Abstract
Treatment of carriers of the CYP2C19*2 allele and ABCB1 TT genotype with clopidogrel is associated with increased ischemic complications after percutaneous coronary intervention (PCI). We sought to evaluate a pharmacogenomic strategy among patients undergoing PCI for ST-elevation myocardial infarction (STEMI), by performing a randomized trial, enrolling 102 patients. Point-of-care genetic testing for CYP2C19*2, ABCB1 TT and CYP2C19*17 was performed with carriers of either the CYP2C19*2 allele or ABCB1 TT genotype randomly assigned to a strategy of prasugrel 10 mg daily or an augmented dosing strategy of clopidogrel (150 mg daily for 6 days then 75 mg daily). The primary end point was the proportion of at-risk carriers exhibiting high on-treatment platelet reactivity (HPR), a marker associated with increased adverse cardiovascular events, after 1 month. Fifty-nine subjects (57.8%) were identified as carriers of at least one at-risk variant. Treatment with prasugrel significantly reduced HPR compared with clopidogrel by P2Y12 reaction unit (PRU) thresholds of >234 (0 vs 24.1%, P=0.0046) and PRU>208 (3.3 vs 34.5%, P=0.0025). The sensitivity of point-of-care testing was 100% (95% CI 88.0–100), 100% (86.3–100) and 96.9% (82.0–99.8) and specificity was 97.0% (88.5–99.5), 97.1% (89.0–99.5) and 98.5% (90.9–99.9) for identifying CYP2C19*2, ABCB1 TT and CYP2C19*17, respectively. Logistic regression confirmed carriers as a strong predictor of HPR (OR=6.58, 95% CI 1.24–34.92; P=0.03). We confirmed that concurrent identification of three separate genetic variants in patients with STEMI receiving PCI is feasible at the bedside. Among carriers of at-risk genotypes, treatment with prasugrel was superior to an augmented dosing strategy of clopidogrel in reducing HPR.
Similar content being viewed by others
Introduction
Inhibition of the platelet P2Y12 receptor is an integral component of therapy for patients with ST-elevation myocardial infarction (STEMI), especially among those receiving early percutaneous coronary intervention (PCI).1, 2, 3 Clopidogrel, the most widely studied P2Y12 inhibitor, has established efficacy in reducing major adverse cardiovascular events (MACE).1, 2, 4, 5, 6 Variability in pharmacodynamic response to clopidogrel is well described, and is associated with increased risk for MACE.7, 8 Prasugrel and ticagrelor provide potent P2Y12 inhibition and decrease MACE in acute coronary syndrome (ACS) when compared to clopidogrel.9, 10 Despite reducing ischemic complications, there is a reluctance for universal adoption of these agents due to their increased risk for bleeding and incremental cost relative to generic clopidogrel.11 Accordingly, there is ongoing interest in investigating the potential benefits of personalized strategies that restrict utilization of novel P2Y12 inhibitors to those at risk for clopidogrel failure.11
Common genetic variants that affect intestinal uptake and biotransformation of clopidogrel alter the levels of its active metabolite and subsequent inhibition of the P2Y12 receptor. Carriers of loss-of-function polymorphisms of the CYP219 gene have reduced clopidogrel-mediated P2Y12 inhibition and an increased risk for MACE after ACS and PCI.12, 13 Of these polymorphisms, the CYP2C19*2 allele (rs4244285) is the most common with a prevalence of up to 30% among those of western European descent and nearly 50% in Asians.14, 15 The ABCB1 gene encodes an intestinal efflux pump and influences clopidogrel absorption. Several studies have shown that patients homozygous for the ABCB1 3435 C→T (rs1045642) variant have an increased propensity for ischemic outcomes after ACS when treated with PCI and clopidogrel.14, 16, 17 In contrast, the CYP2C19*17 allele (rs12248560) upregulates CYP2C19 activity and has been associated with increased bleeding in patients on clopidogrel.18
Previously, our group validated the first point-of-care genetic testing device in clinical medicine and demonstrated the potential clinical utility of CYP2C19*2 genotyping among patients undergoing PCI for stable coronary artery disease and non-ST-elevation ACS.19 Point-of-care genetic testing may be particularly suited to STEMI, given the need for emergent treatment decisions and the prospect that personalized therapy may yield the greatest therapeutic benefit among higher-risk patients. Accordingly, we sought to expand point-of-care genotyping from detection of a single variant to three separate genetic variants and extend its use to a higher acuity population of STEMI patients. We further aimed to compare the pharmacodynamic effects of prasugrel against an augmented dosing strategy of clopidogrel, as validated in the Clopidogrel and Aspirin Optimal Dose Usage to Reduce Recurrent Events−Seventh Organization to Assess Strategies in Ischemic Syndromes (CURRENT–OASIS 7) trial,6 among patients identified as carriers of the CYP2C19*2 allele and/or the ABCB1 TT genotype.
Materials and Methods
The ReAssessment of antiPlatelet therapy using an InDividualized strategy in ST-elevation Myocardial Infarction (RAPID STEMI) trial was a prospective randomized study, which enrolled patients at the University of Ottawa Heart Institute. The study was conducted in accordance with the Declaration of Helsinki and approved by the local human research ethics board. The bedside genetic testing device (Spartan Rx, Spartan Biosciences, Ottawa, Canada) was approved by the Ottawa Hospital Point-of-Care Committee and Health Canada (Investigation Testing Application # 178941). The study was registered with clinicaltrials.gov (NCT01452139).
Participants
Patients were eligible for screening if they were between 18 and 75 years and underwent PCI for STEMI. Patients were excluded if they had: pre-treatment with prasugrel or ticagrelor, requirement for oral anticoagulation, history of stroke or transient ischemic attack, body weight <60 kg, platelet count <100 000 μl−1, known bleeding diathesis, hematocrit <30% or >52%, severe liver dysfunction, renal insufficiency (creatinine clearance <30 ml min−1) or treatment with glycoprotein IIb/IIIa inhibitors in the preceding 24 h. As per the regional STEMI protocol at the time, patients triaged for reperfusion by primary PCI received a 600-mg clopidogrel bolus immediately upon confirmation of STEMI diagnosis. For patients receiving thrombolytic therapy before PCI, 300 mg of clopidogrel was given after initiation of thrombolytic therapy, with further bolus dosing dictated by the treating physician.
Procedure
Point-of-care genetic testing and core laboratory genotyping
All consented patients immediately underwent bedside genetic testing and received separate buccal swabs for each allele (CYP2C19*2, ABCB1 3435 C→T and CYP2C19*17). All the three swab cartridges were inserted concurrently into the bedside genetic testing device, as per our previously described protocol.19 Within 55 min, carrier status for all alleles was available and reported as wild type, heterozygous or homozygous for the minor allele. Blood samples were obtained at consent and underwent genetic analysis in the core laboratory to verify the accuracy of bedside testing. Genomic DNA was extracted using a commercial kit (FlexiGene, Qiagen, Hilden, Germany). Carrier status for the specified alleles was determined using TaqMan single nucleotide polymorphism (SNP) genotyping assays (Life Technologies, Carlsbad, CA, USA). Any discrepancies between point-of-care genotyping and core laboratory analysis were further investigated with direct DNA sequencing using the ABI PRISM dye terminator method (Applied Biosystems, Waltham, MA, USA).
Randomization and masking
Randomization for the study was mandated to occur after PCI in order to avoid delay in revascularization. The randomization sequence was computer generated in randomly selected block sizes of four and six. Serially numbered opaque envelopes were used for concealment. Patients identified as carriers of a CYP2C19*2 allele or the ABCB1 TT genotype were considered to have an at-risk genotype and randomly assigned to either prasugrel (10 mg daily for 4 weeks) or an evidence-based augmented dosing strategy of clopidogrel (150 mg daily for 6 additional days followed by 75 mg daily for 3 weeks; Figure 1). Non-carriers continued with clopidogrel dosing as per the invasive cardiologist. The invasive cardiologists and data analysts were blinded to the genetic results and antiplatelet strategy allocation. Patients and research nurses were not masked to carrier status or P2Y12 inhibitory drug regimen. Physicians and patients are blinded to the platelet function measurements.
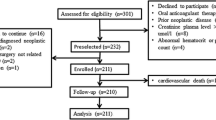
Study flow diagram. PCI, percutaneous coronary intervention; PRU=P2Y12 reaction unit; STEMI=ST-elevation myocardial infarction.
Follow-up and end points
Baseline platelet function testing was conducted at consent using the VerifyNow P2Y12 assay (Accriva, San Diego, CA, USA). Repeat measurements were undertaken at 1-month follow-up. Validated P2Y12 reaction unit (PRU) cutoffs of >234 and >208 were used to define high on-treatment platelet reactivity (HPR), a phenomenon characterized by inadequate P2Y12 inhibition and strongly associated with ischemic outcomes after PCI.8, 20, 21 The primary end point was the proportion of patients with at-risk genotypes assigned to prasugrel with PRU >234 at 1 month compared to those receiving clopidogrel. Secondary end points included a comparison of the two randomized at-risk genotype groups by: HPR cutoff of 208, mean PRU, percentage platelet inhibition, clinical outcomes at 1 month of cardiovascular death, myocardial infarction, urgent revascularization, stent thrombosis (Academic Research Consortium definite and probable)22 and a safety outcome of thrombolysis in myocardial infarction (TIMI) major and minor bleeding.
Statistical analysis
The study was powered for the primary end point of the proportion of at-risk genotype carriers with HPR in the prasugrel group compared to the clopidogrel group. We projected a 40% non-response rate23 among at-risk genotype carriers on clopidogrel and an 85% relative risk reduction with prasugrel.19 We further estimated a 50% prevalence for carriers of either the CYP2C19*2 allele or ABCB1 TT genotype.14 For a power of 80%, 23 carriers were required per arm. Assuming 4% loss to follow-up, we projected enrollment of 96 patients for the study.
Analyses were conducted with the intention-to-treat principle. Fisher’s exact or chi-squared tests were used for comparisons of categorical variables and a t-test was used for continuous variables. Point and 95% confidence interval (CI) estimate for sensitivity and specificity of point-of-care genetic testing were calculated with an exact binomial procedure. Multivariable analysis with logistic regression was conducted to ascertain at-risk genotypes’ association to HPR. Variables in the models included diabetes, smoking status, body mass index and proton-pump inhibitor (PPI) usage. All P-values were two-tailed with an accepted significance level of 0.05. All analyses were performed using SAS (version 9.2).
Results
A total of 102 STEMI patients were enrolled and 59 underwent subsequent randomization following point-of-care identification of an at-risk genotype. Among carriers of at-risk genotypes, 30 were randomized to prasugrel and 29 to clopidogrel. Baseline clinical characteristics did not differ between carriers and non-carriers or among subjects with at-risk genotypes randomized to prasugrel or clopidogrel (Table 1). Follow-up was complete with the exception of one patient from the low-risk genotype group who declined return for follow-up blood work; however, the patient was established to be free of MACE through a telephonic interview. The overall median time from PCI to point-of-care genotyping was 26.8 h (interquartile range: 17.6, 48.8).
Point-of-care genetic testing
The sensitivity of the point-of-care genetic testing device was 100% (95% CI 88.0–100), 100% (86.3–100) and 96.9% (82.0–99.8), and the specificity was 97.0% (88.5–99.5), 97.1% (89.0–99.5) and 98.5% (90.9–99.9) for identifying CYP2C19*2, ABCB1 TT and CYP2C19*17 carrier status, respectively. Of the enrolled patients, 37 (36.3%) were identified as carriers of at least one copy of CYP2C19*2 and 34 (33.7%) as homozygous for the ABCB1 3435T genotype. Distribution of the genetic variants was similar between the randomized groups (Table 2).
High on-treatment platelet reactivity
Baseline platelet function testing revealed that carriers of at-risk genotypes had a higher mean PRU compared to non-carriers (183.5±90.6 vs 147.3±84.7, P=0.040). PRU values among subjects with at-risk genotypes randomized to prasugrel and augmented-dose clopidogrel were similar at baseline (192.6±100.5 vs 174.1±80.6, P=0.4405). Analysis of the primary outcome identified a statistically significant reduction in the proportion of HPR among subjects with at-risk genotypes randomized to prasugrel (0%) compared to clopidogrel (24.1%; P=0.0046). When defined by PRU>208, 3.3% of subjects assigned to prasugrel had HPR compared with 34.5% in the clopidogrel group (P=0.0025). Other measures of platelet inhibition were also increased among prasugrel-treated patients relative to clopidogrel (Table 3). Prasugrel demonstrated consistent trends for reduction in HPR irrespective of the at-risk genotype (Table 4). Among non-carriers treated with clopidogrel, the mean PRU at 1 month was 110.4±85.1, while 4.8 and 9.5% of these subjects had HPR defined by PRU>234 and >208, respectively. Prasugrel-treated carriers, when compared with non-carriers on clopidogrel, did not have an increased risk for HPR at 1 month (P=0.5070 and 0.3932 for PRU>234 and >208, respectively). Patients homozygous for CYP2C19*17 had a 1-month mean PRU value of 87.5±73.2 and had no cases of HPR by either cutoff. The PRU values of carriers of CYP2C19*17 by treatment groups are shown in the Supplementary Table S1.
Multivariate analyses for predictors of 1-month HPR, using a priori determined variables known to influence clopidogrel-mediated platelet reactivity, revealed carrier status for an at-risk genotype to be the strongest independent predictor for HPR, irrespective of cutoffs (Figures 2a and b). PPI use was removed from the models because it perfectly predicted the outcome resulting in questionable validity of the model fit and estimates, particularly for PPI use. The estimates for the other variables in the model were not substantively different when PPI use was excluded.
Multivariate analyses for predictors of high on-treatment platelet reactivity (HPR) at 1 month. Multivariate analyses by logistic regression with dependent variable as HPR at 1 month for patients on clopidogrel. HPR defined as P2Y12 reaction unit (PRU) >234 (a) and >208 (b). Variables in the models included: carriers of at-risk genotypes, diabetes, smoking status and body mass index.
Clinical outcomes and bleeding
During 1-month follow-up, no MACE was observed among randomized patients. No significant differences in bleeding were observed between randomized patients. In the prasugrel arm, 1 (3.3%) patient had a TIMI major and 1 (3.3%) had a TIMI minor bleed. One patient (3.4%) suffered a TIMI major bleed in the clopidogrel arm. In the non-carrier group on clopidogrel, there were 2 (4.7%) TIMI major and 5 (11.6%) TIMI minor bleeds.
Discussion
Our results demonstrate that point-of-care genetic testing at the clinical bedside is capable of concurrent identification of multiple genetic variants. Rapid identification of at-risk genetic variants facilitated a personalized approach to antiplatelet therapy in patients receiving PCI for STEMI. Treatment of CYP2C19*2 and ABCB1 TT carriers with prasugrel resulted in a significant reduction in HPR after 1 month compared to a STEMI dosing of clopidogrel.
Previously, we reported the first successful use of clinical point-of-care genetic testing based on CYP2C19*2 carrier status in a proof-of-concept randomized clinical trial.19 A limitation of this approach, however, was the use of a single genetic variant in isolation when drug response is frequently influenced by multiple variants.14, 16, 24, 25 Notably, in the context of antiplatelet therapy, both CYP2C19 loss-of-function alleles and the ABCB1 TT genotype have been shown to be independent predictors for MACE among patients treated with clopidogrel following PCI for ACS.12, 14, 16, 17 The evidence supporting the clinical importance of CYP2C19 loss-of-function alleles, particularly CYP2C19*2, is derived from numerous clinical trials and observational studies and has been validated by two meta-analyses, each involving ~10 000 subjects.13, 26 Though certain studies have failed to show an association with MACE, these conflicting results have generally emerged from studies which did not restrict analyses to subjects receiving PCI with stenting.27, 28, 29 The data supporting the clinical importance of the ABCB1 TT genotype has been variable with several studies suggesting that carriers have an increased risk for ischemic complications,14, 16, 17 while others failed to identify an association.30, 31 Of note, in the only analysis of data from a randomized controlled trial restricted solely to ACS patients undergoing PCI, both CYP2C19 loss-of-function alleles and the ABCB1 TT genotype were shown to have independent associations with MACE, while carriers of both at-risk genetic variants were observed to be at even greater incremental risk.16
In addition to expanding the number of genetic variants that can be assessed at the bedside, extending the use of point-of-care genetic testing to higher acuity STEMI patients is particularly relevant given the increased risk for subsequent MACE and the need for decisions in an expedient manner. The failure of previous personalized studies guided by platelet function testing, not genotyping, may in part be secondary to the predominant enrollment of non-ACS patients with lower risk for MACE.32, 33, 34 The GRAVITAS (Gauging Responsiveness with A VerifyNow Assay—Impact on Thrombosis and Safety) study, which evaluated the benefits of high-dose clopidogrel among PCI patients with predominantly stable disease, had MACE rates of only 2.3% at 6 months.23 Similarly, the TRIGGER-PCI (Testing platelet Reactivity In patients underGoing elective stent placement on clopidogrel to Guide alternative thErapy with pRasugrel) trial, which only enrolled elective stable patients, was prematurely terminated following only 1 (0.4%) event in 236 patients.33 Indeed, the authors of TRIGGER-PCI attribute a key reason for the failure of the study as the exclusive enrollment of stable coronary artery disease patients, who have less incidence of MACE.33 In contrast, the risk of MACE following ACS approximates 10% at one year, emphasizing the need to improve treatment strategies in this population.9, 10
Several novel strategies have provided additional benefits over standard clopidogrel among patients with non-ST ACS and STEMI.6, 9, 10, 35, 36 The CURRENT–OASIS 7 trial demonstrated that a higher clopidogrel dosing strategy decreased MACE among ACS and STEMI patients undergoing PCI.6 In addition, novel P2Y12 agents, prasugrel and ticagrelor, confer additional ischemic benefits over clopidogrel as shown in the Trial to Assess Improvement in Therapeutic Outcomes by Optimizing Platelet Inhibition with Prasugrel (TRITON–TIMI-38) and the Platelet Inhibition and Patient Outcomes (PLATO) trials.9, 10 However, these strategies were associated with increased fatal and non-fatal bleeding events, which have contributed to reluctance for their universal adoption. Of note, a key finding of the ADAPT-DES study was the importance of both ischemic and hemorrhagic complications to account for all-cause mortality.37 Although there may be proponents for the universal adoption of novel P2Y12 agents, randomized studies of these agents against clopidogrel have consistently demonstrated increased rates of non-CABG-related major bleeding. Consequently, there is an impetus to pursue novel personalized strategies consisting of selective administration of more potent P2Y12 inhibitors that carry the potential to reduce ischemic and hemorrhagic complications concurrently.
In our study, carriers of at-risk genotypes had a significantly higher prevalence of HPR at baseline compared to non-carriers, supporting their classification as an at-risk group for treatment failure with clopidogrel. Moreover, treatment of non-carriers with clopidogrel resulted in only 4.6% of subjects having HPR at 1 month suggesting that clopidogrel may be adequate among the vast majority of subjects who do not possess an at-risk genotype. These findings highlight the potential of a personalized strategy to target more potent therapy to at-risk patients, while sparing the remaining patients from increased bleeding risks of more potent agents; this concept may be particularly well suited for the future development of strategies to achieve a therapeutic window, where ischemic and bleeding risks are optimized simultaneously.38
Within our study, treatment of at-risk genotype carriers with the OASIS-7 dosing of clopidogrel resulted in 24.1% continuing to exhibit HPR at 1 month. In contrast, carriers of at-risk genotypes assigned to prasugrel resulted in complete elimination of HPR. Both augmented-dose clopidogrel and prasugrel have been shown to decrease ischemic complications relative to standard clopidogrel dosing in STEMI patients.6, 9, 35 Of note, both strategies continue to be endorsed in guidelines.39, 40 The Escalating Clopidogrel by Involving a Genetic Strategy (ELEVATE) study showed that clopidogrel dosing at 225 mg daily reduced HPR among heterozygous CYP2C19*2 carriers; however, dosing up to 300 mg daily failed to overcome HPR in homozygotes. Our study design predated publication of ELEVATE,41 but was based on the approach from CURRENT–OASIS 7, which showed clinical benefits when compared to standard dosing of clopidogrel.6 Previous retrospective genetic studies comparing the effects of prasugrel to high-dose clopidogrel in elective PCI patients suggested high-dose clopidogrel was only effective in reducing HPR in non-carriers of CYP2C19*2.42, 43 In contrast, prasugrel effectively reduced HPR irrespective of CYP2C19 *2 status, which is consistent with our trial results. Notably, our study is the first to prospectively demonstrate in a randomized manner that treatment of STEMI patients carrying at-risk genotypes with prasugrel is superior to the OASIS-7 clopidogrel strategy for reducing HPR. These findings should draw caution to the ongoing use of higher dosing of clopidogrel among STEMI patients known to be carriers of at-risk genotypes. This is of particular significance, given that at-risk variants are prevalent and therefore use of an augmented clopidogrel dosing strategy may fail to adequately protect a substantial proportion of the population.
The third variant tested, CYP2C19*17, confers a gain of function.18 Carriers of this SNP have an increased propensity toward bleeding when receiving clopidogrel18 and current pharmacogenetic guidelines recommend standard dosing at 75 mg daily.44 In our study, there were no homozygous carriers of CYP2C19*17 that had HPR after treatment with clopidogrel. Larger prospective studies will be required to further investigate the impact of this SNP among individuals who also carry CYP2C19 loss-of-function alleles and/or the ABCB1 TT genotype.
There are some limitations to our study. First, our study had enrolled patients treated with primary PCI and also a significant proportion of patients who initially had received thrombolytic therapy preceding PCI. However, as a major objective was to provide a proof of concept of being able to identify multiple at-risk variants in patients with STEMI, our enrollment of patients initially treated with thrombolytics should not detract from the fact that the main objective was achieved. Moreover, clopidogrel continues to be the main P2Y12 inhibitor used in patients treated with thrombolytics due previous studies supporting its use in this context.1, 2 Accordingly, we believe that the enrollment of patients treated initially with thrombolytics should not affect the significance of our data. Second, there are other factors, apart from genetics, which may affect clopidogrel response. Consequently, among non-carriers treated with standard-dose clopidogrel in our study, there were some which had HPR at 1 month. Our study was designed to evaluate an intervention using genetics alone. Future strategies will require consideration of a multifactorial approach to address non-genetic causes for HPR.11 Lastly, is our use of HPR as the primary end point. HPR has been extensively investigated and has been shown to be an important predictor of ischemic outcomes after PCI.45, 46, 47, 48, 49 Studies involving antiplatelet agents have routinely utilized HPR to evaluate the efficacy of novel treatment strategies.19, 50 Although we have demonstrated that a gene-guided approach to selective administration of prasugrel was successful in overcoming HPR among carriers of at-risk genotypes, our study was nonetheless small and not powered to determine associations to clinical outcomes. Therefore, future studies powered for evaluation of clinical outcomes will be required to permit integration of a pharmacogenomic approach into routine clinical practice. The Tailored Antiplatelet Therapy Following PCI (TAILOR-PCI) trial, which is powered for clinical outcomes, will be recruiting more than 5000 patients to further evaluate this hypothesis (NCT01742117).
In conclusion, we have extended the use of point-of-care genetic testing to STEMI patients receiving PCI and have shown that concomitant genotyping of three separate SNPs is feasible and accurate. A pharmacogenomic approach with selective administration of prasugrel to carriers of at-risk genetic variants reduces the risk of HPR compared to an augmented dosing strategy of clopidogrel among STEMI patients. These serve as integral steps that will facilitate the execution of large clinical trials capable of definitively evaluating the use of pharmacogenomic strategies in patients with ACS.
References
Chen ZM, Jiang LX, Chen YP, Xie JX, Pan HC, Peto R et al. Addition of clopidogrel to aspirin in 45,852 patients with acute myocardial infarction: randomised placebo-controlled trial. Lancet 2005; 366: 1607–1621.
Sabatine MS, Cannon CP, Gibson CM, Lopez-Sendon JL, Montalescot G, Theroux P et al. Effect of clopidogrel pretreatment before percutaneous coronary intervention in patients with ST-elevation myocardial infarction treated with fibrinolytics: the PCI-CLARITY study. JAMA 2005; 294: 1224–1232.
Kushner FG, Hand M, Smith SC Jr, King SB III, Anderson JL, Antman EM et al. 2009 Focused updates: ACC/AHA guidelines for the management of patients with ST-elevation myocardial infarction (updating the 2004 guideline and 2007 focused update) and ACC/AHA/SCAI guidelines on percutaneous coronary intervention (updating the 2005 guideline and 2007 focused update): a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation 2009; 120: 2271–2306.
Yusuf S, Zhao F, Mehta SR, Chrolavicius S, Tognoni G, Fox KK et al. Effects of clopidogrel in addition to aspirin in patients with acute coronary syndromes without ST-segment elevation. N Engl J Med 2001; 345: 494–502.
Mehta SR, Yusuf S, Peters RJ, Bertrand ME, Lewis BS, Natarajan MK et al. Effects of pretreatment with clopidogrel and aspirin followed by long-term therapy in patients undergoing percutaneous coronary intervention: the PCI-CURE study. Lancet 2001; 358: 527–533.
Mehta SR, Tanguay JF, Eikelboom JW, Jolly SS, Joyner CD, Granger CB et al. Double-dose versus standard-dose clopidogrel and high-dose versus low-dose aspirin in individuals undergoing percutaneous coronary intervention for acute coronary syndromes (CURRENT-OASIS 7): a randomised factorial trial. Lancet 2010; 376: 14233–14243.
Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, Alfonso F, Macaya C, Bass TA et al. Variability in individual responsiveness to clopidogrel: clinical implications, management, and future perspectives. J Am Coll Cardiol 2007; 49: 1505–1516.
Price MJ, Endemann S, Gollapudi RR, Valencia R, Stinis CT, Levisay JP et al. Prognostic significance of post-clopidogrel platelet reactivity assessed by a point-of-care assay on thrombotic events after drug-eluting stent implantation. Eur Heart J 2008; 29: 992–1000.
Wiviott SD, Braunwald E, McCabe CH, Montalescot G, Ruzyllo W, Gottlieb S et al. Prasugrel versus clopidogrel in patients with acute coronary syndromes. N Engl J Med 2007; 357: 2001–2015.
Wallentin L, Becker RC, Budaj A, Cannon CP, Emanuelsson H, Held C et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N Engl J Med 2009; 361: 1045–1057.
So DY, Roberts JD . Overcoming obstacles in pharmacogenomic strategies for antiplatelet drugs: are we RAPID enough? Pharmacogenomics 2012; 13: 1105–1108.
Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med 2009; 360: 354–362.
Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA 2010; 304: 1821–1830.
Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Meneveau N et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med 2009; 360: 363–375.
Xie HG . Genetic variations of S-mephenytoin 4'-hydroxylase (CYP2C19) in the Chinese population. Life Sci 2000; 66: L175–L181.
Mega JL, Close SL, Wiviott SD, Shen L, Walker JR, Simon T et al. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet 2010; 376: 1312–1319.
Cayla G, Hulot JS, O'Connor SA, Pathak A, Scott SA, Gruel Y et al. Clinical, angiographic, and genetic factors associated with early coronary stent thrombosis. JAMA 2011; 306: 1765–1774.
Sibbing D, Koch W, Gebhard D, Schuster T, Braun S, Stegherr J et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation 2010; 121: 512–518.
Roberts JD, Wells GA, Le May MR, Labinaz M, Glover C, Froeschl M et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet 2012; 379: 1705–1711.
Price MJ, Angiolillo DJ, Teirstein PS, Lillie E, Manoukian SV, Berger PB et al. Platelet reactivity and cardiovascular outcomes after percutaneous coronary intervention: a time-dependent analysis of the Gauging Responsiveness with a VerifyNow P2Y12 assay: Impact on Thrombosis and Safety (GRAVITAS) trial. Circulation 2011; 124: 1132–1137.
Bonello L, Tantry US, Marcucci R, Blindt R, Angiolillo DJ, Becker R et al. Consensus and future directions on the definition of high on-treatment platelet reactivity to adenosine diphosphate. J Am Coll Cardiol 2010; 56: 919–933.
Cutlip DE, Windecker S, Mehran R, Boam A, Cohen DJ, van Es GA et al. Clinical end points in coronary stent trials: a case for standardized definitions. Circulation 2007; 115: 2344–2351.
Price MJ, Berger PB, Teirstein PS, Tanguay JF, Angiolillo DJ, Spriggs D et al. Standard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: the GRAVITAS randomized trial. JAMA 2011; 305: 1097–1105.
Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med 2009; 360: 753–764.
Stanulla M, Schaeffeler E, Flohr T, Cario G, Schrauder A, Zimmermann M et al. Thiopurine methyltransferase (TPMT) genotype and early treatment response to mercaptopurine in childhood acute lymphoblastic leukemia. JAMA 2005; 293: 1485–1489.
Hulot JS, Collet JP, Silvain J, Pena A, Bellemain-Appaix A, Barthelemy O et al. Cardiovascular risk in clopidogrel-treated patients according to cytochrome P450 2C19*2 loss-of-function allele or proton pump inhibitor coadministration: a systematic meta-analysis. J Am Coll Cardiol 2010; 56: 134–143.
Pare G, Mehta SR, Yusuf S, Anand SS, Connolly SJ, Hirsh J et al. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med 2010; 363: 1704–1714.
Holmes MV, Perel P, Shah T, Hingorani AD, Casas JP . CYP2C19 genotype, clopidogrel metabolism, platelet function, and cardiovascular events: a systematic review and meta-analysis. JAMA 2011; 306: 2704–2714.
Mega JL, Topol EJ, Sabatine MS . CYP2C19 genotype and cardiovascular events. JAMA 2012; 307: 1482–1483.
Wallentin L, James S, Storey RF, Armstrong M, Barratt BJ, Horrow J et al. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet 2010; 376: 1320–1328.
Price MJ, Murray SS, Angiolillo DJ, Lillie E, Smith EN, Tisch RL et al. Influence of genetic polymorphisms on the effect of high- and standard-dose clopidogrel after percutaneous coronary intervention: the GIFT (Genotype Information and Functional Testing) study. J Am Coll Cardiol 2012; 59: 1928–1937.
Price MJ, Berger PB, Angiolillo DJ, Teirstein PS, Tanguay JF, Kandzari DE et al. Evaluation of individualized clopidogrel therapy after drug-eluting stent implantation in patients with high residual platelet reactivity: design and rationale of the GRAVITAS trial. Am Heart J 2009; 157: 818–824.
Trenk D, Stone GW, Gawaz M, Kastrati A, Angiolillo DJ, Muller U et al. A randomized trial of prasugrel versus clopidogrel in patients with high platelet reactivity on clopidogrel after elective percutaneous coronary intervention with implantation of drug-eluting stents: results of the TRIGGER-PCI (Testing Platelet Reactivity In Patients Undergoing Elective Stent Placement on Clopidogrel to Guide Alternative Therapy With Prasugrel) study. J Am Coll Cardiol 2012; 59: 2159–2164.
Collet JP, Cuisset T, Range G, Cayla G, Elhadad S, Pouillot C et al. Bedside monitoring to adjust antiplatelet therapy for coronary stenting. N Engl J Med 2012; 367: 2100–2109.
Montalescot G, Wiviott SD, Braunwald E, Murphy SA, Gibson CM, McCabe CH et al. Prasugrel compared with clopidogrel in patients undergoing percutaneous coronary intervention for ST-elevation myocardial infarction (TRITON-TIMI 38): double-blind, randomised controlled trial. Lancet 2009; 373: 723–731.
Steg PG, James S, Harrington RA, Ardissino D, Becker RC, Cannon CP et al. Ticagrelor versus clopidogrel in patients with ST-elevation acute coronary syndromes intended for reperfusion with primary percutaneous coronary intervention: a Platelet Inhibition and Patient Outcomes (PLATO) trial subgroup analysis. Circulation 2010; 122: 2131–2141.
Stone GW, Witzenbichler B, Weisz G, Rinaldi MJ, Neumann FJ, Metzger DC et al. Platelet reactivity and clinical outcomes after coronary artery implantation of drug-eluting stents (ADAPT-DES): a prospective multicentre registry study. Lancet 2013; 382: 614–623.
Tantry US, Bonello L, Aradi D, Price MJ, Jeong YH, Angiolillo DJ et al. Consensus and update on the definition of on-treatment platelet reactivity to adenosine diphosphate associated with ischemia and bleeding. J Am Coll Cardiol 2013; 62: 2261–2273.
O'Gara PT, Kushner FG, Ascheim DD, Casey DE Jr, Chung MK, de Lemos JA et al. 2013 ACCF/AHA guideline for the management of ST-elevation myocardial infarction: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol 2013; 61: e78–e140.
Steg PG, James SK, Atar D, Badano LP, Blomstrom-Lundqvist C, Borger MA et al. ESC Guidelines for the management of acute myocardial infarction in patients presenting with ST-segment elevation. Eur Heart J 2012; 33: 2569–2619.
Mega JL, Hochholzer W, Frelinger AL III, Kluk MJ, Angiolillo DJ, Kereiakes DJ et al. Dosing clopidogrel based on CYP2C19 genotype and the effect on platelet reactivity in patients with stable cardiovascular disease. JAMA 2011; 306: 2221–2228.
Sardella G, Calcagno S, Mancone M, Palmirotta R, Lucisano L, Canali E et al. Pharmacodynamic effect of switching therapy in patients with high on-treatment platelet reactivity and genotype variation with high clopidogrel dose versus prasugrel: the RESET GENE trial. Circ Cardiovasc Interv 2012; 5: 698–704.
Saucedo JF, Angiolillo DJ, DeRaad R, Frelinger AL III, Gurbel PA, Costigan TM et al. Decrease in high on-treatment platelet reactivity (HPR) prevalence on switching from clopidogrel to prasugrel: insights from the switching anti-platelet (SWAP) study. Thromb Haemost 2013; 109: 347–355.
Scott SA, Sangkuhl K, Gardner EE, Stein CM, Hulot JS, Johnson JA et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther 2011; 90: 328–332.
Buonamici P, Marcucci R, Migliorini A, Gensini GF, Santini A, Paniccia R et al. Impact of platelet reactivity after clopidogrel administration on drug-eluting stent thrombosis. J Am Coll Cardiol 2007; 49: 2312–2317.
Gurbel PA, Bliden KP, Samara W, Yoho JA, Hayes K, Fissha MZ et al. Clopidogrel effect on platelet reactivity in patients with stent thrombosis: results of the CREST Study. J Am Coll Cardiol 2005; 46: 1827–1832.
Hochholzer W, Trenk D, Bestehorn HP, Fischer B, Valina CM, Ferenc M et al. Impact of the degree of peri-interventional platelet inhibition after loading with clopidogrel on early clinical outcome of elective coronary stent placement. J Am Coll Cardiol 2006; 48: 1742–1750.
Matetzky S, Shenkman B, Guetta V, Shechter M, Bienart R, Goldenberg I et al. Clopidogrel resistance is associated with increased risk of recurrent atherothrombotic events in patients with acute myocardial infarction. Circulation 2004; 109: 3171–3175.
Geisler T, Langer H, Wydymus M, Gohring K, Zurn C, Bigalke B et al. Low response to clopidogrel is associated with cardiovascular outcome after coronary stent implantation. Eur Heart J 2006; 27: 2420–2425.
Alexopoulos D, Dimitropoulos G, Davlouros P, Xanthopoulou I, Kassimis G, Stavrou EF et al. Prasugrel overcomes high on-clopidogrel platelet reactivity post-stenting more effectively than high-dose (150-mg) clopidogrel: the importance of CYP2C19*2 genotyping. JACC Cardiovasc Interv 2011; 4: 403–410.
Acknowledgements
We thank Lyne Stuewe, Cheryl Charlebois, Colleen Chilton and Nitan Garg for their invaluable assistance in conducting the study. DYFS was supported by grants from the Heart and Stroke Foundation of Ontario and the Canadian Institutes of Health Research. The RAPID STEMI study was funded by unrestricted grants from Spartan Biosciences, Inc and the Canadian Institutes of Health Research (FRN 115129).
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
Dr. So holds a physician initiated grant supported by the Canadian Institute of Health Research and Eli Lilly, Canada.
Additional information
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website
Supplementary information
PowerPoint slides
Rights and permissions
About this article
Cite this article
So, D., Wells, G., McPherson, R. et al. A prospective randomized evaluation of a pharmacogenomic approach to antiplatelet therapy among patients with ST-elevation myocardial infarction: the RAPID STEMI study. Pharmacogenomics J 16, 71–78 (2016). https://doi.org/10.1038/tpj.2015.17
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2015.17
- Springer Nature Limited
This article is cited by
-
The need of a multicomponent guiding approach to personalize clopidogrel treatment
The Pharmacogenomics Journal (2021)
-
Comparison of a rapid point-of-care and two laboratory-based CYP2C19*2 genotyping assays for personalisation of antiplatelet therapy
International Journal of Clinical Pharmacy (2016)
-
Gain-of-function single nucleotide variants of the CYP2C19 gene (CYP2C19*17) can identify subtherapeutic voriconazole concentrations in critically ill patients: a case series
Intensive Care Medicine (2015)