of phylogenetic analysis of the dominant operational taxonomic units, representatives of methane oxidizing, sulfate reducing bacteria, as well as heterotrophic bacteria involved in the transformation of organic matter were found.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
INTRODUCTION
The Spitsbergen is a unique region of the Arctic, where fast processes associated with climate warming are recorded. According to meteorological data from Longyearbyen, the average annual air temperature increased during the 20th century from about –9 to –4°С (Humlum et al., 2003). The area of glaciers in the vicinity of Longyearbyen and Barentsburg decreased by about 50% from 1936 to 2017 (Chernov et al., 2018). Spitsbergen permafrost is the warmest within the High Arctic due to the warming influence of currents and air masses carried from the Atlantic with the West Spitsbergen current. In 2016, as part of the creation of a permafrost test site of the Russian Science Center on the eastern bank of Grønfjord Bay, boreholes were drilled to study the structural features of permafrost and to monitor ground temperatures (Demidov et al., 2016b; 2019). Permafrost coastal-marine sediments frozen in the late Pleistocene and Holocene after they emerged from under-sea level, were drilled. This study is a continuation of the ongoing work on a comprehensive study of permafrost in the vicinity of the Barentsburg mine. Comparative microbiological studies of the Arctic systems of sea water-marine sediment, as well as terrestrial-subsea permafrost show the differences between the dominant prokaryotic groups. For the domain Bacteria, studied on the Beaufort Sea shelf (Alaska), a change in the dominant groups was observed from Alpha-, Gammaproteobacteria and Bacteroides in seawater to Deltaproteobacteria, Firmicutes, Chloroflexi, Actinobacteria and Cyanobacteria in the underlying unfrozen marine sediments. A comparison of terrestrial and submarine permafrost about 2500 years old and studied in the area of Cape Mamontov Klyk (western Laptev Sea) showed an increase in the proportion of Deltaproteobacteria, Actinobacteria, Gemmatimonadetes, as well as the KD4-96 group (Chloroflexi), and a decrease in the proportion of phylum Firmicutes (Mitzscherling et al., 2019). The dominant groups of the domain Archaea in permafrost in the Cape Mamontov Klyk area were anaerobic methanogenic, ammonium-oxidizing and methane-oxidizing Archaea. The permafrost in the Buor-Khaya Bay (southern Laptev Sea) was dominated by groups of methanogenic and methane-oxidizing Archaea (Winkel et al., 2018). In turn, according to the DGGE analysis of 16S rRNA genes, marine shelf sediments in the D’Jurville Sea region (East Antarctica) contained mainly Deltaproteobacteria groups, putative sulfide oxidizing classes of Gammaproteobacteria, class Flavobacteria, Planctomycetacia, as well as the domain Archaea (Bowman et al., 2003). For Western Spitsbergen, there are currently few works describing the composition of prokaryotic communities of marine sediments and permafrost. Thus, pure cultures of Actinobacteria, Bacteroidetes, and Proteobacteria phyla (Alpha- and Gamma-) (Srinivas et al., 2009) were isolated from modern marine sediments of Kongsfjord Bay. The method of profiling for the 16S rRNA gene revealed an increase in the proportion of Deltaproteobacteria, Chloroflexi, and Planctomycetes during the transition from sea water to the marine sediment of the Smerinburg Fjord bay (Teske et al., 2011). The presence of 16S rRNA phylotypes forming endospores of thermophilic representatives of the Firmicutes phylum was revealed in unfrozen marine sediments of large fjords of the Western Spitsbergen (Müller et al., 2014). A study of surface and near-surface Holocene sediments in the Adventdalen to a depth of 2 m revealed a change in the phylum of domain Bacteria with Proteobacteria (Alpha-, Beta-, Gamma-), Acidobacteria, Veruccomicrobia, and Actinobacteria (Thermoleophilia order) in the seasonally thawed layer to significantly dominance of the phylum Actinobacteria (family Intrasporangiaceae) in the permafrost layer (Müller et al., 2018). However, studies on the ancient, deep permafrost on the Western Spitsbergen using the new generation sequencing method have not yet been carried out.
The aim of our work was to characterize permafrost coastal marine sediments, covering the entire altitude and age range of the ladder of marine terraces in the Barentsburg region, using the methods of analyzing the V4 region of the 16S rRNA gene of domain Bacteria, as well as cultivating aerobic bacteria at 10°C, in order to gain an idea of the structure of prokaryotic communities and microbiological processes that took place in marine sediments before their freezing for a long time. This is the first part of the study devoted to domain Bacteria.
SAMPLING SITE
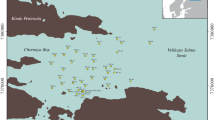
The study used permafrost cores from borehole (Bh) 1 (78.02289° N, 14.29845° E, 2.0 m above sea level, als) drilled at the mouth of the Grøn River, Bh 2 (78.09504° N, 14.24096° E, 75.5 m als) and 5 (78.09856° N, 14.23299° E, 43.0 m als) drilled on the eastern bank of the Isfjord Bay, and Bh 7 (78.04703° N, 14.21962° E, 8.0 m als), drilled at Cape Finneset (Fig. 1). These sediments were represented by sands, sandy loams, loams and clays of coastal-marine genesis (Fig. 2), which accumulated during the middle and late Holocene and then, during a sharp drop in relative sea level, came to the surface, were frozen and covered with a thin cover of continental sediments of various genesis (Forman et al., 2004; Svendsen and Mangerud, 1997). The average annual temperature of Bh 2 (25.09.2018–25.08.2019) at a depth of 5.5 m was ‒2.17°С and a one-time measurement (12.09.2016) of the Bh 7 temperature at a depth of 12.5 m was -0.87°С (Demidov et al., 2020). Despite the fact that in the modern era these sediments are frozen, samples from depths less than 3 m could have been thawed during the Holocene warming, when the seasonal thawing depth exceeded the modern one. In the case of Bh 1, a short-term rise in sea level might caused temporarily thawing of the sediments which nowadays are frozen (Solovieva et al., 2018; Salvigsen and Høgvard, 2005). Samples taken for next generation sequencing were numbered according to Bh numbers and sampling depths (S1-1, S2-4, S5-3, S7-7, S7-9 and S7-12). Soil sample above Bh 1 from a depth of 5 cm was assigned to the type of Gleysols and represented the oxidized gley horizon Gox (Alekseev and Abakumov, 2016).
The region of the study area in the Western Spitsbergen Island (a), permafrost test site in Barentsburg showing the locations of boreholes 1, 2, 5 and 7 (b) and the precise locations of boreholes 2 and 5 (c) (using maps on the websites https://google.ru/maps and https://toposvalbard.npolar.no).
RESEARCH METHODS
Sampling. Drilling was carried out in August and September 2016 using the UKB 12/25 drilling machine (Vorovskiy Head, Ekaterinburg, Russia). The coring was undertaken without washing and without adding chemical reagents. Thin-walled core pipes with an external diameter of 76 to 112 mm were used. After cleaning the surface of the frozen core segments with a sterile scalpel, the samples were placed in sterile bags (Whirl-Pak®, Nasko, USA) and stored at temperatures –4 to –10°С. On 28 October 2016 they were transported to the Laboratory of Climate and Environmental Changes (AARI, St. Petersburg), where they were stored in a freezer at –18°С until analysis in the period from October 2016 to February 2020.
Analysis of carbon monoxide, carbon dioxide, methane and ethylene concentrations. Gas was collected using headspace degassing in 150 mL syringes (Alperin et al., 1985). Special analysis of methane concentrations in the samples was carried out in triplicate on “Kristall 5000.2” gas chromatograph (CJSC Chromatek, Yoshkar-Ola, Russia) with a flame ionization detector. Chromatograph technical parameters and analysis conditions were: chromatographic column length of 3 m, diameter of 2 mm, temperature of 60°С, adsorbent—Hayesep-N 80/100, flame-ionization detector temperature of 150°С. The sampler volume was 0.250 mL, the volume of the introduced gas sample was 3–8 mL. Nitrogen (99.999% purity) with a flow rate of 30 mL / min was used as a carrier gas. The consumption of hydrogen is 20 mL/min, air – 400 mL/min. To calibrate the chromatograph, gas mixtures were used (Monitoring LLC, St. Petersburg, Russia) with the following methane concentrations: 0.49 ± 0.07 ppm, 5.3 ± 0.5 ppm, 10.3 ± 0.6 ppm, 100 ± 5 ppm, 1000 ± 30 ppm.
The isotopic composition of methane and carbon dioxide. The δ13С of methane and carbon dioxide from the VPDB (Vienna Pee Dee Belemnite) standard (Miller and Wheeler, 2012) of a sample from a depth of 2.3 m from Bh 7 was experimentally measured using a Delta V Advantage mass spectrometer combined with a Thermo Science Trace 1310 gas chromatograph (Thermo Fisher Scientific, USA) at FSBI VNIGNI (Aprelevka, Russia).
Total organic carbon measurements and radiocarbon dating. For Bh 2 and 7 a total of 18 samples were analyzed for total organic carbon (TOC) content. Values are given as weight percentage (wt %). The analyses were performed using an elemental analyzer (Elementar Vario MAX C; analytical accuracy ± 0.1 wt %). Hand-picked organic material from two samples of Bh 7 was radiocarbon-dated by Accelerator Mass Spectrometry (AMS) using a Mini Carbon Dating System (MICADAS) at Alfred Wegener Institute (AWI) Helmholtz Center for Polar and Marine Research. Further details on laboratory procedures and sample pre-treatment are given in Opel et al. (2019) and Mollenhauer (2021).
All radiocarbon dates were calibrated using the IntCal13 calibration dataset (Reimer et al., 2013). Ages were given as calibrated years before present (cal BP).
Total cell count. Samples were transferred to sterile 15 mL Falcon tubes (Eppendorf, Germany). After that, 3-5 ml of PBS buffer (composed of 137 mM NaCl, 2.7 mM KCl, 15 mM Na2HPO4, and 1.7 mM KH2PO4) (pH 7.6 in water) as described in Mitzscherling (2017) was added and final sample volume was recorded. Then the samples were vortexed to a homogeneous suspension and treated by low intensity sonication in a 35 KHz ultrasonic bath (Sapfir, Russia) at power setting of 200 W for 30 s. Then the tubes were put in a rack for 10 minutes to separate the coarse grainmineral phase, 100 µL of liquid from the middle of the suspension were transferred to a 0.5 mL tube, mixed with100 µL of 0.01% acridine orange and incubated for 1 min at room temperature. A total 2 µL of the stained suspension was applied to a microscopic slide and covered with 1818 mm cover glass. Fluorescence microscopy was performed with a Zeiss Axio Lab.A1 epifluorescent microscope using the filter cube Fs.09 (BP 450-490, FT 510, LP515) for acridine orange. Cell counting was carried out in triplicate, in field of vision (d = 200 µm) with 10 fields view for each repetition. Cell concentration in the suspension was calculated by multiplying an average cell count per field by 1.032 × 107. Initial cell concentration per gram of the sample was calculated by multiplying the cell count in the suspension by the volume of the suspension and dividing the result by the mass of the initial sample.
Cultivation of aerobic bacteria. To determine the number of aerobic bacteria, frozen samples weighing 0.4–2.7 g from the inner core were taken in the sterile zone of the burner flame and placed in plastic tubes with 4.9–7.4 mL of autoclaved distilled water and manually suspended for 10 minutes until the particles were uniformly distributed throughout the volume. Suspensions were plated on solid nutrient media 1/5TSA (tripticase-soy medium, FlukaTriptic Soy Broth No.2, USA) and R2A (DifcoTM, USA), widely used for permafrost samples (Vishnivetskaya et al., 2000; Gilichinsky et al., 2007; Steven et al., 2007) in 1 : 10 and 1 : 100 dilutions in duplicates and were cultivated for 15 days at an average temperature comfortable for the growth of the psychotolerant component of the community 10°С. The calculation of the number of cells was carried out according to the formula: \(N = \left( {a{{\;}} \times P \times {{\;}}b} \right) - m\), where “a” is the number of colonies on a plate; “P” – is the dilution; “b” is the number of drops in one mL; “m” is the mass of the permafrost sample.
DNA isolation, preparation and sequencing of amplicon libraries. DNA from the samples was isolated using Fast DNA Spin Kit for Soil, according to the manufacturers method (MP Biomedicals, USA). Concentration was measured using a Qubit 2.0 fluorimeter with a dsDNA HS reagent KIT (InvitrogenTM, USA). Amplicon libraries were created using PCR with universal primers for region V4 in accordance with the previously described methodology (Fadrosh et al., 2014) at the Vernadsky Institute of Microbiology. Primers were selected for the most objective ratio of Bacteria and Archaea domains: 515F (5'-GTGBCAGCMGCCGCGGTAA-3') (Hugerth et al., 2014) and Pro-mod-805R (5'-GACTACNVGGGTMTCTAATCC-3') (Merkel et al., 2019). Sequencing was performed on a MiSeq system (Illumina, USA) at “Biospark” (Moscow, Troitsk, Russia) using a reaction MiSeq Reagent Micro KIT v2 that reads 150 nucleotides from each end. Each sample was read in two technical duplicates, including the reagents and air control sample used to subtract the contaminant sequences from the study sample. A total of 164843 sequences were obtained.
Bioinformatic and statistical analysis. Demultiplexing as well as subsequent processing and sequence analysis were performed using the appropriate scripts in QIIME 2 ver2019.1 software (Bolyen et al., 2019). OTUs (Operational Taxonomic Unit) were identified using the SILVAngs 1.4 pipeline (https:// ngs.arb-silva.de/silvangs/) and BLAST (http://blast. ncbi.nlm.nih.gov/Blast.cgi) programs. The coverage index of amplicon libraries was calculated by the formula \(C = {{\;}}1 - {n \mathord{\left/ {\vphantom {n N}} \right. \kern-0em} N}\), where “n” is the number of OTUs represented by one amplicon, “N” is the total number of amplicons (Good, 1953). The Chao1 index (Chao, 1984) was calculated by the formula \({\text{Chao1}} = {{S}_{{{\text{obs}}}}}\) + \(\left( {{{{{a}^{2}}} \mathord{\left/ {\vphantom {{{{a}^{2}}} {2b}}} \right. \kern-0em} {2b}}} \right)\), where “Sobs” is the identified number of phylotypes (OTU), “a” is the number of phylotypes (OTU) represented by one amplicon, “b” is the number of phylotypes represented by two amplicons. The Shannon–Weaver Index (Magurran, 1992)—according to the formula \(H{\kern 1pt} ' =- \sum\nolimits_i {p\ln ~{{p}_{i}}} ~\), where “pi” is the relative abundance of i-th philotype (OTU). The relative abundance of technical replications was combined to indicate the relative content for analysis of the bacterial community using bubble graph diagrams. The analysis of main components and cluster analysis were performed using the Past3 statistical package (Hammer, 2017) using the paired group algorithm (UPGMA) and the Euclidean affinity index.
Identification of isolates by 16S rRNA gene sequences. DNA purification (http://evrogen.ru/kit-user-manuals/BC023.pdf), PCR with standard primers 27f (5'-AGAGTTTGATCMTGGCTCAG-3') and 1492r (5'-TACCTTGTTACGACTT-3') (Lane, 1991) according to the program 96°С—5 min, (96°С—10 s, 60°С—20 s, 72°С—1.5 min) 20 cycles, 72°С—1 min, as well as the determination of 16S rRNA sequences were performed at Evrogen (Moscow, Russia). The sequences were edited in Chromas software (https:// chromas.software.informer.com) and aligned with each other using the Clustal Omega software (https:// www.ebi.ac.uk/Tools/msa/clustalo). The homologues were searched using the BLAST software in GenBank global database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic analysis was performed using Mega X software (Kumar et al., 2016) with the Maximum Likelihood algorithm according to the Tamura-Nei model (Tamura and Nei, 1993).
Deposit at GenBank. The sequence of regions V4 16S rRNA were loaded into the NCBI database as bioproject PRJNA625477 (SRX8122453-SRX8122466). The gene sequences of 16S rRNA isolates were transferred to the NCBI database under the numbers: MN599988-MN599993.
RESULTS
Methane and other carbon-containing gases in samples of Bh 7 near Cape Finneset. The methane content in samples of Bh 7 ranged from 0.00524 ± 0.00007 to 0.75572 ± 0.00426 mL/kg (Fig. 2). The δ13С values for methane from a 2.3 m below surface (bs) sample ranged from –51.71 to –52.03‰ VPDB, and for CO2 they were –17.31 to –18.44‰ VPDB.
Radiocarbon dating and total organic carbon data. The median calibrated age at 8.9 m bs (Bh 7) is 15349 cal BP and at 11.6 m bs 4310 cal BP (Table 1). Taking into account the reservoir effect as estimated for marine sediments on Western Spitsbergen, the true age may be 400 years less than calculated (Mangerud, 1972). The organic carbon content in Bh 2 ranged from 0.85 to 0.92%, and from 0.84 to 1.01% in Bh 7 (Fig. 2).
Total cell count and number of cultivated aerobic bacteria. Total cell number in permafrost samples was 6.73 ± 0.73 × 106 to 3.37 ± 0.19 × 107 cells per g and 8.49 ± 1.31 × 107 cells per g in soil. The number of aerobic bacteria in permafrost horizons ranged from 0 on R2A medium and 1.2 ± 0.56 × 101 on 1/5 TSA medium to 6.20 ± 0.45 × 104 CFU/g on R2A medium, and in the soil from a seasonally thawed layer in the area of Bh 1 on both media was 2.16 ± 0.00 × 105 CFU/g (Table 2). The variety of colonies on the dishes was small and varied from one to three morphological types of colonies having white, beige, yellow and red color.
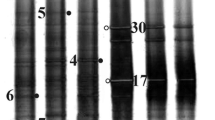
Identification of aerobic bacterial isolates by the 16S rRNA gene. Four of the isolates identified by us turned out to be representatives of the phylum Actinobacteria, family Micrococcaceae, genera Arthrobacter and Pseudarthrobacter, and two were representatives of class Betaproteobacteria, family Comamonadaceae, genus Rhodoferax, and of the phylum Gammaproteobacteria, family Moraxellaceae, genus Psychrobacter (Fig. 3).
Phylogenetic position of isolates from the permafrost of Western Spitsbergen near the Barentsburg mine based on 16S rRNA gene sequences between the nearest type strains from NCBI database (Maximum Likelyhood, Tamura-Nei model, bootstrap method with 500 replicates, very strong branch swap filter). The names of the isolates correspond to the core number and layer depth in cm.
Profiling of the V4 region of 16S rRNA. Genomic DNA was isolated from all samples: in concentrations from 0.1 to 0.8 ng/μl for samples of Bh 1 and 7 and about 0.02 ng/μl for samples of Bh 2 and 5. The content of archaeal DNA in the total prokaryotic DNA did not exceed 1% (Table 3). Among the obtained sequences, from 0.08% (S1-1) to 0.35% (S7-9) could not be identified using SILVAngs 1.4 pipeline.
Taxonomy of bacterial OTUs. In total, 5667 ± 619 OTUs of the Bacteria domain were revealed (3331 ± 427, 159 ± 18, 524 ± 56, 775 ± 60, 95 ± 35, 783 ± 23 for samples S1-1, S2-4, S5-3, S7-7, S7-9, S7-12, respectively), of which 105 were dominant (≥1%) OTUs (18, 13, 19, 23, 7, 25, respectively). A total of 11 phyla were found, and the phyla Actinobacteria and Proteobacteria were dominant in all the studied samples (Fig. 4). The dominant family of the Actinobacteria class was the family of the Actinomycetales order – Intrasporangiaceae. Another widely represented Thermoleophila class included mainly the orders Gaiellales (33–100%) and Solirubrobacterales (3–67%). Acidobacteria phylum was dominated by the classes Acidobacteria (6–100%), Aminicenantales (3–73.5%), Holophaga (2.5–41.5%) and the Subgroup 6 group (8.5–46%). The Gammaproteobacteria class prevailed in sample S1-1 (43%), where it consisted of the methanotrophic family Methylobacteriaceae (76.5%), and in sample S5-3 (71.5%), where 98.5% consisted of the family Pseudomonadales. The Deltaproteobacteria class accounted in sample S2-4 0.8% and Desulfobacteraceae family was represented by 77%. The dominant family in the Betaproteobacteria class in all samples except S7-9 was the Burkholderiaceae family (12.5–99.5%). The Hydrogenophilaceae family (0.0045–1%), 100% represented in samples S1-1, S5-3, S7-7, and S7-12 by the Thiobacillaceae family, also constituted a significant proportion. A smaller proportion was represented by the Sulfuricellaceae family (0.24–38%) except for S7-12, Nitrosomonadaceae (6.5–19%) except for S2-4, Gallionellaceae (4% in S1-1, 1.9% in S7-7 and 6.5% in S7 -9) and Methylophilaceae (8% in S1-1 and 1.4% in S7-7). The Alphaproteobacteria class was dominated by the order Rhizobiales (25.5–63%) and families Acetobacteraceae (1.5–42.5%), and Sphingomonadaceae (2.5–57%). The phylum Bacteroidetes was mainly represented by the class Bacteroidia (in all samples 50–100%). Chlorobi phylum was represented by the class Ignavibacteria (in S1-1—0.9%, S7-7—25.5%, S7-12—8%), and in the S7-12 sample it was completely represented by the OPB56 group, and in the S7-7 sample it was dominated by the group BSV26 (93%). In sample S1-1 with a 50% proportion of the order of Ignavibacteria, the BSV26 group was composed, and the OPB56 and PHOS-HE36 groups—25% each. Phylum Chloroflexi was represented in the studied samples by the classes KD4-96 (0–61%), Gitt-GS136 (S1-1—30% and S2‑4—73.5%), P2-11E (S1-1—16% and S5- 3—1%), OLB14 (S1-1—7%), Chloroflexia (S1-1—3%, S2-4—16% and S7-12—1.5%), Anaerolineae (2.5–81.5%) and Dehalococcoida (S1- 1—0.95%, S7-7—12% and S7-12—7.5%). Phylum Planctomycetes was present in all samples: (0.005–3%). In S1-1, S7-7 and S7-12 it was mainly represented by the class Planctomycetacia (89%, 68.5%, 53.5%), in S2-4—by the class OM190 (100%), in S7-7 by the class SM1A02 (71.5%), and in S7-9—by the class Phycisphaerae (100%). Firmicutes phylum was represented mainly by the classes Bacilli and Clostridia. Bacilli made up 19% of the phylum in sample S1-1, including 16% of the Lactobacillaceae family. The same family made up 3% of Bacilli in sample S2-4, which in turn was 11% of Firmicutes, while sample S5-3 was dominant, where Bacilli accounted for 38% of Firmicutes. The order of Lactobacillales was also represented by the Carnobacteriaceae family (10% for sample S1-1, 13.5% for sample S7-7 and 53% for sample S7-12). The class Clostridia was mainly represented by the order Clostridiales (96.5–100%). Among its representatives, the genus Desulfosporosinus of the family Peptococcaceae was found in all studied samples and amounted to 5–75% of the class. Representatives of the Family XI were present in samples S1-1, S2-4, and S7-12 and amounted to 3, 0.3, and 43%, respectively. In sample S7-12, the Caldicoprobacteriaceae family (14.5% of Clostridia) was found. Phylum Gemmatimonadetes had the largest proportion in sample S1-1, but was also found in Bh 7 (0.25–1%). The class Gemmatimonadetes was 100% in sample S1-1 and 61% in sample S7-12. In sample S7-9, 16.5% of the class was PAUC43f marine benthic group, and in sample S7-12 27% was the Longimicrobiaceae family. Representatives of the phylum Cyanobacteria were found only in samples S1-1, S2-4, S7-7 and S7-12. In samples S2-4 and S7-7, Cyanobacteria were represented by the Gastranaerophilales family, and in sample S7-12 they were also represented by the families Nostocaceae and Leptolyngbyaceae (43.5% each). Phylum Nitrospirae was found in all samples. In S1-1 and S7-12, it was represented mainly by the Nitrospiraceae family (99.5 and 54%). Other representatives of phylum Nitrospirae in these samples were group 4-29-1 (0.45% in S1-1 and 5.5% in S7-12). In sample S2-4, phylum Nitrospirae was represented by one Thermodesulfovibrionia family. Sample S5-3 was also dominated by only one group of BMS9AB35. In samples S7-7 and S7-9, the group 4-29-1 (65 and 15%, respectively) and the Thermodesulfovibrionia family (34 and 85%) dominated. Candidatus Atribacteria was represented by the JS1 group, which dominates in sample S7-12 (12%), and was found in a small proportion in samples S1-1 (0.005%), S2-4 (0.25%), S5-3 (0.065%) and S7-7 (0.075%).
The minor components for samples S1-1, S2-4, S5-3, S7-7, S7-9 and S7-12 were 4.47, 3.46, 2.98, 12.93, 7.80 and 4.70% of the total variety, respectively. They were represented by the phyla Caldeserica, Chlamydiae, Deinococcus-Thermus, Patesibacteria and Veruccomicrobia.
DISCUSSION
The content and origin of methane in Bh 7 near Cape Finneset. The obtained δ13C (VPDB) values for methane correspond to its thermogenic or thermogenic with an admixture of microbial methane (Shakirov et al., 2014). The values for CO2 lie in the range below the values characteristic of a metabolic origin and higher than the values characteristic of mantle and atmospheric CO2 (Yudovich and Ketris, 2010). According to data (Hitchman et al., 1989), these values fall into the ranges of values for biogenic methane (–40 to –109‰ for deep sea sediments, lake sediments, swamps and marches) and thermogenic (–12 to –71‰ for carbon strata) genesis, and according to data (Taranik, 2017) to the ranges obtained by studying gases in the coal mines of Donbass. Due to the absence of methanogenic OTU in samples S7-9 and S7-12 (and the presence of an insignificant proportion of them in sample S7-7), but a significant proportion of anaerobic methane-oxidizing Archaea in these samples, we believe that it cannot be excluded that methane from Bh 7 could be introduced into these Quaternary sediments before freezing along with gases (free or dissolved in groundwater) coming from the Tertiary coal-bearing deposits. This assumption can explain the boundary value of δ13C (VPDB) for methane detected in the sample from a depth of 2.3 m between methane of biochemical genesis (–50 to –80‰) and gas and oil fields (–30 to –50‰) according to Taranik (2017). A gas leak with some liquid hydrocarbons on Cape Finneset was observed in 1911 (Senger et al., 2019), which also supports possibility of thermogenic methane presence in permafrost samples from Bh 7.
Radiocarbon dating and total organic carbon data. The obtained dating of Quaternary permafrost rocks from Bh 7 confirms that the marine terraces in the Barentsburg region have an age range from the late Pleistocene to the Holocene. Total organic carbon amount in Bh 2 and 7 was commensurate with total carbon content, mainly consisting of TOC. The overall TOC content is low barely exceeding 1 wt %.
The results of analysis of the organic carbon content showed its uniform distribution over the Bh, which indicates that the revealed differences in the microbiological characteristics of the permafrost cannot be a consequence of variations in the organic carbon content.
The number of cultivated aerobic bacteria. Total bacterial number of soil and permafrost near Barentsburg was higher than in near-surface 1.8 m layer of permafrost soil in Ny-Ålesund (1.14 × 105–3.98 × 106 cells per g) (Singh et al., 2017) and less than in modern marine sediments (2.1 to 4.7 × 109 cells per mL of wet sediment in Smeerinburgfjorden (Ravenschlang et al., 2001) and 2 × 108 to 4 × 109 cells per mL of wet sediment near Western Spitsbergen (Arctic Ocean) and Tromsø (northern Norway)) (Sahm and Berninger, 1998).
The number of cultivated aerobic bacteria that we obtained in frozen sediments of marine genesis was comparable to that obtained at 10°С in bottom marine sediments in the Ny-Ålesund region, which amounted to 103–104 CFU/g (Srinivas et al., 2009), in turn, our soil values were comparable with those for Ny-Ålesund soils, where it ranged from 103 to 106 CFU/g (Singh et al., 2017). The obtained abundance values for Western Spitsbergen permafrost turned out to be less than those for the previously studied permafrost sediments, using similar nutrient media and temperatures of culturing, of the Kolyma Lowland (Khlebnikova et al., 1990; Vishnivetskaya et al., 2000) and the Gydan Peninsula (Demidov et al., 2016a) and are closer in order to the CFU/g values found in the Antarctic oases (Karaevskaya et al., 2017) (Fig. 5). The formation of rock cryogenic weathering products in the coastal marine zone under the conditions of demolition, the dissected relief of the surrounding land and the presence of glaciers on it—all this brings together the conditions for the formation of permafrost sediments on Western Spitsbergen and in the Antarctic oases. On the contrary, on the Gydan Peninsula and the Kolyma Lowland, precipitation formed on vast plains or on the surrounding shelf. At the same time, the warmest permafrost in Western Spitsbergen has the smallest CFU/g abundance among the Arctic permafrost. Similarly, for Antarctica, the warmest King George Island permafrost has the smallest CFU/g amounts (Karaevskaya et al., 2017), this may be due to the fact that freezing and thawing processes lead to a reduction in bacterial biomass and impoverish diversity of soil microbial communities (Schimel et al., 2007).The morphological diversity of the colonies for the studied samples was small and comparable with the permafrost of the early Pleistocene Olyor and Tomus-Yar suites of the Kolyma lowland (Khlebnikova et al., 1990) (1–3 morphological unites per sample), while in the permafrost of Antarctic oases with comparable values of CFU/g, it varied from 1–3 to 9 morphological unites per sample (Karaevskaya et al., 2017).
Comparison of cultivated aerobic bacteria CFU/g number in Western Spitsbergen permafrost with data from other previously studied regions of the Arctic and Antarctic, obtained using similar nutrient media and temperatures of culturing. Under the name of the district, the average annual temperature of the permafrost is indicated; the medium and cultivation temperatures are indicated in brackets. Data from Kolyma lowland were taken from (Khlebnikova et al., 1990; Vishnivetskaya et al., 2000), from Gydan Peninsula—from (Demidov et al., 2016 a), for Antarctica—from (Karaevskaya et al., 2017).
Isolates of aerobic bacteria. Two of the isolates we obtained, B1-135-1 (MN599988) and B7-1170-1 (MN599990), were 99.8 and 99.0% similar, respectively, to the type strain of Arthrobacter psychrochitiniphilus GP3T isolated from Subantarctic soils which was capable to hydrolyze chitin at 10°С (Wang et al., 2009). Isolate B1-135-2 (MN599992) was 99.8% similar to the type strain of Pseudarthrobacter oxydans DSM 20119T isolated from Subantarctic water, which has proteinase activity at 5°С (Reddy et al., 2009). Isolate B7-1170-2 (MN599991) was 99.8% similar to the type strain of Arthrobacter alpinus S6-3T isolated from alpine soil (Zhang et al., 2010). Isolate B5-310-1 was 97.5% similar to the type strain ED16T of purple non-sulfur bacterium Rhodoferax saidenbachensis isolated from sediments of a reservoir in Saxony (Kaden et al., 2014). Isolate B5-310-2 was 99.1% similar to the type strain of Psychrobacter glaciei Blc20019T, which was isolated both from alpine soil and from soils of the Adelaide Island in Subantarctic (Zeng et al., 2016; Romaniuk et al., 2018). All these genera are found in the corresponding samples according to the analysis of the V4 region of the 16S rRNA gene and make up from the domain Bacteria: 0.65% (Arthrobacter) and 0.1% (Pseudarthrobacter) in S1-1, 0.035% (Psychrobacter and Rhodoferax) in S5-3 and (Arthrobacter) in S7-12 (Fig. 3).
In the literature describing the cultivated aerobic bacterial diversity of marine bottom sediments near the Western Spitsbergen Island, there is a description of the various aerobic genera’s representatives with a predominance of representatives of Pseudomonas, Psychrobacter and Arthrobacter (Srinivas et al., 2009; Konieczna et al., 2011). These species could enter the studied rocks from the ice-free surfaces of the archipelago, or from fresh water bodies.
Characterization of amplicon libraries. The sample S1-1 was characterized by the greatest variety, it’s Shannon–Weaver index was 5.0 ± 0.1, Chao1 index—8540 ± 1156, while the library coverage was 96.3 ± 4.3% (Table 3). The samples of Bh 7 were less diverse, for which the Shannon-Weaver index was 4.1 ± 0.6 to 4.7 ± 0.1, and Chao1—452 ± 169 to 1242 ± 83. Sample S7-9 with the lowest diversity indices and low library coverage, 53.6 ± 9.6%, was knocked out of the general picture as compared with the other two samples S7-7 and S7-12 (96.8 ± 0.4–98.2 ± 0.1%). This phenomena may exist due to unfavorable conditions for the conservation or isolation of DNA in this sample, which differs from others in its high content of plant residues. Samples S2-4 (2.5 ± 0.1) and S5-3 (2.8 ± 0.1) were characterized by the lowest diversity by the Shannon–Weaver index, while the library coverage in them was very good and amounted to 98.3 ± 1.1 and 98.7 ± 0.1, respectively. This may be due to the very low output concentrations of DNA, and this, in turn, to their older age, compared to the samples of the lower marine terraces exposed by Bh 1 and 7.
Taxonomy of dominating OTUs of domain Bacteria. Here we considered the taxonomy of OTUs, comprising >1.5% in the amplicon libraries of domain Bacteria (Fig. 6, Table 4).
Samples S1-1 and S2-4 were characterized by the dominance of the class Actinobacteria of the family Intrasporangiaceae and the genus Oryzihumus, closely related to the type aerobic strain Oryzihumus soli Aerobe-19T (Table 4), growing at 15 and 30°C and pH 5.0 and 7.0, hydrolyzing gelatinous cellulose, esculin, which produces esterase (C4, C8) and lipase (C8), which reduces nitrate to nitrite (Kim et al., 2017).
Sample S1-1 was also distinguished by the dominance of representatives of the class Gammaproteobacteria, most closely related to the type strain of the methylotrophic bacterium Methylobacter tundripaludum SV96T, which bore the greatest similarity to the clones found in waters contaminated with wastes from the coal industry, as well as in hot springs in Japan (Table 4). The phylum Gemmatimonadetes, noted in large numbers in Sample S1-1, includes inhabitants of soil and waste water. In its representatives the center of photosystem II has been found (Zeng et al., 2015, 2017).
The Thermoleophila order, which dominated in samples S2-4, S5-3, S7-7, S7-9, and S7-12, was represented by the family Gaiellaceae, the only type representative of which is Gaiella occulata F2-233T, a strictly aerobe that assimilates some sugars, growing between 15 and 45°С and pH 5.0 and 9.0, NaCl content less than 1%, reducing nitrate to nitrite, was found in deep mineral waters of Portugal (Albuquerque et al., 2011).
The dominant OTUs of a sand sample S5-3 from the class Gammaproteobacteria were represented by the Pseudomonadaceae family, some of which are found in marine habitats (Fendrich, 1988). The closest clones dominating in the OTU sample were found in groundwater, subglacial waters, and subantarctic soils, which were also exposed to pollution by hydrocarbons and heavy metals (Table 4).
The highest proportion in all samples from borehole 7 was the class Deltaproteobacteria, which includes the majority of sulfate-reducing bacteria. The genus Desulfovibrio was a sulfate-reducing aerotolerant bacteria of aquatic ecosystems (Kuever et al., 2015).
The phylum Nitrospirae dominated in samples S7-7 and S7-9, where it was represented by the order Nitrospirales and the family Thermodesulfovibrionia.
Among the representatives of the Clostridia class, it is worth noting the genus Desulfosporosinus, which dominated in samples S2-4, S7-7, and S7-9. Its closest representatives were the inhabitants of fresh and sea waters polluted by hydrocarbons (Table 4), being obligate heterotrophs and using sulfur, thiosulfate and sulfites as an electron acceptor.
The phylum Chloroflexi, including aerobic thermophiles, anaerobic phototrophic bacteria, and halogenophiles (Gupta et al., 2012), dominated in borehole no. 7, being represented by the Anaerolinaceae family of the Anaerolinae class. In turn, the KD4-96 class of the phylum Chloroflexi, which dominated in the sample S7-9, was found in the cold moist meadow soil of the alpine tundra in the Colorado Rocky Mountains (Costello and Schmidt, 2006); it is also often found in soils contaminated with metals (Golebiewski et al. 2014), where its content correlates with the concentrations of iron and aluminum (Wegner and Liesack 2017).
Sample S7-12 was dominated by OTUs belonging to the order Clostridiales. These were representatives of the genus Tissierella, which were closest to the type T. creatinini strain DSM 9508T, which is capable of cleaving creatinine (Farrow et al., 1995; Harms et al., 1998). Also, OTUs of the JS1 group, belonging to the phylum Candidatus Atribacteria, which is widespread in methane-rich anaerobic sediments, including the deep-sea sediments of the Adel Basin in East Antarctica, also constituted a significant portion of it (Carr et al., 2015). In addition, representatives of the phylum Actinobacteria of the WCHB1-81 group, common in fresh waters, including those contaminated with methane (Inceoglu et al., 2015), hydrocarbons, and a chlorinated solvent (Dojka et al., 1998), and cyanotoxins (Dziga et al., 2019).
Thus, as a result of studying the structure of prokaryotic communities in several samples taken from different marine terraces in the area of the Barentsburg mine, we can assume the presence in them before freezing microbiological processes that are generally similar (transformation of organic substances, sulfate reduction) or prevailing mainly on certain terraces: exposed Bh 1, 2 (methanogenesis), Bh 1, 5 (anaerobic ammonium oxidation), Bh 1, 7 (anaerobic oxidation of methane).
Genetic relationship of samples. Based on the analysis of the main components, according to the taxonomic diversity of the Bacteria, the samples S7-7 and S7-12 were closest to each other, the next in similarity was sample S7-9. The isolation of sample S7-9 in Bh 7 can be explained by the increased content of plant material fragments in it, which is most likely due to sedimentation conditions different from the other two samples. Another cluster is formed by samples S1-1 and S5-3 (Fig. 7). In all cases, S2-4 is the most different of all samples. This may be due to different conditions of sediment formation, and with different age of the sediments of Bh 1-7 and Bh 2.
PCA of taxonomical data for domain Bacteria for two technical repetitions (A). Vectors show selected taxonomical factors that are mainly responsible for the variance between samples. Cluster analysis of average abundance values using the paired group algorithm (UPGMA) and the Euclidean affinity index (B).
According to the composition of the dominant OTUs of the domain Bacteria, samples in Bh 5 and 7 were similar to samples of terrestrial and submarine permafrost at Cape Mamontov Klyk, in which phyla Actinobacteria (Thermoleophilia), Chloroflexi (Gitt-GS-136, KD4-96), Firmicutes (Clostridia), Gemmatimonadae prevailed, Alpha-, Betaproteobacteria, as well as Candidatus Atribacteria were found in samples at the boundary of permafrost and the active layers (Mitzscherling et al., 2017, 2019). A special feature of Bh 7 is the presence in samples S7-7 and S7-9 with an abundance of >3% OTU from the phylum Nitrospirae (Thermodesulfovibrionia family), which, apparently, are involved in the sulfate reduction process. In turn, sample S2-4 is similar in bacterial composition to the permafrost layer of late Holocene well-drained loess of Adventdalen Valley (Müller et al., 2018) by the dominance of the phylum Actinobacteria (family Intrasporangiaceae and class Thermoleophilia). According to published data, phyla of Proteobacteria, Firmicutes, Chloroflexi, Acidobacteria, Actinobacteria, and Bacteroidetes are also generally distributed in the Arctic active layer and permafrost layers (Jansson and Taş, 2014; Taş et al., 2018). The phyla Actinobacteria, Chloroflexi, Firmicutes, Gemmatimonadetes, and Proteobacteria were active under freezing temperatures (Coolen and Orsi, 2015; Tuorto et al., 2014). Betaproteobacteria and Firmicutes were previously considered as the most adapted to life at low temperatures and low nutrient content groups, with Firmicutes representatives capable of spore formation, which also explains their significant proportion in frozen ecosystems (Johnson et al., 2007). It has been shown that representatives of Candidatus Atribacteria from gas hydrate sediments survive at high salt concentrations and freezing temperatures (Glass et al., 2019), and are also often found in subseafloor anaerobic ecosystems with low organic matter content (Orsi, 2018).
CONCLUSIONS
For the first time a comparative study of Western Spitsbergen permafrost representing marine terraces located in different sites of the east bank of Grønfjord was carried out, using mutually complementary methods—profiling of the V4 region of 16S rRNA gene, total cell count and aerobic cultivation of psychrotolerant part of the prokaryotic community. Total cell number in permafrost samples was about 107 cells per g that was two orders of magnitude less than in modern marine sediments and one or two orders of magnitude more than in near-surface permafrost layers. The number of cultivated aerobic bacteria of Western Spitsbergen permafrost reached 104 CFU/g was one and a half to three orders of magnitude lower than in other previously studied samples of lower temperature Arctic permafrost.
Decreasing of the CFU’s number and in the diversity of permafrost prokaryotic communities with near zero negative temperatures shows how vulnerable are the ancient permafrost microbial communities during the period of global warming.
Characterized by the predominance of bacterial phyla Actinobacteria, Proteobacteria, Chloroflexi, Nitrospirae, Firmicutes the communities were similar to the communities of modern coastal and marine coastal sediments. Presumably, they were formed mainly under anaerobic, but also under mixed aerobic-anaerobic conditions.
Thus, the functional role of the studied prokaryotic communities seems to be reduced to heterotrophic psychrophilic activity in all samples, anaerobic oxidation of methane by bacteria of the genus Methylobacter (in sample S1-1 from Gren river estuary) and sulfate-reducing activity of bacteria of the phyla Firmicutes and Nitrospirae (in samples from fourth marine terrace of east coast of Isfjord—S2-4 and from first marine terrace of cape Finneset—S7-7 and S7-9). Also, all samples assume the presence of microbiological processes of conversion of hydrocarbons.
The isolates of aerobic bacteria of the genera Arthrobacter, Pseudarthrobacter, Psychrobacter, and Rhodoferax accounted for less than 1% of the total bacterial diversity of the samples. Before freezing these species could probably perform the functions of converting organic matter, including proteins and chitin in cold Spitsbergen ecosystems.
The isotopic composition of methane and carbon dioxide in Bh 7 suggests that the sediments could be formed under conditions of gases’ replenishment from the Tertiary deposits.
REFERENCES
Aissa, F.B., Postec, A., Erauso, G., Payri, C., Pelletier, B., Hamdi, M., Ollivier, B., and Fardeau, M.L., Vallitalea pronyensis sp. nov., isolated from a marine alkaline hydrothermal chimney, Int. J. Syst. Evol. Microbiol., 2014, vol. 64, no. 4, pp. 1160–1165.
Albuquerque, L., França, L., Rainey, F.A., Schumann, P., Nobre, M.F., and da Costa, M.S., Gaiella occulta gen. nov., sp. nov., a novel representative of a deep branching phylogenetic lineage within the class Actinobacteria and proposal of Gaiellaceae fam. nov. and Gaiellales ord. nov., Syst. Appl. Microbiol., 2011, vol. 34, no. 8, pp. 595–599.
Alcántara-Hernández, R.J., Centeno, C.M., Ponce-Mendoza, A., Batista, S., Merino-Ibarra, M., Campo, J., and Falcón, L.I., Characterization and comparison of potential denitrifiers in microbial mats from King George Island, Maritime Antarctica, Polar Virol., 2014, vol. 37, no. 3, pp. 403–416.
Alekseev, I.I. and Abakumov, E.V., Taxonomic and environmental soil diversity of marine terraces of Gronfjord (West Spitsbergen Island), Samar. Luka: Probl. Reg. Global. Ekol., 2016, vol. 25, no. 4, pp. 256–161.
Alperin, M.J. and Reeburgh, W.S., Inhibition experiments on anaerobic methane oxidation, App. Environ. Microbiol., 1985, vol. 50, no. 4, pp. 940–945.
Bajerski, F. and Wagner, D., Bacterial succession in Antarctic soils of two glacier forefields on Larsemann Hills, East Antarctica, FEMS Microbiol. Ecol., 2013, vol. 85, no. 1, pp. 128–142.
Baltrus, D.A., Yourstone, S., Lind, A., Guilbaud, C., Sands, D.C., Jones, C.D., Morris, C.E., and Dangl, J.L., Draft genome sequences of a phylogenetically diverse suite of Pseudomonas syringae strains from multiple source populations, Genome Announce., 2014, vol. 2, no. 1, pp. 1–2. https://mra.asm.org/content/ga/2/1/ e01195-13.full.pdf.
Bolyen, E., Rideout, J.R., Dillon, M.R., Bokulich, N.A., Abnet, C.C., Al-Ghalith, G.A., Alexander, H., Alm, E.J., Arumugam, M., Asnicar, F., Bai, Y., Bisanz, J.E., Bittinger, K., Brejnrod, A., Brislawn, C.J., et al., Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nat. Biotechnol., 2019, vol. 37, no. 8, pp. 852–857. Erratum: Nat. Biotechnol., 2019, vol. 37, no. 9, p. 1091.
Borrel, G., Joblin, K., Guedon, A., Colombet, J., Tardy, V., Lehours, A.C., and Fonty, G., Methanobacterium lacus sp. nov., isolated from the profundal sediment of a freshwater meromictic lake, Int. J. Syst. Evol. Microbiol., 2012, vol. 62, no. 7, pp. 1625–1629.
Bowman, J.P. and McCuaig, R.D., Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment, Appl. Environ. Microbiol., 2003, vol. 69, no. 5, pp. 2463–2483.
Bueno de Mesquita, C.P.B., Schmidt, S.K., and Suding, K.N., Litter-driven feedbacks influence plant colonization of a high elevation early successional ecosystem, Plant Soil, 2019, vol. 444, no. 1, pp. 71–85.
Cabrera-Ospino, M.C., Phylogeny and diversity of genes for poorly characterized type of arsenite oxidase involved in anaerobic arsenic oxidation, Ph.D. Thesis, Japan, 2018.
Carr, S.A., Orcutt, B.N., Mandernack, K.W., and Spear, J.R., Abundant Atribacteria in deep marine sediment from the Adélie Basin, Antarctica, Front. Microbiol., 2015, vol. 6, id 872.
Chao, A., Nonparametric estimation of the number of classes in a population, Scand. J. Stat, 1984, vol. 11, pp. 265–270.
Chen, M.-Y., Wu, Sh.-H., Lin, G.-H., Lu, Ch.-P., Lin, Y.-T., Chang, W.-Ch., and Tsay, S.-S., Rubrobacter taiwanensis sp. nov., a novel thermophilic, radiation-resistant species isolated from hot springs, Int. J. Syst. Evol. Microbiol., 2004, vol. 54, no. 5, pp. 1849–1855.
Chernov, R.A. and Murav’ev, A.Ya., Contemporary changes in the area of glaciers in the western part of the Nordenskjold Land (Svalbard), Led Sneg, 2018, vol. 58, no. 4, pp. 462–472.
Coolen, M.J. and Orsi, W.D., The transcriptional response of microbial communities in thawing Alaskan permafrost soils, Front. Microbiol., 2015, vol. 6, id 197.
Coolen, M.J., Talbot, H.M., Abbas, B.A., Ward, C., Schouten, S., Volkman, J.K., and Damste, J.S., Sources for sedimentary bacteriohopanepolyols as revealed by 16S rDNA stratigraphy, Environ. Microbiol., 2008, vol. 10, no. 7, pp. 1783–1803.
Costello, E.K. and Schmidt, S.K., Microbial diversity in alpine tundra wet meadow soil: Novel Chloroflexi from a cold, water-saturated environment, Environ. Microbiol., 2006, vol. 8, no. 8, pp. 1471–1486.
Cravo-Laureau, C., Matheron, R., Joulian, C., Cayol, J.L., and Hirschler-Rea, A., Desulfatibacillum alkenivorans sp. nov., a novel n-alkene-degrading, sulfate-reducing bacterium, and emended description of the genus desulfatibacillum, Int. J. Syst. Evol. Microbiol., 2004, vol. 54, no. 5, pp. 1639–1642.
Demidov, N.E., Baranskaya, A.V., Durdenko, E.V., Zanina, O.G., Karaevskaya, E.S., Pushina, Z.V., Rivkina, E.M., Spirina, E.V., and Spenser, R., Biogeochemistry of frozen waters of the Arctic coast of the Gydan Peninsula, Probl. Arkt. Antarkt., 2016a, vol. 1, no. 3, pp. 34–49.
Demidov, N.E., Karaevskaya, E.S., Verkulich, S.R., Nikulina, A.L., and Savatyugin, L.M. First results of permafrost monitoring in the cryospheric polygon of the Russian Scientific Center on Spitsbergen (RSCS), Probl. Arkt. Antarkt., 2016b, vol. 1, no. 4, pp. 67–79.
Demidov, N., Wetterich, S., Verkulich, S., Ekaykin, A., Meyer, H., Anisimov, M., Schirrmeister, L., Demidov, V., and Hodson, A.J., Geochemical signatures of pingo ice and its origin in Grøndalen, West Spitsbergen, Cryosphere, 2019, vol. 13, pp. 3155–3169.
Demidov, N.E., Borisik, A.L., Verkulich, S.R., Wetterich, S., Gunar, A.Yu., Demidov, V.E., Zheltenkova, N.V., Koshurnikov, A.V., Miloslavskii, M.Yu., Nikulina, A.L., Novikov, A.L., Savatyugin, L.M., Sirotkin, A.N., Terekhov, A.V., Ugryumov, Yu.V., et al., Geocryological and hydrogeological conditions of the western part of Nordenskiold Land (Spitsbergen Archipelago), Izv., Atmos. Ocean. Phys., 2020, vol. 56, no. 8, pp. 1376–1400.
De Wever, A., Muylaert, K., Van der Gucht, K., Pirlot, S., Cocquyt, C., Descy, J.P., Plisnier, P.-D., and Vyverman, W., Bacterial community composition in Lake Tanganyika: Vertical and horizontal heterogeneity, Appl. Environ. Microbiol., 2005, vol. 71, no. 9, pp. 5029–5037.
Dojka, M.A., Hugenholtz, P., Haack, S.K., and Pace, N.R., Microbial diversity in a hydrocarbon and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation, Appl. Environ. Microbiol., 1998, no. 10, pp. 3869–3877.
Donachie, S.P., Hou, S., Lee, K.S., Riley, C.W., Pikina, A., Belisle, C., Kempe, S., Gregory, T.S., Bossuyt, A., Boerema, J., Liu, J., Freitas, T.A., Malahoff, A., and Alam, M., The Hawaiian Archipelago: A microbial diversity hotspot, Microb. Ecol., 2014, vol. 48, no. 4, pp. 509–520.
Drahota, P., Mikutta, C., Falteisek, L., Duchoslav, V., and Klementova, M., Biologically induced formation of realgar deposits in soil, Geochim. Cosmochim. Acta, 2017, vol. 218, pp. 237–256.
Dziga, D., Kokocinski, M., Barylski, J., Nowicki, G., Maksylewicz, A., Antosiak, A., Banaś, A.K., and Strzałka, W., Correlation between specific groups of heterotrophic bacteria and microcystin biodegradation in freshwater bodies of central Europe, FEMS Microbiol. Ecol., 2019, vol. 95, no. 11, id 162.
Elshahed, M.S., Youssef, N.H., Spain, A.M., Sheik, C., Najar, F.Z., Sukharnikov, L.O., Roe, B.A., Davis, J.P., Schloss, P.D., Bailey, V.L., and Krumholz, L.R., Novelty and uniqueness patterns of rare members of the soil biosphere, Appl. Environ. Microbiol., 2008, vol. 74, no. 17, pp. 5422–5428.
Fabisch, M., Freyer, G., Johnson, C.A., Buchel, G., Akob, D.M., Neu, T.R., and Kusel, K., Dominance of ‘Gallionella capsiferriformans’ and heavy metal association with Gallionella-like stalks in metal-rich pH 6 mine water discharge, Geobiology, 2016, vol. 14, no. 1, pp. 68–90.
Fadrosh, D.W., Ma, B., Gajer, P., Sengamalay, N., Ott, S., Brotman, R.M., and Ravel, J., An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform, Microbiome, 2014, vol. 2, no. 1, id 6.
Farrow, J.A.E., Lawson, P.A., Hippe, H., Gauglitz, U., and Collins, M.D., Phylogenetic evidence that the Gram-negative nonsporulating bacterium Tissierella (Bacteroides) praeacuta is a member of the Clostridium subphylum of the Gram-positive bacteria and description of Tissierella creatinini sp. nov., Syst. Evol. Microbiol., 1995, vol. 45, pp. 436–440.
Fendrich, C., Halovibrio variabilis gen. nov. sp. nov., Pseudomonas halophila sp. nov. and a new halophilic aerobic coccoid Eubacterium from Great Salt Lake, Utah, USA, Syst. Appl. Microbiol., 1988, vol. 11, no. 1, pp. 36–43.
Flynn, T.M., Sanford, R.A., Ryu, H., Bethke, C.M., Levine, A.D., Ashbolt, N.J., and Santo Domingo, J.W., Functional microbial diversity explains groundwater chemistry in a pristine aquifer, BMC Microbiol., 2013, vol. 13, no. 1, pp. 1–15.
Forman, S.L., Lubinski, D.J., Ingolfsson, O., Zeeberg, J.J., Snyder, J.A., Siegert, M.J., and Matishov, G.G., A review of postglacial emergence on Svalbard, Franz Josef Land and Novaya Zemlya, Northern Eurasia, Quat. Sci. Rev., 2004, vol. 23, pp. 1391–1434.
Gallego, V., Sanchez-Porro, C., Garcia, M.T., and Ventosa, A., Massilia aurea sp. nov., isolated from drinking water, Int. J. Syst. Appl. Microbiol., 2006, vol. 56, no. 10, pp. 2449–2453.
Ganzert, L., Bajerski, F., and Wagner, D., Bacterial community composition and diversity of five different permafrost-affected soils of Northeast Greenland, FEMS Microbiol. Ecol., 2014, vol. 89, no. 2, pp. 426–441.
Gilichinsky, D.A., Wilson, G.S., Friedmann, E.I., McKay, C.P., Sletten, R.S., Rivkina, E.M., Vishnivetskaya, T.A., Erokhina, L.G., Ivanushkina, N.E., Kochkina, G.A., Shcherbakova, V.A., Soina, V.S., Spirina, E.V., Vorobyova, E.A., Fyodorov-Davydov, D.G., et al., Microbial populations in Antarctic permafrost: Biodiversity, state, age and implication for astrobiology, Astrobiology, 2007, vol. 7, no. 2, pp. 275–311.
Glass, J.B., Ranjan, P., Kretz, C.B., Nunn, B.L., Johnson, A.M., McManus, J., and Stewart, F.J., Adaptations of Atribacteria to life in methane hydrates: Hot traits for cold life, bioRxiv, 2019, 536078. https://www. biorxiv.org/content/10.1101/536078v1.
Gleeson, D.F., Williamson, C., Grasby, S.E., Pappalardo, R.T., Spear, J.R., and Templeton, A.S., Low temperature S0 biomineralization at a supraglacial spring system in the Canadian High Arctic, Geobiology, 2011, vol. 9, no. 4, pp. 360–375.
Gołębiewski, M., Deja-Sikora, E., Cichosz, M., Tretyn, A., and Wróbel, B., 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils, Microb. Ecol., 2014, vol. 67, no. 3, pp. 635–647.
Good, I.J., The population frequencies of species and the estimation of population parameters, Biometrika, 1953, vol. 40, pp. 237–264.
Gupta, R.S., Chander, P., and George, S., Phylogenetic framework and molecular signatures for the class Chloroflexi and its different clades; proposal for division of the class Chloroflexi class. nov. into the suborder Chloroflexineae subord. nov., consisting of the emended family Oscillochloridaceae and the family Chloroflexaceae fam. nov., and the suborder Roseiflexineae subord. nov., containing the family Roseiflexaceae fam. Nov., Antonie van Leeuwenhoek, 2012, vol. 103, no. 1, pp. 99–119.
Gustave, W., Yuan, Z.F., Sekar, R., Ren, Y.X., Liu, J.Y., Zhang, J., and Chen, Z., Soil organic matter amount determines the behavior of iron and arsenic in paddy soil with microbial fuel cells, Chemosphere, 2019, vol. 237, id 124459.
Hammer, Ø., Harper, D.A.T., and Ryan, P.D., PAST: Paleontological statistics software package for education and data analysis, Palaeontol. Electron., 2001, vol. 4, no. 1.
Hanson, B.T., Molecular microbial ecology of sediments and subsurface groundwater at a coal tar-contaminated waste site, Ph.D. Dissertation, New York: Cornell Univ., 2013.
Harms, C., Ludwig, U., and Andreesen, J.R., Sarcosine reductase of Tissierella creatinophila: Purification and characterization of its components, Arch. Microbiol., 1998, vol. 170, no. 6, pp. 442–450.
Hitchman, S.P., Darling, W.G., and Williams, G.M., Stable isotope ratios in methane containing gases in the United Kingdom, Tech. Rep. WE/89/30, 1990. http:// nora.nerc.ac.uk/id/eprint/502528/1/WE89030.pdf.
Hugerth, L.W., Wefer, H.A., Lundin, S., Jakobsson, H.E., Lindberg, M., Rodin, S., Engstrand, L., and Andersson, A.F., DegePrime, a program for degenerate primer design for broad-taxonomic-range PCR in microbial ecology studies, Appl. Environ. Microbiol., 2014, vol. 80, no. 16, pp. 5116–5123.
Humlum, O., Instanes, A., and Sollid, J.L., Permafrost in Svalbard: A review of research history, climatic background and engineering challenges, Polar Res., 2003, vol. 22, pp. 191–215.
Inceoglu, B., Bettaieb, A., Da Silva, C.A.T., Lee, K.S.S., Haj, F.G., and Hammock, B.D., Endoplasmic reticulum stress in the peripheral nervous system is a significant driver of neuropathic pain, Proc. Natl. Acad. Sci. U. S. A., 2015, vol. 112, no. 29, pp. 9082–9087.
Jansson, J.K. and Taş, N., The microbial ecology of permafrost, Nat. Rev. Microbiol., 2014, vol. 12, no. 6, pp. 414–425.
Jeon, C.O., Park, M., Ro, H.S., Park, W., and Madsen, E.L., The naphthalene catabolic (nag) genes of Polaromonas naphthalenivorans CJ2: Evolutionary implications for two gene clusters and novel regulatory control, Appl. Environ. Microbiol., 2006, vol. 72, no. 2, pp. 1086–1095.
Johnson, S.S., Hebsgaard, M.B., Christensen, T.R., Mastepanov, M., Nielsen, R., Munch, K., Brand, T., Gilbert, M.T.P., Zuber, M.T., Bunce, M., Rønn, R., Gilichinsky, D., Froese, D., and Willerslev, E., Ancient bacteria show evidence of DNA repair, Proc. Natl. Acad. Sci. U. S. A., 2007, vol. 104, pp. 14401–14405.
Jørgensen, B.B., Beulig, F., Egger, M., Petro, C., Scholze, C., and Røy, H., Organoclastic sulfate reduction in the sulfate-methane transition of marine sediments, Geochim. Cosmochim. Acta, 2019, vol. 254, pp. 231–245.
Kaden, R., Spröer, C., Beyer, D., and Krolla-Sidenstein, P., Rhodoferax saidenbachensis sp. nov., a psychrotolerant, very slowly growing bacterium within the family Comamonadaceae, proposal of appropriate taxonomic position of Albidiferax ferrireducens strain T118T in the genus Rhodoferax and emended description of the genus Rhodoferax, Int. J. Syst. Evol. Microbiol., 2014, vol. 64, no. 4, pp. 1186–1193.
Kaminski, M.A., Furmanczyk, E.M., Sobczak, A., Dziembowski, A., and Lipinski, L., Pseudomonas silesiensis sp. nov. strain A3T isolated from a biological pesticide sewage treatment plant and analysis of the complete genome sequence, Syst. and Appl. Microbiol., 2018, vol. 41, no. 1, pp. 13–22.
Karaevskaya, E.S., Demchenko, L.S., Demidov, N.E., Rivkina, E.M., Bulat, S.A., and Gilichinsky, D.A., Archaeal diversity in permafrost sediments of Bunger Hills Oasis and King George Island (Antarctica) according to the 16S rRNA gene sequencing, Microbiology, 2014, vol. 83, no. 4, pp. 398–406.
Karaevskaya, E.S., Demidov, N.E., Shmelev, D.G., Rivkina, E.M., and Bulat, S.A., Study of the bacterial communities in the Antarctic oases’ permafrost by means of culturing, Probl. Arkt. Antarkt., 2017, vol. 1, no. 2, pp. 27–42.
Khlebnikova, G.M., Gilichinskii, D.A., Fedorov-Davydov, D.G., and Vorob’eva, E.A., Quantitative evaluation of microorganisms in permafrost deposits and buried soils, Microbiology, 1990, vol. 59, no. 1, pp. 106–112.
Kielak, A., Pijl, A.S., Van Veen, J.A., and Kowalchuk, G.A., Differences in vegetation composition and plant species identity lead to only minor changes in soil-borne microbial communities in a former arable field, FEMS Microbiol. Ecol., 2008, vol. 63, no. 3, pp. 372–382.
Kim, D.U., Kim, S.G., Lee, H., Park, A.Y., and Ka, J.O., Oryzihumus soli sp. nov., isolated from soil and emended description of the genus Oryzihumus, Int. J. Syst. Evol. Microbiol., 2017, vol. 67, no. 10, pp. 3960–3964.
Konieczna, I., Wojtasik, B., Kwinkowski, M., Burska, D., Nowiński, K., Żarnowiec, P., and Kaca, W., Analysis of cultivable aerobic bacteria isolated from bottom sediments in the Wijdefjorden region, Spitsbergen, Pol. Polar Res., 2011, vol. 32, no. 2, pp. 181–195.
Korehi, H., Blöthe, M., and Schippers, A., Microbial diversity at the moderate acidic stage in three different sulfidic mine tailings dumps generating acid mine drainage, Res. Microbiol., 2014, vol. 165, no. 9, pp. 713–718.
Kuever, J., Rainey, F.A., and Widdel, F., Desulfovibrio, in Bergey’s Manual of Systematics of Archaea and Bacteria, New York: Wiley, 2015, pp. 1–17.
Kumar, S., Stecher, G., and Tamura, K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets, Mol. Biol. Evol., 2016, vol. 33, no. is. 7, pp. 1870–1874.
Kumar, S., Stecher, G., Knyaz, C., and Tamura, K., MEGA X: Molecular evolutionary genetics analysis across computing platforms, Mol. Biol. Evol., 2018, vol. 35, pp. 1547–1549.
Kwon, M., Kim, M., Priscu, J.C., Hong, S.G., Kim, S.J., and Kim, O.S., Bacterial biodiversity in permanently ice-covered lakes of the McMurdo Dry Valleys, Antarctica, in 15th International Symposium on Microbial Ecology, Seoul, 2014.
Lane, D.J., 16S/23S rRNA sequencing, in Nucleic Acid Techniques in Bacterial Systematics, Stackebrandt, E. and Goodfellow, M., Eds., Chichester, U.K.: Wiley, 1991, pp. 115–175.
Lee, Y.J., Romanek, C.S., and Wiegel, J., Desulfosporosinus youngiae sp. nov., a spore-forming, sulfate-reducing bacterium isolated from a constructed wetland treating acid mine drainage, Int. J. Syst. Evol. Microbiol., 2009, vol. 59, no. 11, pp. 2743–2746.
Magurran, A.E., Ecological Diversity and Its Measurement, London: Croom Helm, 1983; Moscow: Mir, 1992.
Mangerud, J., Radiocarbon dating of marine shells, including a discussion of apparent age of recent shells from Norway, Boreas, 1972, vol. 1, no. 2, pp. 143–172.
Merkel, A.Yu., Tarnovetskii, I.Yu., Podosokorskaya, O.A., and Toshchakov, S.V., Analysis of 16S rRNA primer systems for profiling of thermophilic microbial communities, Microbiology, 2019, vol. 88, no. 6, pp. 671–680.
Miller, C.B. and Wheeler, P.A., Biological Oceanography, New York: Wiley, 2012.
Mitzscherling, J., Winkel, M., Winterfeld, M., Horn, F., Yang, S., Grigoriev, M.N., Wagner, D., Overduin, P.P., and Liebner, S., The development of permafrost bacterial communities under submarine conditions, J. Geophys. Res.: Biogeosci., 2017, vol. 122, no. 7, pp. 1689–1704.
Mitzscherling, J., Horn, F., Winterfeld, M., Mahler, L., Kallmeyer, J., Overduin, P.P., Schirrmeister, L., Winkel, M., Grigoriev, M.N., Wagner, D., and Liebner, S., Microbial community composition and abundance after millennia of submarine permafrost warming, Biogeosciences, 2019, vol. 16, pp. 3941–3958.
Mollenhauer, G., Grotheer, H., Gentz, T., Bonk, E., and Hefter, J., Standard operation procedures and performance of the MICADAS radiocarbon laboratory at Alfred Wegener Institute (AWI), Germany, Nucl. Instrum. Methods Phys. Res., Sect. B, 2021, vol. 496, pp. 45–51.
Müller, A.L., De Rezende, J.R., Hubert, C.R., Kjeldsen, K.U., Lagkouvardos, I., Berry, D., Jørgensen, B.B., and Loy, A., Endospores of thermophilic bacteria as tracers of microbial dispersal by ocean currents, ISME J., 2014, vol. 8, no. 6, pp. 1153–1165.
Müller, O., Bang-Andreasen, T., White, IIIR.A., Elberling, B., Taş, N., Kneafsey, T., Jansson, J.K., and Øvreås, L., Disentangling the complexity of permafrost soil by using high resolution profiling of microbial community composition, key functions and respiration rates, Environ. Microbiol., 2018, vol. 20, no. 12, pp. 4328–4342.
Niu, J., Deng, J., Xiao, Y., He, Z., Zhang, X., Van Nostrand, J.D., Liang, Y., Deng, Y., Liu, X., and Yin, H., The shift of microbial communities and their roles in sulfur and iron cycling in a copper ore bioleaching system, Sci. Rep., 2016, vol. 6, id 34744.
Nunoura, T., Hirai, M., Miyazaki, M., Kazama, H., Makita, H., Hirayama, H., Furushima, Y., Yamamoto, H., Imachi, H., and Takai, K., Isolation and characterization of a thermophilic, obligately anaerobic and heterotrophic marine Chloroflexi bacterium from a Chloroflexi-dominated microbial community associated with a Japanese shallow hydrothermal system, and proposal for Thermomarinilinea lacunofontalis gen. nov., sp. nov, Microbes Environ., 2013, vol. 28, no. 2, pp. 228–235.
Opel, T., Murton, J.B., Wetterich, S., Meyer, H., Ashastina, K., Günther, F., Grotheer, H., Mollenhauer, G., Danilov, P.P., Boeskorov, V., Savvinov, G.N., and Schirrmeister, L., Past climate and continentality inferred from ice wedges at Batagay megaslump in the Northern Hemisphere’s most continental region, Yana Highlands, interior Yakutia, Clim. Past, 2019, vol. 15, pp. 1443–1461.
Orsi, W.D., Ecology and evolution of seafloor and subseafloor microbial communities, Nat. Rev. Microbiol., 2018, vol. 16, no. 11, pp. 671–683.
Pascual, J., Blanco, S., García-López, M., García-Salamanca, A., Bursakov, S.A., Genilloud, O., Bills, J.F., Ramos, J.L., and van Dillewijn, P., Assessing bacterial diversity in the rhizosphere of Thymus zygis growing in the Sierra Nevada National Park (Spain) through culture-dependent and independent approaches, PLoS One, 2016, vol. 11, no. 1, id e0146558.
Pavlov, M.S., Lira, F., Martinez, J.L., Olivares-Pacheco, J., and Marshall, S.H., Pseudomonas fildesensis sp. nov., a psychrotolerant bacterium isolated from Antarctic soil of King George Island, South Shetland Islands, Int. J. Syst. Evol. Microbiol., 2020, vol. 70, no. 5, pp. 3255–3263. https://doi.org/10.1099/ijsem.0.004165
Peipoch, M., Jones, R., and Valett, H.M., Spatial patterns in biofilm diversity across hierarchical levels of river-floodplain landscapes, PLoS One, 2015, vol. 10, no. 12, id e0144303.
Perini, L., Gostinčar, C., and Gunde-Cimerman, N., Fungal and bacterial diversity of Svalbard subglacial ice, Sci. Rep., 2019, vol. 9, no. 1, pp. 1–15.
Pozzato, L., Cathalot, C., Berrached, C., Toussaint, F., Stetten, E., Caprais, J.C., Pastor, L., Olu, K., and Rabouille, C., Early diagenesis in the Congo deep-sea fan sediments dominated by massive terrigenous deposits: Part I. Oxygen consumption and organic carbon mineralization using a micro-electrode approach, Deep Sea Res., Part II, 2017, vol. 142, pp. 125–138.
Quéméneur, M., Erauso, G., Frouin, E., Zeghal, E., Vandecasteele, C., Ollivier, B., Tamburini, Ch., Garel, M., Menez, B., and Postec, A., Hydrostatic pressure helps to cultivate an original anaerobic bacterium from the Atlantis Massif subseafloor (IODP Expedition 357): Petrocella atlantisensis gen. nov. sp. nov., Front. Microbiol., 2019, vol. 10, id 1497.
Ravenschlag, K., Sahm, K., and Amann, R., Quantitative molecular analysis of the microbial community in marine Arctic sediments (Svalbard), Appl. Environ. Microbiol., 2001, vol. 67, no. 1, pp. 387–395.
Reddy, P.V.V., Rao, S.S.S.N., Pratibha, M.S., Sailaja, B., Kavya, B., Manorama, R.R., and Shivaji, S., Bacterial diversity and bioprospecting for cold-active enzymes from culturable bacteria associated with sediment from a melt water stream of Midtre Lovénbreen glacier, an Arctic glacier, Res. Microbiol., 2009, vol. 160, no. 8, pp. 538–546.
Reimer, P.J., Bard, E., Bayliss, A., Beck, J.W., Blackwell, P.G., Bronk Ramsey, C., Buck, C.E., Cheng, H., Edwards, R.L., Friedrich, M., Grootes, P.M., Guilderson, T.P., Haflidason, H., Hajdas, I., Hatté, C., et al., IntCal13 and MARINE13 radiocarbon age calibration curves 0–50000 years cal BP, Radiocarbon, 2013, vol. 55, no. 4, pp. 1869–1887.
Romaniuk, K., Ciok, A., Decewicz, P., Uhrynowski, W., Budzik, K., Nieckarz, M., and Dziewit, L., Insight into heavy metal resistome of soil psychrotolerant bacteria originating from King George Island (Antarctica), Polar Biol., 2018, vol. 41, no. 7, pp. 1319–1333.
Sahm, K. and Berninger, U.G., Abundance, vertical distribution, and community structure of benthic prokaryotes from permanently cold marine sediments (Svalbard, Arctic Ocean), Mar. Ecol. Prog. Ser., 1998, vol. 165, pp. 71–80.
Salvigsen, O. and Høgvard, K., Glacial history, Holocene shoreline displacement and palaeoclimate based on radiocarbon ages in the area of Bockfjorden, north-western Spitsbergen, Svalbard, Polar Res., 2005, vol. 25, no. 1, pp. 15–24.
Sánchez-Andrea, I., Stams, A.J., Amils, R., and Sanz, J.L., Enrichment and isolation of acidophilic sulfate-reducing bacteria from Tinto River sediments, Environ. Microbiol. Rep., 2013, vol. 5, no. 5, pp. 672–678.
Schimel, J., Balser, T.C., and Wallenstein, M., Microbial stress-response physiology and its implications for ecosystem function, Ecology, 2007, vol. 88, no. 6, pp. 1386–1394.
Senger, K., Brugmans, P., Grundvåg, S.-A., Jochmann, M., Nøttvedt, A., Olaussen, S., Skotte, A., and Smyrak-Sikora, A., Petroleum, coal and research drilling onshore Svalbard: A historical perspective, Norw. J. Geol., 2019, vol. 99, no. 3.
Shakirov, R.B., Chemical and isotopic characteristics of hydrocarbon gases from Mendeleev and Golovnin volcanoes, Kunashir Island, Geochem. Int., 2014, vol. 52, no. 3, pp. 247–259.
Singh, P., Singh, S.M., Singh, R.N., Naik, S., Roy, U., Srivastava, A., and Bölter, M., Bacterial communities in ancient permafrost profiles of Svalbard, Arctic, J. Basic Microbiol., 2017, vol. 57, no. 12, pp. 1018–1036.
Singh, S.K., Verma, P., Ramaiah, N., Chandrashekar, A.A., and Shouche, Y.S., Phylogenetic diversity of archaeal 16s rRNA and ammonia monooxygenase genes from tropical estuarine sediments on the central west coast of India, Res. Microbiol., 2010, vol. 161, no. 3, pp. 177–186.
Solov’eva, D.A., Savel’eva, L.A., Verkulich, S.R., and Zazovskaya, E.P., Postglacial changes in the natural environment in the area of the Barentsburg mine (West Spitsbergen Island), in Theory and Methods of Polar Science: Proc. of Int. Youth Sci. Conf. on Polar Geodesy, Glaciology, Hydrology and Geophysics, St. Petersburg, 2018, pp. 213–222.
Srinivas, T.N.R., Rao, S.N., Reddy, P.V.V., Pratibha, M.S., Sailaja, B., Kavya, B., and Shivaji, S., Bacterial diversity and bioprospecting for cold-active lipases, amylases and proteases, from bacteria of Kongsfjorden and Ny-Ålesund, Svalbard, Arctic, Curr. Microbiol., 2009, vol. 59, no. 5, pp. 537–547.
Steven, B., Briggs, G., McKay, Ch.P., Pollard, W.H., Greer, C.W., and Whyte, L.G., Characterization of the microbial diversity in permafrost sample from the Canadian high Arctic using culture-dependent and culture-independent methods, FEMS Microbiol. Ecol., 2007, pp. 513–523.
Steven, B., Pollard, W.H., Greer, C.W., and Whyte, L.G., Microbial diversity and activity through a permafrost/ground ice core profile from the Canadian high Arctic, Environ. Microbiol., 2008, vol. 10, no. 12, pp. 3388–3403.
Stieglmeier, M., Klingl, A., Alves, R.J., Rittmann, S.K.M., Melcher, M., Leisch, N., and Schleper, C., Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota, Int. J. Syst. Evol. Microbiol., 2014, vol. 64, no. 8, id 2738.
Svendsen, J.I. and Mangerud, J., Holocene glacial and climatic variations on Spitsbergen, Svalbard, Holocene, 1997, vol. 7, no. 1, pp. 45–57.
Szpak, M.T., Monteys, X., O’Reilly, S., Simpson, A.J., Garcia, X., Evans, R.L., Allen, C.C.R., McNally, D.J., Countier-Murias, D., and Kelleher, B.P., Geophysical and geochemical survey of a large marine pockmark on the Malin Shelf, Ireland, Geochem., Geophys., Geosyst., 2012, vol. 13, no. 1.
Tamura, K. and Nei, M., Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees, Mol. Biol. Evol., 1993, vol. 10, pp. 512–526.
Taranik, A.A., Geochemical indicators of long-term coalbed methane production areas as an independent source of energy on the example of Donbas deposits, Gazov. Prom-st., 2017, vol. 755, no. 7, pp. 24–27.
Taş, N., Prestat, E., Wang, S., Wu, Y., Ulrich, C., Kneafsey, T., Tringe, S.G., Torn, M.S., Hubbard, S.S., and Jansson, J.K., Landscape topography structures the soil microbiome in Arctic polygonal tundra, Nat. Commun., 2018, vol. 9, no. 1, pp. 1–13.
Teske, A., Durbin, A., Ziervogel, K., Cox, C., and Arnosti, C., Microbial community composition and function in permanently cold seawater and sediments from an Arctic fjord of Svalbard, Appl. Environ. Microbiol., 2011, vol. 77, no. 6, pp. 2008–2018.
Tischer, K., Kleinsteuber, S., Schleinitz, K.M., Fetzer, I., Spott, O., Stange, F., Lohse, U., Franz, J., Neumann, F., Gerling, S., Schmind, Ch., Hasselwander, E., Harms, H., and Wendeberg, A., Microbial communities along biogeochemical gradients in a hydrocarbon-contaminated aquifer, Environ. Microbiol., 2013, vol. 15, no. 9, pp. 2603–2615.
Tuorto, S.J., Darias, P., McGuinness, L.R., Panikov, N., Zhang, T., Häggblom, M.M., Kerkhof, L.J., Bacterial genome replication at subzero temperatures in permafrost, ISME J., 2014, vol. 8, no. 1, pp. 139–149.
Vázquez, S., Monien, P., Minetti, R.P., Jürgens, J., Curtosi, A., Primitz, J.V., Frikenhaus, S., Abele, D., Mac Cormack,W., and Helmke, E., Bacterial communities and chemical parameters in soils and coastal sediments in response to diesel spills at Carlini Station, Antarctica, Sci. Total Environ., 2017, vol. 605, pp. 26–37.
Vishnivetskaya, T., Kathariou, S., McGrath, J., Gilichinsky, D., and Tiedje, J.M., Low-temperature recovery strategies for the isolation of bacteria from ancient permafrost sediments, Extremophiles, 2000, vol. 4, no. 3, pp. 165–173.
Wang, F., Gai, Y., Chen, M., and Xiao, X., Arthrobacter psychrochitiniphilus sp. nov., a psychrotrophic bacterium isolated from Antarctica, Int. J. Syst. Evol. Microbiol., 2009, vol. 59, pp. 2759–2762.
Wartiainen, I., Hestnes, A.G., McDonald, I.R., and Svenning, M.M., Methylobacter tundripaludum sp. nov., a methane-oxidizing bacterium from Arctic wetland soil on the Svalbard islands, Norway (78°N), Int. J. Syst. Evol. Microbiol., 2006, vol. 56, no. 1, pp. 109–113.
Watanabe, T., Kojima, H., and Fukui, M., Identity of major sulfur-cycle prokaryotes in freshwater lake ecosystems revealed by a comprehensive phylogenetic study of the dissimilatory adenylylsulfate reductase, Sci. Rep., 2016, vol. 6, id 36262.
Weber, Y., Damsté, J.S.S., Zopfi, J., De Jonge, C., Gilli, A., Schubert, C.J., Lepori, F., Lehmann, M.F., and Niemann, H., Redox-dependent niche differentiation provides evidence for multiple bacterial sources of glycerol tetraether lipids in lakes, Proc. Natl. Acad. Sci. U. S. A., 2018, vol. 115, no. 43, pp. 10926–10931.
Wegner, C.E. and Liesack, W., Unexpected dominance of elusive Acidobacteria in early industrial soft coal slags, Front. Microbiol., 2017, vol. 8, id 1023.
Wei, N. and Finneran, K.T., Microbial community analyses of three distinct, liquid cultures that degrade methyl tert-butyl ether using anaerobic metabolism, Biodegradation, 2009, vol. 20, no. 5, pp. 695–707.
Wilhelm, R.C., Hanson, B.T., Chandra, S., and Madsen, E., Community dynamics and functional characteristics of naphthalene-degrading populations in contaminated surface sediments and hypoxic/anoxic groundwater, Environ. Microbiol., 2018, vol. 20, no. 10, pp. 3543–3559.
Yamada, T., Imachi, H., Ohashi, A., Harada, H., Hanada, S., Kamagata, Y., and Sekiguchi, Y., Bellilinea caldifistulae gen. nov., sp. nov. and Longilinea arvoryzae gen. nov., sp. nov., strictly anaerobic, filamentous bacteria of the phylum Chloroflexi isolated from methanogenic propionate-degrading consortia, Int. J. Syst. Evol. Microbiol., 2007, vol. 57, no. 10, pp. 2299–2306.
Yudovich Yu.E. and Ketris, M.P., Carbon isotope ratios in the stratosphere and biosphere, Biosfera, 2010, vol. 2, no. 2, pp. 231–247.
Zeglin, L.H., Wang, B., Waythomas, C., Rainey, F., and Talbot, S.L., Organic matter quantity and source affects microbial community structure and function following volcanic eruption on Kasatochi Island, Alaska, Environ. Microbiol., 2016, vol. 18, no. 1, pp. 146–158.
Zeng, Y., Selyanin, V., Lukeš, M., Dean, J., Kaftan, D., Feng, F., and Koblížek, M., Characterization of the microaerophilic, bacteriochlorophyll a-containing bacterium Gemmatimonas phototrophica sp. nov., and emended descriptions of the genus Gemmatimonas and Gemmatimonas aurantiaca, Int. J. Syst. Evol. Microbiol., 2015, vol. 65, no. 8, pp. 2410–2419.
Zeng, Y.-X., Yu, Y., Liu, Y., and Li, H.-R., Psychrobacter glaciei sp. nov., isolated from the ice core of an Arctic glacier, Int. J. Syst. Evol. Microbiol., 2016, vol. 66, pp. 1792–1798.
Zeng, Y. and Koblížek, M., Phototrophic Gemmatimonadetes: A new “purple” branch on the bacterial tree of life, in Modern Topics in the Phototrophic Prokaryotes, Springer, 2017, pp. 163–192.
Zhang, D.C., Schumann, P., Liu, H.C., Xin, Y.H., Zhou, Y.G., Schinner, F., and Margesin, R., Arthrobacter alpinus sp. nov., a psychrophilic bacterium isolated from Alpine soil, Int. J. Syst. Evol. Microbiol., 2010, vol. 60, no. 9, pp. 2149–2153.
ACKNOWLEDGMENTS
The authors thank E.A. Vorobyova (Lomonosov Moscow State University) for help in organizing experimental work on the cultivation of permafrost samples; A.Yu. Merkel (Vinogradsky Institute of Microbiology, RAS) for performing NGS analysis and consultation on the method; M.Y. Cherbunina and D.G. Shmelev for consultation on the properties of methane in permafrost.
Funding
The research has been supported by the grant from the Russian Science Foundation for the project no. 19-77-10066 (to Nikita Demidov) with respect to the next generation sequencing and by the Deutsche Forschungsgemeinschaft (DFG grant no. WE4390/7-1 to Sebastian Wetterich) with respect to the radiocarbon dating and TOC content. Field work at the cryosphere test polygon near Barentsburg was carried out as part of the Russian Arctic Expedition on Spitsbergen (RAES).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Karaevskaya, E.S., Demidov, N.E., Kazantsev, V.S. et al. Bacterial Communities of Frozen Quaternary Sediments of Marine Origin on the Coast of Western Spitsbergen. Izv. Atmos. Ocean. Phys. 57, 895–917 (2021). https://doi.org/10.1134/S000143382108003X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S000143382108003X