Abstract
Integration of advances in understanding of gene regulation through epigenetic mechanisms with the study of the influence of social environments on behavior has generated the evolving field of social and behavioral epigenetics. This field provides novel perspectives on how social adversity impacts biology and raises issues regarding the relationship between DNA and identity, multigenerational biological impacts of social environments, and intervention as a strategy to target epigenetic plasticity. In this chapter I highlight studies within the field of social and behavioral epigenetics, discuss the changing scientific and societal views contributed to by these studies, and speculate regarding the future implications of this field of study for our evolving understanding of the gene, individuals, and society.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Introduction
Though there has been long-standing division between considerations of the role of nature vs. nurture in determining the origins of variation in personality, behavior, health, and well-being, this traditional view has been revised in light of demonstrated gene-environment interactions (GxE) and their influence on these outcomes. A classic example of this interaction is in the prediction of depression based on stress and genotype: individuals with a specific polymorphism within the gene encoding the serotonin transporter (SLC6A4) are at higher risk for depression only when they have experienced elevated lifetime stress (Caspi et al. 2003). Under conditions of low stress, no effects of genotype are observed. Thus, the impact of genes/nature on the traits of an individual is tempered by the environmental experiences of that individual. This shift in understanding of nature and nurture has important implications for how we think about genes and their influences. In particular, gene-environment interactions provide evidence of plasticity and an ability to overcome the constraints of genetic determinism. However, the occurrence of a gene-environment interaction is derived primarily from statistical relationships—the presence of a statistical interaction between genotype and environmental exposure. These interactions suggest a phenomenon but do not provide a mechanism.
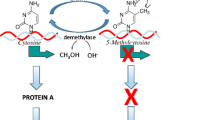
In the past decade there has been a rapidly growing literature focused on the biological mechanisms through which interactions between genes and the environment occur (Champagne 2012; Meaney 2010). At the core of these mechanistic studies is epigenetics. The term “epigenetics” was coined by Conrad Waddington, a developmental biologist, in the 1940s to refer to the interplay between genes and their products that account for variation in phenotype. From this perspective, genes were viewed as being “organized” or “induced” in their activity with resulting consequences for development (Waddington 1940). By the 1980s, biologists had identified possible molecular processes to account for variation in gene regulation through studies of DNA methylation (Razin and Riggs 1980). DNA methylation is the chemical modification of a cytosine within the DNA sequence, resulting in 5-methylcytosine (Culp et al. 1970). Early studies of DNA methylation indicated that the activity of genes can be altered in this way and that this alteration is fundamental to driving diversity of phenotype—albeit at a cellular/molecular level accounting for cellular differentiation (Jones et al. 1983). However, the notion that these molecular epigenetic mechanisms could be modified by the environment to account for the phenomenon of nature-nurture interactions has only been the focus of epigenetic research in the past decade (Weaver et al. 2004; Dolinoy et al. 2006).
The field of social and behavioral epigenetics explores the relationship between the quality of the social environment, epigenetic variation, and behavioral variation and is part of the broader study of how environments (i.e. nutritional, toxicological, social) come to induce phenotypic variation at the level of the organism (i.e. growth, metabolism, health, behavior) via epigenetic mechanisms. Though the initial studies linking social experiences to epigenetic changes in the brain with consequences for behavior were conducted in model organisms, such as rats, there is growing support for the relevance of these mechanisms for humans. Both individual-level social experiences, such as psychosocial stress (Monk et al. 2016), trauma (Yehuda et al. 2014), and exposure to adverse parent-offspring interactions (McGowan et al. 2009), and group-level experiences, such as poverty (Lam et al. 2012) and racial discrimination (Brody et al. 2016a), may exert lasting biological influences through epigenetic variation. Epigenetic studies illustrate the integration of biology and the social world in unprecedented ways by demonstrating the direct effect on DNA function of the social environment. Moreover, there is increasing focus on the transmission of environmentally induced molecular changes across generations. This multigenerational perspective has forced a reconsideration of the narrowness with which we view the biology of inheritance (Danchin et al. 2011) and suggests a broader and more dynamic process of evolution (Laland et al. 2015). Given the scientific revolution that this body of work has triggered, social and behavioral epigenetics raises many issues of societal relevance, including the biology of social adversity, the relationship between DNA and identity, and intervention as a strategy to target epigenetic plasticity (Brody et al. 2016a, b; Swartz et al. 2016). This chapter will highlight studies within the field of social and behavioral epigenetics, discuss the changing scientific and societal views contributed to by these studies, and speculate about the future implications of this field of study for our evolving understanding of the gene, individuals, and society.
A Primer of Modern Epigenetics
Advances in the methodological tools available to interrogate biology at a molecular level have enabled rapid scientific discovery within the field of epigenetics. In particular, these advances have revealed the dynamic process of gene regulation—involving multiple types of epigenetic modifications occurring within a temporal-spatial context. DNA methylation is perhaps the most fully explored modification of cytosines within the DNA sequence (Razin and Riggs 1980). The addition of a methyl-group to cytosines within DNA is generally an epigenetic mechanism of gene silencing when occurring within the promoter—the regulatory region of a gene (Razin 1998). This chemical modification of DNA does not alter the DNA sequence. The gene silencing occurring as a consequence of DNA methylation is contributed to by the accumulation of methyl-binding proteins within the methylated genomic region which serves to limit accessibility to the DNA (Fan and Hutnick 2005). DNA methylation patterns are mitotically heritable such that when cells divide they transmit this pattern to daughter cells (Jones et al. 1983). This transmission process is critical to the phenomenon of cellular differentiation, where all cells descend from an omnipotent stem cell that generates more lineage-specific cell types.
In addition to direct chemical modifications to DNA, there are two other main classes of epigenetic mechanisms: post-translational histone modifications and non-coding RNA. Within the cell nucleus, DNA is physically wrapped around a cluster of proteins called histones (e.g. H3, H4). Histone proteins can, like DNA, be modified through the addition of a variety of chemicals, leading, for example, to acetylation, methylation, and ubiquitination (Cheung et al. 2000). Histone chemical modifications serve to either create a more densely packed chromatin structure associated with gene silencing or loosen interactions between DNA and histones to promote gene activation. The type of chemical added, the location within the histone where the chemical has been added, and the genomic location where the modified histone interacts with the DNA are collectively predictive of the impact of post-translational histone modifications on gene expression (Jenuwein and Allis 2001). Finally, there is increasing understanding of the role of non-coding RNAs—RNA that does not produce a protein product—in gene regulation (Sato et al. 2011). The product of “junk DNA” (Ohno 1972), non-coding RNA molecules can alter the function of a gene by interacting with proteins and mRNA produced from coding regions of the genome (i.e. genes) and may also interact directly with DNA. The function of non-coding RNA molecules in gene regulation has challenged the way in which we define “functional” with regard to the genome—producing a protein may be one of many functions that a genome can have (Tragante et al. 2014; Graur et al. 2015).
Overall, though epigenetics is often described as a molecular “on/off” switch, the complexity of these biological processes is immense. Each of type of epigenetic modification operates within a genomic context and has spatial and temporal features that contribute to their predicted effects. Beyond that initial complexity, there is interaction between different types of epigenetic modifications in the prediction of gene expression (Molina-Serrano et al. 2013). Thus, increasing understanding of epigenetics reveals how highly complex, multilayered, and contextually sensitive these biological mechanisms are, as a first step in the process of generating phenotype from genotype. Though developing simple analogies to communicate the basic principle of epigenetics is important for transmitting emerging scientific ideas, the complexity involved in epigenetics should not be lost. Organisms are complex and epigenetics builds an infinitely complex and dynamic layer of biological information within the genome.
Mothering the Epigenome
The role of epigenetics in gene regulation and cellular differentiation has been accepted for decades; however, a relatively novel concept to emerge is that these mechanisms can be shaped or “induced” by the environment. Certainly, the cellular environment is important in setting epigenetic state of DNA as it is through cell-signaling and cell-cell interactions that cellular differentiation occurs. However, the question that has moved the study of epigenetics into the realm of the social world is whether the experiences of an individual can shape epigenetic variation within the genome (see Fig. 10.1). Theoretical discussions regarding epigenetic plasticity have existed within the literature for decades—such as the idea that memories might be encoded or “ticketed” within DNA through cytosine modifications (Griffith and Mahler 1969). However, a theoretical stumbling block to a wider appreciation of epigenetic plasticity was present in the hypothesized role of stable epigenetic patterns in defining cell types. How can a mechanism confer both stability of phenotype (i.e. maintenance of a muscle cell type vs. a neuron) and plasticity in response to a lifetime of environmental signals? Though the solution to this dilemma has yet to be elucidated, evidence of epigenetic plasticity and stability exists and is demonstrated by the impact of early life mother-infant interactions.
Illustration of the complex interplay between genotype and social environment in predicting phenotype within and across generations. Epigenetic variation is a mechanism through which divergent phenotypes can arise through interactions of genotype with different environmental conditions across development. This epigenetic variation can be transmitted across generations leading to the inheritance of phenotypic variation
Mammalian development is characterized by a high level of investment in the care of offspring from the prenatal period through to young adulthood. Though biparental care is present in some species, including humans, mothers are the primary caregivers in most reproductive contexts and invest significant energetic resources through placentation, lactation, and offspring-directed behaviors (Fowden and Moore 2012; Jenkins et al. 2016). Maternal reproductive investment is essential to offspring growth and development. However, there is significant within-species variation in maternal behavior (Hane et al. 2010; Maestripieri et al. 1997; Champagne et al. 2003). Studies of natural variations in maternal behavior reveal the critical role of mothers in shaping epigenetic outcomes. Offspring of female laboratory rats that engage in low vs. high levels of postpartum maternal licking/grooming (LG) during the first week of life differ significantly on physiological, neurobiological, and behavioral outcomes (Meaney 2001). These effects persist into adulthood. Adult offspring that have experienced low levels of LG during infancy have heightened stress reactivity, behavioral inhibition within novel environments, increased aggressiveness in social interactions, impaired learning/memory capacity, and altered reproductive behavior (Meaney 2001; Cameron et al. 2005). These functional outcomes are associated with altered gene expression within specific neural systems associated with the hypothalamic-pituitary-adrenal (HPA) response to stress, fear, cognition, and maternal/sexual behavior. What is particularly notable about the observed association between early life mother-infant interactions and gene expression is its persistence. The activity of genes within the brain is stably altered by the quality of the social environment occurring early in development. Analyses of DNA methylation levels and post-translational histone modifications within the brain of offspring that differ in their experience of postnatal maternal care reveal the role of maternal behavior in shaping these epigenetic mechanisms (Weaver et al. 2004; Suderman et al. 2012; McGowan et al. 2011). The epigenetic effects of maternal care occur at a broad range of genomic locations, including specific gene promoters involved in stress reactivity. The regulatory region of the gene encoding the glucocorticoid receptor (NR3C1) is hypermethylated in the hippocampus of offspring of low-LG compared to high-LG mothers (Weaver et al. 2004). In concert with increased DNA methylation are decreased levels of histone acetylation which collectively accounts for the decreased NR3C1 gene expression and protein observed in the hippocampus of low-LG offspring (Liu et al. 1997; Francis et al. 1999). The result of this gene regulatory state is to reduce the capacity of low-LG offspring to adapt to stress.
The determination of an epigenetic consequence of maternal behavior has been the launching point for studies of social and behavioral epigenetics. Moreover, further exploration of the dynamics of epigenetic change within the NR3C1 gene has revealed important principles of environmental interplay within the genome. First, epigenetic variation emerges in response to the cues in the environment. At birth, there are no epigenetic differences in DNA methylation of NR3C1 in the hippocampus of low- compared to high-LG rat offspring (Weaver et al. 2004). After several days of differential maternal care, group differences in DNA methylation are observed and persist into adulthood. Second, though the epigenetic effects of maternal care persist into adulthood, there is continued epigenetic plasticity in the adult brain whereby NR3C1 gene activity can be “reset” resulting in a shift in the stress reactivity of offspring. Pharmacological manipulations in adulthood that decrease DNA methylation or increase histone acetylation can be used to shift the phenotype of a low-LG rat toward that of a high-LG rat, and the converse can be achieved by increasing DNA methylation (Weaver et al. 2004; Weaver et al. 2005). Thus, reversibility of both epigenetic variation and the phenotype associated with this variation is possible, even when stability has been maintained throughout infancy and adolescence. Finally, studies exploring the link between maternal behavior and NR3C1 DNA methylation have revealed the cascade of sensory, neural circuit, hormonal, and transcriptional events that link this particular aspect of the social environment to a change in DNA methylation (Hellstrom et al. 2012). Somatosensory stimulation features prominently in this cascade as a way through which an organism senses the quality of caregiving (Ferber et al. 2008; Hellstrom et al. 2012).
Though natural variations in maternal behavior have served as the starting point for studies examining epigenetic interplay with the social environment, subsequent studies have examined a broad range of “nurture” cues, including the experience of abuse and neglect. In rodents, disruptions to the postnatal environment result in an increased incidence of abusive caregiving, resulting in altered DNA methylation, histone acetylation, and gene expression within the brain of offspring (Roth et al. 2009; Blaze et al. 2015; Doherty et al. 2016). In humans, a history of childhood abuse is predictive of increased hippocampal DNA methylation within the NR3C1 gene and similar overall patterns of epigenetic variation to what has been observed in the rodent model comparing low- and high-LG offspring (Suderman et al. 2012). Global increases in DNA methylation have been observed in blood samples from institution-reared orphans (Naumova et al. 2012) and analyses of buccal cells indicate hypomethylation in the SLC6A4 gene as a function of increased exposure to institutional care (Non et al. 2016). The ability to detect epigenetic signatures of early life adversity in tissues outside the brain is an important methodological step in translating laboratory-based findings into the real-world analyses of human biobehavioral processes and to field studies of animals exposed to ecological pressures meaningful in discussions of fitness and evolution. Though there is ongoing debate about the relevance of these “peripheral” epigenetic changes in understanding the brain and behavior, there is increasing evidence of epigenetic concordance across different tissue types in response to environmental cues (Nemoda et al. 2015; Farré et al. 2015; Kundakovic et al. 2015).
Evidence for the profound impact of maternal care on offspring development that extends to epigenetic outcomes has placed increased emphasis on the development of parenting interventions. Despite a recognized need to provide additional support and education to parents (Shuman and Masterpasqua 1981), these interventions have not typically been implemented at a global or national level. However, family-based programs that focus on developing attachment security, managing stress, and treating parental and child psychiatric illness have promise in reducing mental illness and improving child and parent well-being (Cicchetti et al. 2006; Lowell et al. 2011). Though parental neglect or abuse can exert significant “wear and tear” on the biology and behavior of children, it may be possible to shift developmental trajectories through intervention. Moreover, this plasticity may manifest at the level of epigenetic variation. One epigenetic metric that delves into the biological “wear and tear” experienced by an individual is referred to as “epigenetic age” (Horvath 2013). Analyses of DNA methylation from virtually any cell in the body can give an approximate estimate of our chronological age. Thus, our cells have a memory of time. However, in some cases, the epigenetic estimate of chronological age suggests we may be biologically “older” than our chronological age. This phenomenon is referred to as “age acceleration” and is thought to reflect a process of “wear and tear” (Horvath 2013). Epigenetic age acceleration has been observed in response to disease (Horvath and Levine 2015), prenatal adversity (Simpkin et al. 2016), and exposure to parental depression (Brody et al. 2016b). Within intervention studies, programs that reduce harsh parenting can reduce epigenetic age acceleration with potential for improved physical and mental health outcomes (Brody et al. 2016b). Intervention studies have significant potential to “reset” epigenetic outcomes. However, it is important within the context of intervention studies to not lose sight of the cascade of events within the social environment that influence parent-offspring interactions. Studies of maternal behavior in laboratory rodents and in primates provide empirical support for the influence of social stress and social support on the quality of mother-infant interactions (Ruppenthal et al. 1976; Curley et al. 2009; Champagne and Meaney 2007; Champagne and Meaney 2006). Similarly, human parenting occurs within a broader context of socioeconomic pressures, family dynamics, community well-being, and exposures to nutritional and toxicological factors that may alter reproductive systems and stress physiology. Integrating context into the discourse of the impact of mother-infant interactions on the epigenome will be particularly important for identifying the distal predictors of parenting, identifying society/community level targets for intervention, and lessening the “blame the mother” sentiment that may arise from the focus on the more proximal influences of child development (Winett et al. 2016).
Psychosocial Stress and Epigenetic Plasticity
Stress is a highly conserved process of coordinating the biology of an organism in response to threat. Psychosocial stress and mood during pregnancy can have a lasting impact on offspring development with consequences for psychiatric risk (Koubovec et al. 2005; Weinstock 2008). These psychological states are associated with heightened HPA activation, resulting in increased glucocorticoid levels within the mother—a classic physiological response to threat (Kane et al. 2014; O’Connor et al. 2014). In humans, objective stress exposure, maternal perceived stress, anxiety, and depression can epigenetically alter offspring via three distinct yet interactive routes (Monk et al. 2012). The first pathway is through epigenetic variation within the placenta. During pregnancy, the placenta acts as a critical interface between the mother and the fetus (Burton and Jauniaux 2015). Gene expression and epigenetic profiles within the placenta change during the course of pregnancy (Novakovic et al. 2010; Sitras et al. 2012), and variation in these profiles is predictive of fetal growth restriction (Jensen et al. 2014; Roifman et al. 2016). Among mothers that report elevated perceived stress during pregnancy, there is increased placental DNA methylation within the 11HSD2B gene—a gene encoding an enzyme that buffers the fetus from maternal stress hormone (Monk et al. 2016). Moreover, increased 11HSD2B DNA methylation within the placenta is predictive of impaired neurodevelopment in the fetus. Variation in 11HSD2B DNA methylation is also observed as a consequence of socioeconomic status (SES)—though this association suggests decreased DNA methylation of 11HSD2B in response to stress (Appleton et al. 2013). Epigenetic variation in several other placental gene targets is predictive of stress responsivity, self-regulation, and sensory development (Paquette et al. 2014; Conradt et al. 2015). A second route of prenatal epigenetic influence is the direct impact of maternal psychosocial stress on fetal tissues—including the brain. In humans, analyses of epigenetic effects in offspring who have experienced prenatal stress have primarily relied on blood, buccal cells, or saliva. Altered DNA methylation within stress-related genes such as NR3C1 and neural plasticity-related genes such as brain-derived neurotrophic factor (BDNF) has been detected in these tissues associated with prenatal exposure to maternal stress (Radtke et al. 2011; Hompes et al. 2013; Braithwaite et al. 2015; Unternaehrer et al. 2016). Moreover, studies in laboratory rodents provide experimental support for the presence of these epigenetic effects within the brain (Mueller and Bale 2008; Peña et al. 2012). A third route through which prenatal epigenetic effects may be mediated is via alterations in the quality of postnatal mother-infant interactions. Stress during pregnancy may alter mental health of the mother during the postpartum period, and there is a heightened risk of impaired mother-infant interactions associated with postpartum depression (Brummelte and Galea 2016; Dollberg et al. 2016). Influence of prenatal stress on the quality of the postnatal environment highlights the interplay between experiences occurring at different developmental time points.
Epigenetic plasticity in response to stress continues during postnatal development and persists into adulthood. The deprivation of maternal care during infancy can be perceived as a threat and activate the HPA response to stress with epigenetic consequences. In laboratory rodents, prolonged postnatal maternal separation, often referred to as early life stress, leads to increased activity of stress-related genes (Murgatroyd et al. 2009; Chen et al. 2012) and epigenetic silencing of genes involved in moderating stress responses (Kember et al. 2012; Kundakovic et al. 2013) within the hypothalamus and hippocampus. Moreover, the effects of maternal separation occurring during infancy can be ameliorated if offspring are placed on a diet that alters DNA methylation in adulthood (Paternain et al. 2016). Histone modifications and non-coding RNA expression are also altered by early life stress. For example, activity of the BDNF gene is decreased by maternal separation, and this effect coincides with decreased histone acetylation within the hippocampus (Seo et al. 2016). Altered expression of microRNA—a small non-coding RNA—is observed in the frontal cortex of offspring exposed to maternal separation (Uchida et al. 2010). Early life stress-associated epigenetic variation may account for increased stress vulnerability in response to subsequent stressors as both behavioral and epigenetic variations are exacerbated when maternal separation is combined with adult chronic stress exposure (Seo et al. 2016). Finally, studies in primates illustrate the integration of environment, genetics, and epigenetics in the study of stress vulnerability. Among rhesus macaques that possess the risk SLC6A4 gene variant, DNA methylation of the SLC6A4 gene rather than SLC6A4 gene sequence predicts heightened effects of maternal separation on behavioral stress reactivity in infants (Kinnally et al. 2010). Putative risk genotypes may thus mediate their effects via altered epigenetic variation, suggesting that the phenotypic effects of genes may be shifted through targeting of the epigenome.
Plasticity is typically a phenomenon associated with being young. However, it is apparent that, despite the potential stability of epigenetic effects of early life experiences, epigenetic plasticity can persist across the life span in response to social stress. In humans, adult trauma exposure is associated with epigenetic age acceleration (Boks et al. 2015), and altered DNA methylation is associated with adult SES (Subramanyam et al. 2013). Studies of SES have typically focused on the link between childhood SES and health outcomes; however, given the plasticity of the epigenome, a lifecourse perspective may be more informative in predicting, for example, indices of biological weathering such as epigenetic age acceleration (Simons et al. 2016). Studies in laboratory rodents indicate that a variety of social stressors in adulthood, including social exclusion (Krause et al. 2015) and exposure to aggressive social interactions (Jung et al. 2015; Kenworthy et al. 2014), can impact the epigenome, and pharmacological targeting of histones may ameliorate the effects of social stress (Covington et al. 2015). Moreover, resilience to stress can be described from an epigenetic perspective. Among adult mice exposed to social stress, there is decreased DNA methylation within the corticotropin-releasing factor (CRF) gene—a key player within the HPA response to stress (Elliott et al. 2010). However, among individual mice that are resilient to social stress (i.e. do not display social avoidance or depressive-like behaviors following social stress exposure), there is no alteration in DNA methylation of CRF—the gene remains epigenetically silent. Overall, increasing evidence for epigenetic plasticity in adulthood suggests that intervention and reversal of both genetic and environmentally mediated effects may be possible long after the sensitive period of early development. Further, it may be possible to “re-open” plasticity beyond classic critical periods occurring prenatally or in childhood, leading to improved biobehavioral functioning (Takesian and Hensch 2013).
Revisiting the Inheritance of Acquired Characteristics
The discovery of DNA canalized the gene-centric view of inheritance. This view was inconsistent with the notion of the inheritance of acquired characteristics that was historically integrated into theories of inheritance (Zirkle 1935) and was developed further by Jean Baptiste Lamarck into a theory of evolution (Lamarck 1809). Lamarck posited that the characteristics of an organism were driven by the “habits of life”—a statement describing the dynamic developmental process whereby environments shape the individual. Lamarck also described a process whereby if the environmental exposures that are driving the “habits of life” were to be sustained over chronological time and repeated across several generations, the phenotypes that emerged would be passed to descents and preserved by heritability. The notion of the inheritance of acquired characteristics, within the context of Lamarckian theory, rests heavily on the idea that the phenotypic adaptations that emerge within an individual are important for the development and survival of that individual. To lose those adaptations from one generation to the next was to compromise the development and survival of generations to come. As Paul Kammerer, a biologist and proponent of Lamarckian theory, once wrote: “If acquired characteristics cannot be passed on … then no true organic progress is possible. Man lives and suffers in vain. Whatever he might have acquired in the course of his lifetime dies with him. His children and his children’s children must ever and again start from the bottom” (Kammerer 1914). However, without mechanistic support, the idea of the inheritance of acquired characteristics failed to flourish and was supplanted by the more rapidly developing ideas within quantitative and subsequently molecular genetics.
Within epigenetics, the inheritance of acquired characteristics has gained new momentum, primarily due to experimental studies illustrating the transmission of environmentally induced phenotypes from one generation to the next (see Fig. 10.1).
Critical examples of epigenetic inheritance come from studies within social and behavioral epigenetics. For example, male mice exposed to social instability (i.e. changing social groups repeatedly to prevent the establishment of stable social groups) are altered in their phenotype—this manipulation leads to increased indices of stress (Saavedra-Rodríguez and Feig 2013). Grand-offspring and great-grand-offspring of stressed males exhibit increased indices of anxiety. This transmission is remarkable given that laboratory male mice have no postnatal contact with their offspring and that the transmission to great-grand-offspring occurs exclusively through the patriline (i.e. via male descendants). Though this transmission does not reveal a biological mechanism, it strongly suggests a germline inheritance of an environmentally induced effect. Analyses of sperm from males exposed to stress in early life or in adulthood indicate epigenetic variation, including altered DNA methylation and increased microRNA expression (Franklin et al. 2010; Gapp et al. 2014; Rodgers et al. 2013). Further, these epigenetic changes are also observed in the offspring of exposed males, and the phenotypes observed in offspring can be generated by manipulating epigenetic variation in the developing embryo (Rodgers et al. 2015; Gapp et al. 2014). Though the issue of how these epigenetic marks survive the epigenetic reprogramming that is occurring post-fertilization remains, there is increasing support for the hypothesis that epigenetic inheritance is possible and may have adaptive consequences for offspring (Zeybel et al. 2012).
Though developmental studies of social and behavioral epigenetics have focused primarily on mothers, it is notable that studies of epigenetic inheritance focus almost exclusively on fathers. The rational for this parental divide is in the relative role of mothers vs. fathers in mammalian reproduction. While mothers create the context of development during prenatal and postnatal life, the role of fathers is limited to fertilization. Thus, for an epigenetic inheritance to occur via the patriline, it is assumed that the only route possible is via sperm/seminal fluid (Curley et al. 2011). However, mothers are also capable of transmitting traits across generations via epigenetic mechanisms. In contrast to fathers, mothers achieve this transmission through their interactions with offspring (Champagne 2011). For example, variation in maternal LG is transmitted across generations via the matriline, such that offspring and grand-offspring of low-LG mothers also engage in low levels of LG. This transmission occurs in response to the effects of postnatal LG on epigenetic regulation of the estrogen receptor alpha gene (ESR1) within the developing hypothalamus. The experience of low levels of LG results in epigenetic silencing of ESR1, and this effect persists into adulthood, rendering female offspring less sensitive to estrogens and less primed to engage in maternal behavior (Champagne et al. 2006; Peña et al. 2013). As a consequence, the LG phenotype persists in the next generation. Similar cycles have been observed in laboratory rats in response to abusive maternal care mediated through epigenetic regulation of BDNF (Roth et al. 2009). Maternal transmission of epigenetic effects across generations is entirely experience-dependent and can be modified by stress or social support (Champagne and Meaney 2007; Champagne and Meaney 2006) allowing for heightened responsiveness to intervention and changing environmental conditions. Finally, though paternal and maternal inheritance systems are often dissociated—either experimentally or theoretically—it is important to take an integrative perspective when considering how parents can epigenetically influence their offspring. Much like genes and environments, parents interact to produce phenotypic outcomes.
Epigenetics and the Gene
Given changing views of development and inheritance contributed to by advances in the study of epigenetics—how should we think about the gene? Genetics is certainly thriving and is central to many new health initiatives including the Precision Medicine Initiative in the United States (Goodman et al. 2016) and the global Human Variome Project (Burn and Watson 2016). These initiatives focus on genome-wide sequencing of DNA as a strategy for improved diagnosis and treatment of disease. There is also increased availability of direct-to-consumer genetic sequencing resources aimed at defining the origins of individual traits or characterizing an individual’s ancestry (Niemiec and Howard 2016; Phillips 2016). Thus, “identity” is still largely linked to DNA despite growing acceptance of the role of gene regulatory processes in shaping development and inheritance. A significant barrier to a better integration of epigenetics and genetics is likely methodological. DNA is stable and identical across tissues. Epigenetic variation is tissue specific and can vary within and across days. The divergent properties of these two molecular features within our cells create challenges when trying to generate a cohesive predictive model of phenotypic outcomes. Overcoming these challenges will be essential to better understand how knowledge of DNA and knowledge of epigenetic profiles can be better used in the design of interventions and to shape public views on the plasticity vs. stability of our biology in response to the social environment.
Future Directions in Social and Behavioral Epigenetics
Research within the field of social and behavioral epigenetics is rapidly evolving through incorporation of novel methods in the analyses of epigenetic variation and broader application of these analyses to humans. Though DNA is still the primary focus of much of the diagnostic work in the biomedical sciences, within the social and behavioral sciences, there has been more substantial integration of epigenetics. Behavior is complex and dynamic—much like the epigenome—and it is perhaps this complexity that has motivated biological explanations to span beyond the constraints of DNA sequence. Epigenetic variation provides a molecular context to DNA, and there is increasing evidence that the phenomenon of GxE interactions is accounted for by epigenetic mechanisms. One of the many challenges ahead for social and behavioral epigenetics is in the integration of multiple levels of the social environment. The tactile interactions between a human mother and infant that trigger epigenetic effects are the consequence of a cascade of individual- and group-level factors that characterize the environment of families, communities, institutions, and nations. Though animal studies can be used to strip away that context to examine the proximal influences on development, translating these studies to humans requires a better understanding of the relationship between proximal and distal influences. A second challenge to the field involves the integration of genetics and epigenetics. The goal of studies within the field of epigenetics is not to replace the study of DNA. Rather, the goal is to integrate these molecular factors into a more comprehensive theory of the origins and inheritance of phenotype. This integrative approach will be necessary to avoid perpetuating nature vs. nurture dichotomies and to create a framework for understanding the coexistence of stability and plasticity of phenotypic variation.
References
Appleton, Allison A., David A. Armstrong, Corina Lesseur, Joyce Lee, James F. Padbury, Barry M. Lester, and Carmen J. Marsit. 2013. Patterning in Placental 11-B Hydroxysteroid Dehydrogenase Methylation According to Prenatal Socioeconomic Adversity. PloS One 8 (9): e74691.
Blaze, Jennifer, Arun Asok, and Tania L. Roth. 2015. Long-Term Effects of Early-Life Caregiving Experiences on Brain-Derived Neurotrophic Factor Histone Acetylation in the Adult Rat mPFC. Stress 18 (6): 607–615.
Boks, Marco P., Hans C. van Mierlo, Bart P.F. Rutten, Timothy R.D.J. Radstake, Lot De Witte, Elbert Geuze, Steve Horvath, et al. 2015. Longitudinal Changes of Telomere Length and Epigenetic Age Related to Traumatic Stress and Post-Traumatic Stress Disorder. Psychoneuroendocrinology 51: 506–512.
Braithwaite, E.C., M. Kundakovic, P.G. Ramchandani, S.E. Murphy, and F.A. Champagne. 2015. Maternal Prenatal Depressive Symptoms Predict Infant NR3C1 1F and BDNF IV DNA Methylation. Epigenetics 10 (5): 408–417.
Brody, Gene H., Gregory E. Miller, Tianyi Yu, Steven R.H. Beach, and Edith Chen. 2016a. Supportive Family Environments Ameliorate the Link Between Racial Discrimination and Epigenetic Aging: A Replication Across Two Longitudinal Cohorts. Psychological Science 27 (4): 530–541.
Brody, Gene H., Tianyi Yu, Edith Chen, Steven R.H. Beach, and Gregory E. Miller. 2016b. Family-Centered Prevention Ameliorates the Longitudinal Association between Risky Family Processes and Epigenetic Aging. Journal of Child Psychology and Psychiatry 57 (5): 566–574.
Brummelte, Susanne, and Liisa A.M. Galea. 2016. Postpartum Depression: Etiology, Treatment and Consequences for Maternal Care. Hormones and Behavior 77: 153–166.
Burn, John, and Michael Watson. 2016. The Human Variome Project. Human Mutation 37 (6): 505–507.
Burton, Graham J., and Eric Jauniaux. 2015. What Is the Placenta? American Journal of Obstetrics and Gynecology 213 (4 Suppl): S6.e1–SS6-8.
Cameron, Nicole M., Frances A. Champagne, Carine Parent, Eric W. Fish, Kumi Ozaki-Kuroda, and Michael J. Meaney. 2005. The Programming of Individual Differences in Defensive Responses and Reproductive Strategies in the Rat through Variations in Maternal Care. Neuroscience and Biobehavioral Reviews 29 (4–5): 843–865.
Caspi, Avshalom, Karen Sugden, Terrie E. Moffitt, Alan Taylor, Ian W. Craig, HonaLee Harrington, Joseph McClay, et al. 2003. Influence of Life Stress on Depression: Moderation by a Polymorphism in the 5-HTT Gene. Science (New York, N.Y.) 301 (5631): 386–389.
Champagne, Frances A. 2011. Maternal Imprints and the Origins of Variation. Hormones and Behavior 60 (1): 4–11.
––––——. 2012. Interplay between Social Experiences and the Genome: Epigenetic Consequences for Behavior. Advances in Genetics 77: 33–57.
Champagne, Frances A., Darlene D. Francis, Adam Mar, and Michael J. Meaney. 2003. Variations in Maternal Care in the Rat as a Mediating Influence for the Effects of Environment on Development. Physiology & Behavior 79 (3): 359–371.
Champagne, Frances A., and Michael J. Meaney. 2006. Stress during Gestation Alters Postpartum Maternal Care and the Development of the Offspring in a Rodent Model. Biological Psychiatry 59 (12): 1227–1235.
––––——. 2007. Transgenerational Effects of Social Environment on Variations in Maternal Care and Behavioral Response to Novelty. Behavioral Neuroscience 121 (6): 1353–1363.
Champagne, Frances A., Ian C.G. Weaver, Josie Diorio, Sergiy Dymov, Moshe Szyf, and Michael J. Meaney. 2006. Maternal Care Associated with Methylation of the Estrogen Receptor-alpha1b Promoter and Estrogen Receptor-Alpha Expression in the Medial Preoptic Area of Female Offspring. Endocrinology 147 (6): 2909–2915.
Chen, J., A.N. Evans, Y. Liu, M. Honda, J.M. Saavedra, and G. Aguilera. 2012. Maternal Deprivation in Rats Is Associated with Corticotrophin-Releasing Hormone (CRH) Promoter Hypomethylation and Enhances CRH Transcriptional Responses to Stress in Adulthood. Journal of Neuroendocrinology 24 (7): 1055–1064.
Cheung, P., C.D. Allis, and P. Sassone-Corsi. 2000. Signaling to Chromatin through Histone Modifications. Cell 103 (2): 263–271.
Cicchetti, Dante, Fred A. Rogosch, and Sheree L. Toth. 2006. Fostering Secure Attachment in Infants in Maltreating Families through Preventive Interventions. Development and Psychopathology 18 (3): 623–649.
Conradt, Elisabeth, Mary Fei, Linda LaGasse, Edward Tronick, Dylan Guerin, Daniel Gorman, Carmen J. Marsit, and Barry M. Lester. 2015. Prenatal Predictors of Infant Self-Regulation: The Contributions of Placental DNA Methylation of NR3C1 and Neuroendocrine Activity. Frontiers in Behavioral Neuroscience 9: 130.
Covington, H.E., I. Maze, V. Vialou, and E.J. Nestler. 2015. Antidepressant Action of HDAC Inhibition in the Prefrontal Cortex. Neuroscience 298: 329–335.
Culp, L.A., E. Dore, and G.M. Brown. 1970. Methylated Bases in DNA of Animal Origin. Archives of Biochemistry and Biophysics 136 (1): 73–79.
Curley, James P., Stephanie Davidson, Patrick Bateson, and Frances A. Champagne. 2009. Social Enrichment during Postnatal Development Induces Transgenerational Effects on Emotional and Reproductive Behavior in Mice. Frontiers in Behavioral Neuroscience 3: 25.
Curley, James P., Rahia Mashoodh, and Frances A. Champagne. 2011. Epigenetics and the Origins of Paternal Effects. Hormones and Behavior 59 (3): 306–314.
Danchin, Étienne, Anne Charmantier, Frances A. Champagne, Alex Mesoudi, Benoit Pujol, and Simon Blanchet. 2011. Beyond DNA: Integrating Inclusive Inheritance into an Extended Theory of Evolution. Nature Reviews Genetics 12 (7): 475–486.
Doherty, Tiffany S., Amy Forster, and Tania L. Roth. 2016. Global and Gene-Specific DNA Methylation Alterations in the Adolescent Amygdala and Hippocampus in an Animal Model of Caregiver Maltreatment. Behavioural Brain Research 298 (Pt A): 55–61.
Dolinoy, Dana C., Jennifer R. Weidman, Robert A. Waterland, and Randy L. Jirtle. 2006. Maternal Genistein Alters Coat Color and Protects Avy Mouse Offspring from Obesity by Modifying the Fetal Epigenome. Environmental Health Perspectives 114 (4): 567–572.
Dollberg, Daphna G., Tamir Rozenfeld, and Michael Kupfermincz. 2016. Early Parental Adaptation, Prenatal Distress, and High-Risk Pregnancy. Journal of Pediatric Psychology 41 (8): 915–929.
Elliott, Evan, Gili Ezra-Nevo, Limor Regev, Adi Neufeld-Cohen, and Alon Chen. 2010. Resilience to Social Stress Coincides with Functional DNA Methylation of the Crf Gene in Adult Mice. Nature Neuroscience 13 (11): 1351–1353.
Fan, Guoping, and Leah Hutnick. 2005. Methyl-CpG Binding Proteins in the Nervous System. Cell Research 15 (4): 255–261.
Farré, Pau, Meaghan J. Jones, Michael J. Meaney, Eldon Emberly, Gustavo Turecki, and Michael S. Kobor. 2015. Concordant and Discordant DNA Methylation Signatures of Aging in Human Blood and Brain. Epigenetics & Chromatin 8: 19.
Ferber, Sari Goldstein, Ruth Feldman, and Imad R. Makhoul. 2008. The Development of Maternal Touch across the First Year of Life. Early Human Development 84 (6): 363–370.
Fowden, A.L., and T. Moore. 2012. Maternal-Fetal Resource Allocation: Co-Operation and Conflict. Placenta 33 (Suppl 2): e11–e15.
Francis, D., J. Diorio, D. Liu, and M.J. Meaney. 1999. Nongenomic Transmission across Generations of Maternal Behavior and Stress Responses in the Rat. Science 286 (5442): 1155–1158.
Franklin, Tamara B., Holger Russig, Isabelle C. Weiss, Johannes Gräff, Natacha Linder, Aubin Michalon, Sandor Vizi, and Isabelle M. Mansuy. 2010. Epigenetic Transmission of the Impact of Early Stress across Generations. Biological Psychiatry 68 (5): 408–415.
Gapp, Katharina, Ali Jawaid, Peter Sarkies, Johannes Bohacek, Pawel Pelczar, Julien Prados, Laurent Farinelli, Eric Miska, and Isabelle M. Mansuy. 2014. Implication of Sperm RNAs in Transgenerational Inheritance of the Effects of Early Trauma in Mice. Nature Neuroscience 17 (5): 667–669.
Goodman, Deborah, Catherine O. Johnson, Lari Wenzel, Deborah Bowen, Celeste Condit, and Karen L. Edwards. 2016. Consent Issues in Genetic Research: Views of Research Participants. Public Health Genomics 19: 220–228.
Graur, Dan, Yichen Zheng, and Ricardo B.R. Azevedo. 2015. An Evolutionary Classification of Genomic Function. Genome Biology and Evolution 7 (3): 642–645.
Griffith, J.S., and H.R. Mahler. 1969. DNA Ticketing Theory of Memory. Nature 223 (5206): 580–582.
Hane, Amie Ashley, Heather A. Henderson, Bethany C. Reeb-Sutherland, and Nathan A. Fox. 2010. Ordinary Variations in Human Maternal Caregiving in Infancy and Biobehavioral Development in Early Childhood: A Follow-up Study. Developmental Psychobiology 52 (6): 558–567.
Hellstrom, Ian C., Sabine K. Dhir, Josie C. Diorio, and Michael J. Meaney. 2012. Maternal Licking Regulates Hippocampal Glucocorticoid Receptor Transcription through a Thyroid Hormone-Serotonin-NGFI-A Signalling Cascade. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 367 (1601): 2495–2510.
Hompes, Titia, Benedetta Izzi, Edith Gellens, Maarten Morreels, Steffen Fieuws, Anne Pexsters, Ganel Schops, et al. 2013. Investigating the Influence of Maternal Cortisol and Emotional State during Pregnancy on the DNA Methylation Status of the Glucocorticoid Receptor Gene (NR3C1) Promoter Region in Cord Blood. Journal of Psychiatric Research 47 (7): 880–891.
Horvath, Steve. 2013. DNA Methylation Age of Human Tissues and Cell Types. Genome Biology 14 (10): R115.
Horvath, Steve, and Andrew J. Levine. 2015. HIV-1 Infection Accelerates Age According to the Epigenetic Clock. The Journal of Infectious Diseases 212 (10): 1563–1573.
Jenkins, Jennifer M., Patrick McGowan, and Ariel Knafo-Noam. 2016. Parent-Offspring Transaction: Mechanisms and the Value of within Family Designs. Hormones and Behavior 77: 53–61.
Jensen, A.B., S.J. Tunster, and R.M. John. 2014. The Significance of Elevated Placental PHLDA2 in Human Growth Restricted Pregnancies. Placenta 35 (8): 528–532.
Jenuwein, T., and C.D. Allis. 2001. Translating the Histone Code. Science 293 (5532): 1074–1080.
Jones, P.A., S.M. Taylor, and V. Wilson. 1983. DNA Modification, Differentiation, and Transformation. The Journal of Experimental Zoology 228 (2): 287–295.
Jung, Seung Ho, Yufen Wang, Taewan Kim, Andrew Tarr, Brenda Reader, Nicole Powell, and John F. Sheridan. 2015. Molecular Mechanisms of Repeated Social Defeat-Induced Glucocorticoid Resistance: Role of microRNA. Brain, Behavior, and Immunity 44: 195–206.
Kammerer, Paul. 1914. The Inheritance of Acquired Characteristics. New York: Boni and Liveright.
Kane, Heidi S., Christine Dunkel Schetter, Laura M. Glynn, Calvin J. Hobel, and Curt A. Sandman. 2014. Pregnancy Anxiety and Prenatal Cortisol Trajectories. Biological Psychology 100 (July): 13–19.
Kember, R.L., E.L. Dempster, T.H.A. Lee, L.C. Schalkwyk, J. Mill, and C. Fernandes. 2012. Maternal Separation Is Associated with Strain-Specific Responses to Stress and Epigenetic Alterations to Nr3c1, Avp, and Nr4a1 in Mouse. Brain and Behavior: A Cognitive Neuroscience Perspective 2 (4): 455–467.
Kenworthy, C.A., A. Sengupta, S.M. Luz, E.S. Ver Hoeve, K. Meda, S. Bhatnagar, and T. Abel. 2014. Social Defeat Induces Changes in Histone Acetylation and Expression of Histone Modifying Enzymes in the Ventral Hippocampus, Prefrontal Cortex, and Dorsal Raphe Nucleus. Neuroscience 264: 88–98.
Kinnally, E.L., J.P. Capitanio, R. Leibel, L. Deng, C. LeDuc, F. Haghighi, and J.J. Mann. 2010. Epigenetic Regulation of Serotonin Transporter Expression and Behavior in Infant Rhesus Macaques. Genes, Brain, and Behavior 9 (6): 575–582.
Koubovec, D., L. Geerts, H.J. Odendaal, Dan J. Stein, and B. Vythilingum. 2005. Effects of Psychologic Stress on Fetal Development and Pregnancy Outcome. Current Psychiatry Reports 7 (4): 274–280.
Krause, Linda, Bernhard Haubold, and Angelika G. Börsch-Haubold. 2015. Social Exclusion Changes Histone Modifications H3K4me3 and H3K27ac in Liver Tissue of Wild House Mice. PloS One 10 (8): e0133988.
Kundakovic, Marija, Sean Lim, Kathryn Gudsnuk, and Frances A. Champagne. 2013. Sex-Specific and Strain-Dependent Effects of Early Life Adversity on Behavioral and Epigenetic Outcomes. Frontiers in Psychiatry 4: 78.
Kundakovic, Marija, Kathryn Gudsnuk, Julie B. Herbstman, Deliang Tang, Frederica P. Perera, and Frances A. Champagne. 2015. DNA Methylation of BDNF as a Biomarker of Early-Life Adversity. Proceedings of the National Academy of Sciences of the United States of America 112 (22): 6807–6813.
Laland, Kevin N., Tobias Uller, Marcus W. Feldman, Kim Sterelny, Gerd B. Müller, Armin Moczek, Eva Jablonka, and John Odling-Smee. 2015. The Extended Evolutionary Synthesis: Its Structure, Assumptions and Predictions. Proceedings. Biological Sciences/The Royal Society 282 (1813): 20151019.
Lam, Lucia L., Eldon Emberly, Hunter B. Fraser, Sarah M. Neumann, Edith Chen, Gregory E. Miller, and Michael S. Kobor. 2012. Factors Underlying Variable DNA Methylation in a Human Community Cohort. Proceedings of the National Academy of Sciences of the United States of America 109 (Suppl 2): 17253–17260.
Lamarck, J.-B. 1809. Philosophie Zoologique. France: National Museum of Natural History.
Liu, D., J. Diorio, B. Tannenbaum, C. Caldji, D. Francis, A. Freedman, S. Sharma, D. Pearson, P.M. Plotsky, and M.J. Meaney. 1997. Maternal Care, Hippocampal Glucocorticoid Receptors, and Hypothalamic-Pituitary-Adrenal Responses to Stress. Science 277 (5332): 1659–1662.
Lowell, Darcy I., Alice S. Carter, Leandra Godoy, Belinda Paulicin, and Margaret J. Briggs-Gowan. 2011. A Randomized Controlled Trial of Child FIRST: A Comprehensive Home-Based Intervention Translating Research into Early Childhood Practice. Child Development 82 (1): 193–208.
Maestripieri, D., K. Wallen, and K.A. Carroll. 1997. Infant Abuse Runs in Families of Group-Living Pigtail Macaques. Child Abuse & Neglect 21 (5): 465–471.
McGowan, Patrick O., Aya Sasaki, Ana C. D’Alessio, Sergiy Dymov, Benoit Labonté, Moshe Szyf, Gustavo Turecki, and Michael J. Meaney. 2009. Epigenetic Regulation of the Glucocorticoid Receptor in Human Brain Associates with Childhood Abuse. Nature Neuroscience 12 (3): 342–348.
McGowan, Patrick O., Matthew Suderman, Aya Sasaki, Tony C.T. Huang, Michael Hallett, Michael J. Meaney, and Moshe Szyf. 2011. Broad Epigenetic Signature of Maternal Care in the Brain of Adult Rats. PloS One 6 (2): e14739.
Meaney, M.J. 2001. Maternal Care, Gene Expression, and the Transmission of Individual Differences in Stress Reactivity across Generations. Annual Review of Neuroscience 24: 1161–1192.
Meaney, Michael J. 2010. Epigenetics and the Biological Definition of Gene X Environment Interactions. Child Development 81 (1): 41–79.
Molina-Serrano, Diego, Vassia Schiza, and Antonis Kirmizis. 2013. Cross-Talk among Epigenetic Modifications: Lessons from Histone Arginine Methylation. Biochemical Society Transactions 41 (3): 751–759.
Monk, Catherine, Tianshu Feng, Seonjoo Lee, Izabela Krupska, Frances A. Champagne, and Benjamin Tycko. 2016. Distress during Pregnancy: Epigenetic Regulation of Placenta Glucocorticoid-Related Genes and Fetal Neurobehavior. The American Journal of Psychiatry 173 (7): 705–713.
Monk, Catherine, Julie Spicer, and Frances A. Champagne. 2012. Linking Prenatal Maternal Adversity to Developmental Outcomes in Infants: The Role of Epigenetic Pathways. Development and Psychopathology 24 (4): 1361–1376.
Mueller, Bridget R., and Tracy L. Bale. 2008. Sex-Specific Programming of Offspring Emotionality after Stress Early in Pregnancy. Journal of Neuroscience 28 (36): 9055–9065.
Murgatroyd, Chris, Alexandre V. Patchev, Yonghe Wu, Vincenzo Micale, Yvonne Bockmühl, Dieter Fischer, Florian Holsboer, Carsten T. Wotjak, Osborne F.X. Almeida, and Dietmar Spengler. 2009. Dynamic DNA Methylation Programs Persistent Adverse Effects of Early-Life Stress. Nature Neuroscience 12 (12): 1559–1566.
Naumova, Oksana Yu, Maria Lee, Roman Koposov, Moshe Szyf, Mary Dozier, and Elena L. Grigorenko. 2012. Differential Patterns of Whole-Genome DNA Methylation in Institutionalized Children and Children Raised by Their Biological Parents. Development and Psychopathology 24 (1): 143–155.
Nemoda, Z., R. Massart, M. Suderman, M. Hallett, T. Li, M. Coote, N. Cody, et al. 2015. Maternal Depression Is Associated with DNA Methylation Changes in Cord Blood T Lymphocytes and Adult Hippocampi. Translational Psychiatry 5: e545.
Niemiec, Emilia, and Heidi Carmen Howard. 2016. Ethical Issues in Consumer Genome Sequencing: Use of Consumers’ Samples and Data. Applied & Translational Genomics 8: 23–30.
Non, Amy L., Brittany M. Hollister, Kathryn L. Humphreys, Ainash Childebayeva, Kyle Esteves, Charles H. Zeanah, Nathan A. Fox, Charles A. Nelson, and Stacy S. Drury. 2016. DNA Methylation at Stress-Related Genes is Associated with Exposure to Early Life Institutionalization. American Journal of Physical Anthropology. doi:10.1002/ajpa.23010.
Novakovic, Boris, Nick C. Wong, Mandy Sibson, Hong-Kiat Ng, Ruth Morley, Ursula Manuelpillai, Thomas Down, et al. 2010. DNA Methylation-Mediated down-Regulation of DNA Methyltransferase-1 (DNMT1) Is Coincident With, but Not Essential For, Global Hypomethylation in Human Placenta. The Journal of Biological Chemistry 285 (13): 9583–9593.
O’Connor, Thomas G., Tang Wan, Michelle A. Gilchrist, Jan A. Moynihan, Eva K. Pressman, and Emma Robertson Blackmore. 2014. Diurnal Cortisol Patterns and Psychiatric Symptoms in Pregnancy: Short-Term Longitudinal Study. Biological Psychology 96 (February): 35–41.
Ohno, S. 1972. So much ‘junk’ DNA in Our Genome. Brookhaven Symposia in Biology 23: 366–370.
Paquette, Alison G., Barry M. Lester, Devin C. Koestler, Corina Lesseur, David A. Armstrong, and Carmen J. Marsit. 2014. Placental FKBP5 Genetic and Epigenetic Variation Is Associated with Infant Neurobehavioral Outcomes in the RICHS Cohort. PloS One 9 (8): e104913.
Paternain, Laura, Eva Martisova, Campión Javier, J. Alfredo Martínez, Maria J. Ramírez, and Fermin I. Milagro. 2016. Methyl Donor Supplementation in Rats Reverses the Deleterious Effect of Maternal Separation on Depression-like Behaviour. Behavioural Brain Research 299: 51–58.
Peña, Catherine Jensen, Y. Dana Neugut, and Frances A. Champagne. 2013. Developmental Timing of the Effects of Maternal Care on Gene Expression and Epigenetic Regulation of Hormone Receptor Levels in Female Rats. Endocrinology 154 (11): 4340–4351.
Peña, Catherine Jensen, Catherine Monk, and Frances A. Champagne. 2012. Epigenetic Effects of Prenatal Stress on 11β-Hydroxysteroid Dehydrogenase-2 in the Placenta and Fetal Brain. PloS One 7 (6): e39791.
Phillips, Andelka M. 2016. Only a Click Away—DTC Genetics for Ancestry, Health, Love…and More: A View of the Business and Regulatory Landscape. Applied & Translational Genomics 8: 16–22.
Radtke, K.M., M. Ruf, H.M. Gunter, K. Dohrmann, M. Schauer, A. Meyer, and T. Elbert. 2011. Transgenerational Impact of Intimate Partner Violence on Methylation in the Promoter of the Glucocorticoid Receptor. Translational Psychiatry 1: e21.
Razin, A. 1998. CpG Methylation, Chromatin Structure and Gene Silencing-a Three-Way Connection. The EMBO Journal 17 (17): 4905–4908.
Razin, A., and A.D. Riggs. 1980. DNA Methylation and Gene Function. Science 210 (4470): 604–610.
Rodgers, Ali B., Christopher P. Morgan, N. Adrian Leu, and Tracy L. Bale. 2015. Transgenerational Epigenetic Programming via Sperm microRNA Recapitulates Effects of Paternal Stress. Proceedings of the National Academy of Sciences of the United States of America 112 (44): 13699–13704.
Rodgers, Ali B., Christopher P. Morgan, Stefanie L. Bronson, Sonia Revello, and Tracy L. Bale. 2013. Paternal Stress Exposure Alters Sperm microRNA Content and Reprograms Offspring HPA Stress Axis Regulation. Journal of Neuroscience 33 (21): 9003–9012.
Roifman, Maian, Sanaa Choufani, Andrei L. Turinsky, Sascha Drewlo, Sarah Keating, Michael Brudno, John Kingdom, and Rosanna Weksberg. 2016. Genome-Wide Placental DNA Methylation Analysis of Severely Growth-Discordant Monochorionic Twins Reveals Novel Epigenetic Targets for Intrauterine Growth Restriction. Clinical Epigenetics 8: 70.
Roth, Tania L., Farah D. Lubin, Adam J. Funk, and J. David Sweatt. 2009. Lasting Epigenetic Influence of Early-Life Adversity on the BDNF Gene. Biological Psychiatry 65 (9): 760–769.
Ruppenthal, G.C., G.L. Arling, H.F. Harlow, G.P. Sackett, and S.J. Suomi. 1976. A 10-Year Perspective of Motherless-Mother Monkey Behavior. Journal of Abnormal Psychology 85 (4): 341–349.
Saavedra-Rodríguez, Lorena, and Larry A. Feig. 2013. Chronic Social Instability Induces Anxiety and Defective Social Interactions across Generations. Biological Psychiatry 73 (1): 44–53.
Sato, Fumiaki, Soken Tsuchiya, Stephen J. Meltzer, and Kazuharu Shimizu. 2011. MicroRNAs and Epigenetics. The FEBS Journal 278 (10): 1598–1609.
Seo, Mi Kyoung, Nguyen Ngoc Ly, Chan Hong Lee, Hye Yeon Cho, Cheol Min Choi, Le Hoa Nhu, Jung Goo Lee, et al. 2016. Early Life Stress Increases Stress Vulnerability through BDNF Gene Epigenetic Changes in the Rat Hippocampus. Neuropharmacology 105: 388–397.
Shuman, B.J., and F. Masterpasqua. 1981. Preventive Intervention during the Perinatal and Infancy Periods: Overview and Guidelines for Evaluation. Prevention in Human Services 1 (1–2): 41–57.
Simons, Ronald L., Man Kit Lei, Steven R.H. Beach, Robert A. Philibert, Carolyn E. Cutrona, Frederick X. Gibbons, and Ashley Barr. 2016. Economic Hardship and Biological Weathering: The Epigenetics of Aging in a U.S. Sample of Black Women. Social Science & Medicine (1982) 150: 192–200.
Simpkin, Andrew J., Gibran Hemani, Matthew Suderman, Tom R. Gaunt, Oliver Lyttleton, Wendy L. Mcardle, Susan M. Ring, et al. 2016. Prenatal and Early Life Influences on Epigenetic Age in Children: A Study of Mother-Offspring Pairs from Two Cohort Studies. Human Molecular Genetics 25 (1): 191–201.
Sitras, Vasilis, Christopher Fenton, Ruth Paulssen, Åse Vårtun, and G. Acharya. 2012. Differences in Gene Expression between First and Third Trimester Human Placenta: A Microarray Study. PloS One 7 (3): e33294.
Subramanyam, Malavika A., Ana V. Diez-Roux, J. Richard Pilsner, Eduardo Villamor, Kathleen M. Donohue, Yongmei Liu, and Nancy S. Jenny. 2013. Social Factors and Leukocyte DNA Methylation of Repetitive Sequences: The Multi-Ethnic Study of Atherosclerosis. PloS One 8 (1): e54018.
Suderman, Matthew, Patrick O. McGowan, Aya Sasaki, Tony C.T. Huang, Michael T. Hallett, Michael J. Meaney, Gustavo Turecki, and Moshe Szyf. 2012. Conserved Epigenetic Sensitivity to Early Life Experience in the Rat and Human Hippocampus. Proceedings of the National Academy of Sciences of the United States of America 109 (Suppl 2): 17266–17272.
Swartz, J. R., A. R. Hariri, and D. E. Williamson. 2016. An Epigenetic Mechanism Links Socioeconomic Status to Changes in Depression-Related Brain Function in High-Risk Adolescents. Molecular Psychiatry, May.
Takesian, Anne E., and Takao K. Hensch. 2013. Balancing Plasticity/stability across Brain Development. Progress in Brain Research 207: 3–34.
Tragante, Vinicius, Jason H. Moore, and Folkert W. Asselbergs. 2014. The ENCODE Project and Perspectives on Pathways. Genetic Epidemiology 38 (4): 275–280.
Uchida, Shusaku, Kumiko Hara, Ayumi Kobayashi, Hiromasa Funato, Teruyuki Hobara, Koji Otsuki, Hirotaka Yamagata, Bruce S. McEwen, and Yoshifumi Watanabe. 2010. Early Life Stress Enhances Behavioral Vulnerability to Stress through the Activation of REST4-Mediated Gene Transcription in the Medial Prefrontal Cortex of Rodents. Journal of Neuroscience 30 (45): 15007–15018.
Unternaehrer, Eva, Margarete Bolten, Irina Nast, Simon Staehli, Andrea H. Meyer, Emma Dempster, Dirk H. Hellhammer, Roselind Lieb, and Gunther Meinlschmidt. 2016. Maternal Adversities during Pregnancy and Cord Blood Oxytocin Receptor (OXTR) DNA Methylation. Social Cognitive and Affective Neuroscience 11 (9): 1460–1470.
Waddington, C.H. 1940. Organisers & Genes. Cambridge: Cambridge University Press.
Weaver, Ian C.G., Nadia Cervoni, Frances A. Champagne, Ana C. D’Alessio, Shakti Sharma, Jonathan R. Seckl, Sergiy Dymov, Moshe Szyf, and Michael J. Meaney. 2004. Epigenetic Programming by Maternal Behavior. Nature Neuroscience 7 (8): 847–854.
Weaver, Ian C.G., Frances A. Champagne, Shelley E. Brown, Sergiy Dymov, Shakti Sharma, Michael J. Meaney, and Moshe Szyf. 2005. Reversal of Maternal Programming of Stress Responses in Adult Offspring through Methyl Supplementation: Altering Epigenetic Marking Later in Life. Journal of Neuroscience 25 (47): 11045–11054.
Weinstock, Marta. 2008. The Long-Term Behavioural Consequences of Prenatal Stress. Neuroscience and Biobehavioral Reviews 32 (6): 1073–1086.
Winett, Liana B., Alyssa B. Wulf, and Lawrence Wallack. 2016. Framing Strategies to Avoid Mother-Blame in Communicating the Origins of Chronic Disease. American Journal of Public Health 106 (8): 1369–1373.
Yehuda, Rachel, Nikolaos P. Daskalakis, Amy Lehrner, Frank Desarnaud, Heather N. Bader, Iouri Makotkine, Janine D. Flory, Linda M. Bierer, and Michael J. Meaney. 2014. Influences of Maternal and Paternal PTSD on Epigenetic Regulation of the Glucocorticoid Receptor Gene in Holocaust Survivor Offspring. The American Journal of Psychiatry 171 (8): 872–880.
Zeybel, Müjdat, Timothy Hardy, Yi K. Wong, John C. Mathers, Christopher R. Fox, Agata Gackowska, Fiona Oakley, et al. 2012. Multigenerational Epigenetic Adaptation of the Hepatic Wound-Healing Response. Nature Medicine 18 (9): 1369–1377.
Zirkle, Conway. 1935. The Inheritance of Acquired Characters and the Provisional Hypothesis of Pangenesis. The American Naturalist 69 (724): 417–445.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Copyright information
© 2018 The Author(s)
About this chapter
Cite this chapter
Champagne, F.A. (2018). Social and Behavioral Epigenetics: Evolving Perspectives on Nature-Nurture Interplay, Plasticity, and Inheritance. In: Meloni, M., Cromby, J., Fitzgerald, D., Lloyd, S. (eds) The Palgrave Handbook of Biology and Society. Palgrave Macmillan, London. https://doi.org/10.1057/978-1-137-52879-7_10
Download citation
DOI: https://doi.org/10.1057/978-1-137-52879-7_10
Publisher Name: Palgrave Macmillan, London
Print ISBN: 978-1-137-52878-0
Online ISBN: 978-1-137-52879-7
eBook Packages: Social SciencesSocial Sciences (R0)