Abstract
Goosegrass (Eleusine indica) is one of the most serious annual grassy weeds worldwide, and its evolved herbicide-resistant populations are more difficult to control. Quantitative real-time PCR (qPCR) is a common technique for investigating the resistance mechanism; however, there is as yet no report on the systematic selection of stable reference genes for goosegrass. This study proposed to test the expression stability of 9 candidate reference genes in goosegrass in different tissues and developmental stages and under stress from three types of herbicide. The results show that for different developmental stages and organs (control), eukaryotic initiation factor 4 A (eIF-4) is the most stable reference gene. Chloroplast acetolactate synthase (ALS) is the most stable reference gene under glyphosate stress. Under glufosinate stress, eIF-4 is the best reference gene. Ubiquitin-conjugating enzyme (UCE) is the most stable reference gene under quizalofop-p-ethyl stress. The gene eIF-4 is the recommended reference gene for goosegrass under the stress of all three herbicides. Moreover, pairwise analysis showed that seven reference genes were sufficient to normalize the gene expression data under three herbicides treatment. This study provides a list of reliable reference genes for transcript normalization in goosegrass, which will facilitate resistance mechanism studies in this weed species.
Similar content being viewed by others
Introduction
Weeds are one of the major biotic threats to crops and can result in a 34% loss of crop yield worldwide1. Herbicides are a class of chemicals that can control weeds efficiently and economically, enhance crop yield and liberate the workforce. Unfortunately, over-reliance on herbicides has resulted in the evolution of resistance in weed species2. To date, 251 weed species in 66 countries have been found to have evolved resistance to 160 different herbicides3. Goosegrass (Eleusine indica) is considered one of the top ten worst weeds, distributed worldwide and affecting crops4. Acetyl-CoA carboxylase (ACCase)-inhibiting herbicides and non-selective herbicides such as glyphosate, glufosinate and paraquat are widely used to control the goosegrass5. After many years of control by herbicides, goosegrass has evolved resistance to many of them, including glyphosate6,7, diclofop8, fluazifop9, glufosinate10 and paraquat11,12.
Clarifying the resistance mechanism for weeds is essential to control them, ensuring crop yield and the sustainable application of herbicides13. In general, resistance mechanisms can be categorized as target and non-target14. Gene expression analysis is commonly applied to explore target-site gene amplification and the over-expression of detoxifying enzyme genes, which are related to herbicide resistance15,16,17. Quantitative real-time PCR (qPCR) is necessarily in wide use18. Normalization during qPCR analysis is usually performed using a reference gene that must be expressed at stable levels regardless of the experimental conditions to ensure the veracity of the qPCR analysis19,20. Several studies have been published with the aim of identifying suitable reference genes for expression analysis under different conditions in plants, including weeds21,22,23,24. However, few studies focus on the stress under herbicide treatment for weeds25,26,27,28. With the development of next-generation sequencing technologies that can be used to investigate resistance mechanisms for weeds, the expression levels of increasing numbers of genes should be validated. Thus, the selection of a stably expressed reference gene is important for the accuracy of the results.
Studies of the resistance mechanism of goosegrass to herbicides have been conducted at both the phenotypic character and the molecular levels7,29,30. Acetyl-CoA carboxylase (ACCase), 5-enolpyruvylshikimate-3 phosphate synthase (EPSPS) and glutamine synthetase (GS) are three common genes whose expression levels are affected in resistant goosegrass and constitute the target-set of quizalofop-p-ethyl glyphosate and glufosinate8,15. However, no systematic analysis of reference gene selection for normalization in various development stages of goosegrass and the herbicide stress has previously been performed. In this study, 9 selected reference genes were cloned and identified in goosegrass, and then the stability of gene expression was assessed in different developmental stages, different organs, and different times after treatment with three herbicides. The aim of this study is to provide useful guidelines for future gene expression studies in goosegrass.
Results
RNA isolation and primer selection
The RNA quality of all samples was detected by both 1% agarose gel electrophoresis and A260/A280 values. All selected samples in this study showed a specificity band, and the values of A260/A280 ranged from 1.90 to 2.11 (Supplementary Figure S1). The 9 candidate reference genes in this study were retrieved from the transcriptome database of E. indica. Partial sequences of the 9 reference genes were reconfirmed by the PCR results and submitted to the NCBI database (Table 1). At least three primer pairs were designed, and the selected primers showed amplification specificity by gel electrophoresis. Then, the candidate primer pairs were detected by qPCR based on a standard curve. The results showed that all the appropriate primers exhibited a single band with the expected size by gel electrophoresis (Fig. 1) and a single peak in the melting curve (Supplementary Figure S2). In addition, all the primer pairs showed acceptable amplification efficiency (E), which ranged from 90.12% to 97.25%, and the correlation coefficients (R2) varied from 0.990 to 0.998 (Table 1; Supplementary Figure S3).
Expression profiles of candidate reference genes
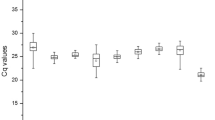
The expression levels of the 9 candidate reference genes in goosegrass under different conditions were determined by the ABI7500, and the results were shown as quantification cycles31. According to the box-plot, the Cq values of the 9 reference genes displays a diversity ranging from 16.52 to 32.86, and their expression patterns are similar under different treatment (Fig. 2). The gene elongation factor 1-alpha (EF) showed the low Cq values ranging from 16.52 to 24.37, indicating high expression. In contrast, the gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) displayed high Cq values ranging from 23.57 to 32.86, corresponding to low expression. Compared across the different developmental stages and organs (control), the expression of the gene beta-tubulin 4 (TUB) was affected significantly under herbicide stress. The standard deviation (SD) of the samples for the control was 1.42, while the SD value can reach 3.55 under herbicide treatment.
GeNorm analysis
This software evaluated the stability level of the reference genes by two parameters, stability value (M) and pairwise variation (Vn/Vn+1). The lowest M value means the most stable gene expression. The reference gene was accepted if the M value was below 1.5. In our research, eIF-4 and EF ranked as the most stable genes under the control experiment set. Alpha tubulin 1 (TUA) is considered the least stable gene, with an M value of 1.48, and two genes (TUB and heat shock protein 70 (HSP) were not accepted by geNorm because the M values were greater than 1.5 (Table 2; Supplementary Figure S4). Under the stress of glyphosate, glufosinate and the total set, the most stable genes were the same (eukaryotic initiation factor 4A (eIF-4) and chloroplast acetolactate synthase (ALS), while eIF-4 and ubiquitin-conjugating enzyme (UCE) were ranked as the most stable genes under quizalofop-p-ethyl stress. In addition, to define the best pair using geNorm, we also estimated the pairwise variation (Vn/Vn+1) to determine the minimal number of genes for reliable normalization with a cut-off value of 0.15. The pairwise variation was more than 0.15 in all the experiment sets (Supplementary Figure S5). As recommended in this case32, we used the lowest Vn/n+1 value to determine the number of reference genes adequate for normalization. Under the stress of the three types of herbicide, using the most seven stable reference genes is considered to be a valid normalization strategy. Under the control and total conditions, the eight most stable reference genes should be used.
NormFinder analysis
The stability value is the parameter used to evaluate the appropriate reference genes by NormFinder, where the lower stability value indicates higher expression stability20. In this research, the results analysed by NormFinder were similar to the analysis by geNorm. Under glufosinate stress, quizalofop-p-ethyl stress and for all samples, the most and least stable reference genes were the same as in the results evaluated by the geNorm, i.e., eIF-4 and TUB, respectively (Table 2). The gene eIF-4 was ranked as the most stable (0.24) under the control experiment set, while HSP was the most varied gene (1.33). ALS was the most stable gene (0.71) under glyphosate stress, while the least stable gene was TUB (1.79).
BestKeeper analysis
The algorithm of the BestKeeper software is different from geNorm and NormFinder, depending on the SD and coefficient of variance (CV). The lowest SD and CV indicate the most stable reference gene. In addition, the parameter SD for the accepted reference gene must be lower than 1.0. The results analysis by BestKeeper differed from the results analysis by geNorm and NormFinder, with no more than five accepted reference genes in the entire experimental set (Table 2). Under the control set, total samples and quizalofop-p-ethyl stress conditions, the TUA was ranked as the most stable reference gene, while the most stable genes under glyphosate and glufosinate stress were HSP and UCE, respectively. The gene ACTIN was acceptable under all the experimental set.
ReFinder analysis
To address the heterogeneity of the results evaluated by the geNorm, NormFinder and BestKeeper, another web tool ReFinder was used, which can compare the data generated by the other three software programs to give a comprehensive ranking confirming the stability ranking via different programs. The results show that the most stable reference genes were eIF-4 (1.41), ALS (1.50), eIF-4 (1.63), UCE (1.32) and eIF-4 (1.50) for the control, glyphosate, glufosinate, quizalofop-p-ethyl and total set (Table 2).
Expression profiles of EPSPS, GS and ACCase
The expression of the target-site genes of glyphosate (EPSPS), glufosinate (GS) and quizalofop-p-ethyl (ACCase) were determined in the leaf tissue after treatment at different times. Both the most stable reference genes (eIF-4, ALS) and the most varying reference gene (TUB) were chosen for this study. ALS is the most stable reference gene for goosegrass under glyphosate stress. Using ALS as the reference gene, the expression of EPSPS at 72 h and 96 h is 3.7 and 2.8 times higher than at 48 h after glyphosate treatment, respectively. When using the TUB, it was only 1.2 times and 0.7 times, respectively (Fig. 3). For GS, the expression at 96 h is 1.3 times higher than at 48 h using the best reference gene eIF-4. The most variable reference gene, TUB, can produce a result of up to 5.6 times. The expression of ACCase in goosegrass after quizalofop-p-ethyl treatment was altered only slightly. However, a difference can still be found between the more and less stable reference genes. The expression at 48 h is 1.5 times the expression at 72 h when using eIF-4, while the result is 2.4 using the least stable gene, TUB.
Discussion
Herbicide-resistant weeds are a significant problem for crop production worldwide. Countering the resistance mechanism can provide a beneficial way to control them and ensure food safety. The qPCR method is the most common way to determine the expression profiles for genes related to herbicide resistance in weeds33,34, especially with the application of the technology of next-generation sequencing to investigate the resistance mechanism15,35,36. The selection of a stable reference gene is crucial for the accuracy of the qPCR results. However, little research has focused on finding reference gene for weeds, including goosegrass, under herbicide stress27,28. In this research, we selected stable reference genes from 9 candidate reference genes under the stress of three herbicides. The results showed that the most stable reference gene is ALS under glyphosate stress, eIF-4 under glufosinate stress, and UCE under quizalofop-p-ethyl stress. The gene eIF-4 is the best candidate reference gene in goosegrass under stress from all three herbicides.
The first report to identify reference genes under the herbicide stress of ACCase inhibitor is on Alopecurus myosuroides, and ubiquitin (UBQ), TUB, and GAPDH were selected for normalization27. In our research, the most stable gene under the ACCase inhibitor quizalofop-p-ethyl was UCE, followed by eIF-4 and EF, while TUB was the least stable gene for goosegrass. Another study found that the most stable genes for Avena fatuaunder the stress of ACCase and ALS inhibitor herbicides were TATA-binding protein (TBP) and GAPDH26. Compared to this research, the most stable reference genes under the stress of the ALS inhibitor herbicide tribenuron were ACT7 and UBC for Descurainia sophia25. These results indicated that the selection of the stable gene differed for different species. Herbicide stress is a type of abiotic stress, and the gene eIF-4 showed stable expression in our research. This result was similar to other findings in grasses under different stresses37,38,39,40. Glyphosate is a very important herbicide due to the development of transgenic technology. Unfortunately, 36 weed species have evolved resistance to glyphosate thus far3. Many studies focus on investigating the resistance mechanism using qPCR; however, fewer studies systematically select the reference gene34,41,42. In our research, ALS is the most stable reference in E. indica under glyphosate stress, followed by TUA and eIF-4. The same results were found for Amaranthus palmeri and Kochia scoparia, where ALS was selected as the reference gene33,42. In our previous study, ACTIN was found to be more stable than GAPDH, as calculated by the software Bestkeeper16. This finding is consistent with the results in this study. However, the results of the stability ranking calculated by the software geNorm and NormFinder were different from the results calculated by Bestkeeper. This heterogeneity was also found in other plant species and may be due to the different algorithms for the software geNorm and NormFinder24. Fortunate, the software RefFinder can generate a comprehensive ranking of reference genes for all the other software.
Many research also focus on the genes expression in different tissues and developmental stages. We selected the stable expression reference genes in goosegrass under three different tissues and growing stages, respectively. Both the geNorm and NormFinder calculated the eIF-4 as the most stabile reference. While, TUA was ranking as the most stabile gene used the Bestkeeper. All the three softwares ranked HSP as the least stabile reference gene. The RefFinder got a comprehensive result that ranking the eIF-4 as the most stable reference gene. In Camellia sinensis, the eIF-4 was also selected as the best stable reference gene by the geNorm under the developmental stage24. And the phenomenon that different results were got using the different software was also found in Pennisetum glaucum23.
EPSPS is the target-set of glyphosate, from the shikimate pathway43. We determined the expression of EPSPS in the plants of a GR goosegrass population, caused by the amplification of the target set16. The expression of EPSPS shows a signification increase from 48 h to 72 h after treatment with glyphosate, as detected using the stable reference genes ALS and eIF-4. However, this phenomenon is not detected using the reference gene TUB. This result indicates that an appropriate reference is important for the precision of the result. Glufosinate is a popular non-selective herbicide with a target-site of GS and useful to control the GR weed species15. It is necessary to select the stable reference gene for the expression study due to the importance of this herbicide. In our research, the expression level of the reference genes eIF-4 and ALS is similar to the GS in goosegrass. And these two reference genes show a higher stable than the TUB after glufosinate treated at different times. Several reports have identified ACCase inhibitor herbicide-resistant goosegrass worldwide. The expression level of ACCase is relatively similar to the reference gene 18S rRNA9. We also observed this phenomenon in our research, although the reference is different.
In summary, we found stables reference genes in goosegrass under the stress of three herbicides. Across different tissues and developmental stages, eIF-4 is the best reference gene. TUA can serve as reference gene under glyphosate stress, whereas under glufosinate stress, eIF-4 is the most stable reference gene. UCE can be used as reference gene under quizalofop-p-ethyl stress. Under stress from the three herbicides overall, eIF-4 is the best reference gene for gene expression analysis.
Methods
Plant material and treatments
Seed samples of the goosegrass populations for this study were collected from the roadside of turf farms in Chengdu, Sichuan Province. The goosegrass seeds were cultured using the method in our previous research, with a single plant per pot7. When the plants reached the 4–5 leaves stage, tillering stage, and flowering fruit-bearing stage, the three types of herbicides were applied for stress treatment. The doses of these three herbicide were as follows: 840 g ae ha−1 of glyphosate (Roundup, isopropylamine salt of glyphosate, 410 g ae L−1, Monsanto Company, St. Louis, USA), 200 g a.i. ha−1 of Glufosinate (Basta®, 200 g L−1 glufosinate ammonium, soluble concentration (SL), Bayer CropScience, Frankfurt, Germany), and 37.5 g a.i. ha−1 of quizalofop-p-ethyl (50 g L−1 EC, Nissan chemical industry, Tokyo, Japan). The samples were collected and described as summarized in Table 3. For the tissues study, three organ types were harvested from the flowering fruit bearing stage. The fully emerged leaves were collected from three different developmental stages for the study of developmental stage. In the time treatment, the leaves were collected at the 4–5 leaf stage. Three biological replicates were performed and all samples were immediately frozen in liquid nitrogen and stored at −80 °C prior to RNA extraction.
Total RNA isolation and cDNA synthesis
The frozen tissue was ground in liquid nitrogen using a mortar and pestle. Total RNA was extracted using the RNAprep Pure Plan Kit (Tiangen Biotech Beijing CO., LTD, Beijing, China). Genomic DNA was completely eliminated using RNase-Free DNase I (Tiangen Biotech Beijing CO., LTD, Beijing, China), according to the manufacturer’s instructions. The integrity was checked by 1.0% agarose gel electrophoresis. RNA and genomic DNA concentrations were determined using the NanoDrop-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). First-strand complementary DNA was synthesized using the FastQuant RT kit (Tiangen Biotech Beijing CO., LTD, Beijing, China) with 1 μg of total RNA and oligo (dT) in a total volume of 20 μL. The samples were stored at −80 °C until used.
Candidate reference gene selection, primer design and gene cloning
9 common reference genes, ACTIN, GAPDH, ALS, EF, eIF-4, TUA, TUB, UCE and HSP, were selected for this study. The genes TUA and TUB were retrieved from the database of the National Centre for Biotechnology (NCBI). The other reference genes were obtained by querying the E. indica transcriptome database (SRR3614245) in our previous study. To confirm the sequences of the 9 candidate reference genes, all of them were cloned using polymerase chain reaction (PCR) and sequenced using the method described in our previous study16.
qPCR assay
Primer pair sets for the confirmed candidate reference genes were designed using the software Oligo 7.0 (Table 1). The qPCR was performed in an ABI 7500 PCR machine utilizing SYBR Green detection chemistry (Applied Biosystems, Foster City, USA). The reaction mixture (25 μL) contained 12.5 μL 2 × Power SYBR Green PCR Master Mix, 1 μL of cDNA, 1 μL of each primer and 9.5 μL of ddH2O. The qPCR cycling conditions were as follows: 10 min at 95 °C, 40 cycles of 95 °C for 20 s and 60 °C for 1 min, then increasing the temperature by 0.5 °C every 5 s to obtain the product melt curve. All the qPCR assays included three technical and biological replicates. Standard curves were drawn to determine the amplification efficiency (E) and correlation coefficient (R2) of the diluted series based on the diluted (2×) cDNA series.
Data analysis for expression stability
The software programs geNorm32, NormFinder44 and Bestkeeper45 were selected to calculate the expression stability of the 9 candidate reference genes. The web tool ReFinder46 was then selected to generate a comprehensive ranking for the reference genes by comparing the results calculated by these three software programs. The Cq values of all reference genes used in geNorm and NormFinder were converted into relative quantities according to the formula2−∆Ct (∆Ct = the corresponding Cq value − minimum Cq)47. The geNorm program calculated the expression stability value (M), and reference genes with M < 1.5 were considered stable. The geNorm was also used to calculate the pairwise variation (Vn/Vn+1) between the two sequential normalization factors (NFn and NFn+1) in order to determine the minimum number of reference genes for optimal normalization and the recommended cut-off value was 0.1548. NormFinder calculated the expression stabilities of the candidate reference genes by combining the intra- and inter-group variations in a sample set containing any number of samples organized in any number of groups. Raw Cq values were directly analysed by BestKeeper.
The expression characteristics of EPSPS, GS and ACCase in goosegrass
To validate the stability of the selected reference genes for qPCR analysis under different herbicide stress, the expression of the target-site genes of glyphosate (EPSPS), glufosinate (GS) and quizalofop-p-ethyl (ACCase) were determined at different times (48 h, 72 h and 96 h) for the leaf tissues (4–5 leaves stage) after treatment. The plant culture and the herbicide treatment were performed using the methods described above. Both the most stable reference genes (eIF-4, ALS) and the most varying reference gene (TUB) were chosen for this study. The sequences of these three genes were retrieved from the transcript data in our previous study and re-confirmed49. The primer designing and qPCR methods were similar to the ones described above. EPSPS, GS and ACCase were amplified using the primer pairs (EP-F: 5′ GGTGGCAAGGTTAAGTTATCTGG 3′, EP-R: 5′ TCAACATAAGGGATGGAGATCAG 3′), (GS-F: 5′ GCGTGAAGATGGTGGATATGAAG 3′, GS-R: 5′ TTCGTGTTTCCCTGTCAACCTC 3′) and (ACC-F: 5′ TCATCATTCACATCAAACAGCATTCTC 3′, ACC-R: 5′ TGCCCATCACAATCCAACCAAAG 3′), respectively. The amount of transcript accumulated for these three genes normalized to the reference genes were analyzed using 2−∆∆Ct method47. Three technical and biological replicates were performed in this study, and the data were statistically compared by one-way ANOVA.
Additional Information
How to cite this article: Chen, J. et al. Selection of relatively exact reference genes for gene expression studies in goosegrass (Eleusine indica) under herbicide stress. Sci. Rep. 7, 46494; doi: 10.1038/srep46494 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Oerke, E.-C. Crop losses to pests. The Journal of Agricultural Science 144, 31–43 (2006).
Heap, I. In Integrated Pest Management (eds Pimentel Dayid & Peshin Rajinder ) 281–301 (Springer, 2014).
Heap, I. The International Survey of Herbicide Resistant Weeds http://www.weedscience.org (2016).
Holm, L. G., Plucknett, D. L., Pancho, J. V. & Herberger, J. P. The world’s worst weeds: Distribution and Biology. p, (609) (University Press of Hawaii, Hawaii, 1977).
Corbett, J. L., Askew, S. D., Thomas, W. E. & Wilcut, J. W. Weed Efficacy Evaluations for Bromoxynil, Glufosinate, Glyphosate, Pyrithiobac, and Sulfosate1. Weed Technology 18, 443–453 (2004).
Lee, L. J. & Ngim, J. A first report of glyphosate‐resistant goosegrass (Eleusine indica (L) Gaertn) in Malaysia. Pest Management Science 56, 336–339 (2000).
Chen, J., Huang, H., Wei, S., Zhang, C. & Huang, Z. Characterization of glyphosate-resistant goosegrass (Eleusine indica) populations in China. Journal of Integrative Agriculture 14, 919–925 (2015).
McCullough, P. E., Yu, J., Raymer, P. & Chen, Z. First Report of ACCase-Resistant Goosegrass (Eleusine indica) in the United States. Weed Science 64, 399–408 (2016).
San Cha, T., Najihah, M. G., Sahid, I. B. & Chuah, T. S. Molecular basis for resistance to ACCase-inhibiting fluazifop in Eleusine indica from Malaysia. Pesticide Biochemistry and Physiology 111, 7–13 (2014).
Jalaludin, A., Ngim, J., Bakar, B. H. & Alias, Z. Preliminary findings of potentially resistant goosegrass (Eleusine indica) to glufosinate‐ammonium in Malaysia. Weed Biology and Management 10, 256–260 (2010).
Jalaludin, A., Yu, Q. & Powles, S. Multiple resistance across glufosinate, glyphosate, paraquat and ACCase‐inhibiting herbicides in an Eleusine indica population. Weed Research 55, 82–89 (2015).
Seng, C. T., Van Lun, L., San, C. T. & SAHID, I. B. Initial report of glufosinate and paraquat multiple resistance that evolved in a biotype of goosegrass (Eleusine indica) in Malaysia. Weed Biology and Management 10, 229–233 (2010).
Délye, C., Jasieniuk, M. & Le Corre, V. Deciphering the evolution of herbicide resistance in weeds. Trends in Genetics 29, 649–658 (2013).
Powles, S. B. & Yu, Q. Evolution in action: plants resistant to herbicides. Annual Review of Plant Biology 61, 317–347 (2010).
Gaines, T. A. et al. RNA-Seq transcriptome analysis to identify genes involved in metabolism-based diclofop resistance in Lolium rigidum . The Plant Journal 78, 865–876 (2014).
Chen, J. et al. Mutations and amplification of EPSPS gene confer resistance to glyphosate in goosegrass (Eleusine indica). Planta 242, 1–10 (2015).
Cummins, I. et al. Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proceedings of the National Academy of Sciences 110, 5812–5817 (2013).
Derveaux, S., Vandesompele, J. & Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 50, 227–230 (2010).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55, 611–622 (2009).
Bustin, S. A. et al. MIQE precis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Molecular Biology 11, 74, 10.1186/1471-2199-11-74 (2010).
Martins, P. K. et al. Selection of reliable reference genes for RT-qPCR analysis during developmental stages and abiotic stress in Setaria viridis . Scientific Reports 6, 28348, 10.1038/srep28348 (2016).
Zhang, S., Zeng, Y., Yi, X. & Zhang, Y. Selection of suitable reference genes for quantitative RT-PCR normalization in the halophyte Halostachys caspica under salt and drought stress. Scientific Reports 6, 30363, 10.1038/srep30363 (2016).
Shivhare, R. & Lata, C. Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Scientific Reports 6, 23036; 10.1038/srep23036 (2016).
Wu, Z. J., Tian, C., Jiang, Q., Li, X. H. & Zhuang, J. Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis). Scientific Reports 6, 19748; 10.1038/srep19748 (2016).
Xu, X. et al. Selection of relatively exact reference genes for gene expression studies in flixweed (Descurainia sophia) by quantitative real-time polymerase chain reaction. Pesticide Biochemistry and Physiology 127, 59–66 (2016).
Wrzesińska, B., Kierzek, R. & Obrępalska‐Stęplowska, A. Evaluation of six commonly used reference genes for gene expression studies in herbicide‐resistant Avena fatua biotypes. Weed Research 56, 284–292 (2016).
Petit, C., Pernin, F., Heydel, J. M. & Délye, C. Validation of a set of reference genes to study response to herbicide stress in grasses. BMC Research Notes 5, 18; 10.1186/1756-0500-5-18 (2012).
Duhoux, A. & Délye, C. Reference genes to study herbicide stress response in Lolium sp.: up-regulation of P450 genes in plants resistant to acetolactate-synthase inhibitors. PLoS One 8, e63576; 10.1371/journal.pone.0063576 (2013).
ZHANG, C. et al. Investigating the mechanisms of glyphosate resistance in goosegrass (Eleusine indica) population from South China. Journal of Integrative Agriculture 14, 909–918 (2015).
Huffman, J. L., Riggins, C. W., Steckel, L. E. & Tranel, P. J. The EPSPS Pro106Ser substitution solely accounts for glyphosate resistance in a goosegrass (Eleusine indica) population from Tennessee, United States. Journal of Integrative Agriculture 15, 1304–1312 (2016).
Gomes, M. P. et al. Alteration of plant physiology by glyphosate and its by-product aminomethylphosphonic acid: an overview. Journal of Experimental Botany 65, 4691–4703 (2014).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3, 1; 10.1186/gb-2002-3-7-research0034 (2002).
Gaines, T. A. et al. Gene amplification confers glyphosate resistance in Amaranthus palmeri . Proceedings of the National Academy of Sciences 107, 1029–1034 (2010).
Salas, R. A., Scott, R. C., Dayan, F. E. & Burgos, N. R. EPSPS Gene Amplification in Glyphosate-Resistant Italian Ryegrass (Lolium perenne ssp. multiflorum) Populations from Arkansas, USA. Journal of Agricultural and Food Chemistry 63, 5885–5893 (2015).
Peng, Y. et al. Characterization of the horseweed (Conyza canadensis) transcriptome using GS-FLX 454 pyrosequencing and its application for expression analysis of candidate non-target herbicide resistance genes. Pest Management Science 66, 1053–1062 (2010).
Riggins, C. W., Peng, Y. Jr, C. N. S. & Tranel, P. J. Characterization of de novo transcriptome for waterhemp (Amaranthus tuberculatus) using GS-FLX 454 pyrosequencing and its application for studies of herbicide target-site genes. Pest Management Science 66, 1042–1052 (2010).
Dombrowski, J. E. & Martin, R. C. Evaluation of reference genes for quantitative RT-PCR in Lolium temulentum under abiotic stress. Plant Science 176, 390–396 (2008).
Jain, M., Nijhawan, A., Tyagi, A. K. & Khurana, J. P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and Biophysical Research Communications 345, 646–651 (2006).
Hong, S. Y., Seo, P. J., Yang, M. S., Xiang, F. & Park, C. M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biology 8, 112; 10.1186/1471-2229-8-112 (2008).
Faccioli, P. et al. A combined strategy of “in silico” transcriptome analysis and web search engine optimization allows an agile identification of reference genes suitable for normalization in gene expression studies. Plant Molecular Biology 63, 679–688 (2007).
Malone, J. M., Morran, S., Shirley, N., Boutsalis, P. & Preston, C. EPSPS gene amplification in glyphosate-resistant Bromus diandrus . Pest Management Science 72, 81–88 (2015).
Wiersma, A. T. et al. Gene amplification of 5-enol-pyruvylshikimate-3-phosphate synthase in glyphosate-resistant Kochia scoparia . Planta, 1–12 (2014).
Duke, S. O. & Powles, S. B. Glyphosate: a once-in-a-century herbicide. Pest Management Science 64, 319–325 (2008).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research 64, 5245–5250 (2004).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnology Letters 26, 509–515 (2004).
Xie, F., Xiao, P., Chen, D., Xu, L. & Zhang, B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Molecular Biology 80, 75–84 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408 (2001).
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F. & Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology 8, R19; 10.1186/gb-2007-8-2-r19 (2007).
Chen, J. et al. Investigating the mechanisms of glyphosate resistance in goosegrass (Eleusine indica (L.) Gaertn.) by RNA Sequencing technology. The Plant Journal 89, 407–415 (2017)
Acknowledgements
This research was funded by Special Fund for Agro-scientific Research in the Public Interest (201303022), and The National Natural Science Foundation of China (No. 31601666).
Author information
Authors and Affiliations
Contributions
C.X.Z. and J.C.C. designed the research; J.C.C. performed research; J.C.C., Z.F.H. and C.L.J. analyzed data; J.C.C. and C.X.Z. wrote the manuscript. H.J.H., S.H.W., Y.L., J.Z. and C.X.Z. revised the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chen, J., Huang, Z., Huang, H. et al. Selection of relatively exact reference genes for gene expression studies in goosegrass (Eleusine indica) under herbicide stress. Sci Rep 7, 46494 (2017). https://doi.org/10.1038/srep46494
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep46494
- Springer Nature Limited
This article is cited by
-
Systematic screening and validation of reliable reference genes for qRT-PCR analysis in Okra (Abelmoschus esculentus L.)
Scientific Reports (2022)
-
Identification of the most suitable reference gene for gene expression studies with development and abiotic stress response in Bromus sterilis
Scientific Reports (2021)
-
Identification and evaluation of suitable reference genes for gene expression analysis in rubber tree leaf
Molecular Biology Reports (2020)
-
Identification and evaluation of reference genes for quantitative real-time PCR analysis in Polygonum cuspidatum based on transcriptome data
BMC Plant Biology (2019)
-
Selection of reference genes for qPCR normalization in buffalobur (Solanum rostratum Dunal)
Scientific Reports (2019)