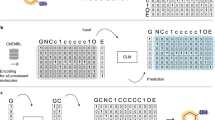
Abstract
Machine learning has provided a means to accelerate early-stage drug discovery by combining molecule generation and filtering steps in a single architecture that leverages the experience and design preferences of medicinal chemists. However, designing machine learning models that can achieve this on the fly to the satisfaction of medicinal chemists remains a challenge owing to the enormous search space. Researchers have addressed de novo design of molecules by decomposing the problem into a series of tasks determined by design criteria. Here we provide a comprehensive overview of the current state of the art in molecular design using machine learning models as well as important design decisions, such as the choice of molecular representations, generative methods and optimization strategies. Subsequently, we present a collection of practical applications in which the reviewed methodologies have been experimentally validated, encompassing both academic and industrial efforts. Finally, we draw attention to the theoretical, computational and empirical challenges in deploying generative machine learning and highlight future opportunities to better align such approaches to achieve realistic drug discovery end points.



Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.References
Delaney, J. S. ESOL: estimating aqueous solubility directly from molecular structure. J. Chem. Inf. Comput. 44, 1000–1005 (2004).
Gillette, J. R., Mitchell, J. R. & Brodie, B. B. Biochemical mechanisms of drug toxicity. Annu. Rev. Pharmacol. 14, 271–288 (1974).
Gibaldi, M. & Perrier, D. Pharmacokinetics (CRC Press, 1982).
Bohacek, R. S., McMartin, C. & Guida, W. C. The art and practice of structure-based drug design: a molecular modeling perspective. Med. Res. Rev. 16, 3–50 (1996).
Stumpfe, D. & Bajorath, J. Exploring activity cliffs in medicinal chemistry: miniperspective. J. Med. Chem. 55, 2932–2942 (2012).
Scannell, J. W., Blanckley, A., Boldon, H. & Warrington, B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 11, 191–200 (2012).
Berdigaliyev, N. & Aljofan, M. An overview of drug discovery and development. Future Med. Chem. 12, 939–947 (2020).
Ringel, M. S., Scannell, J. W., Baedeker, M. & Schulze, U. Breaking Eroom’s law. Nat. Rev. Drug Discov. 19, 833–834 (2020).
Aparoy, P., Kumar Reddy, K. & Reddanna, P. Structure and ligand based drug design strategies in the development of novel 5-LOX inhibitors. Curr. Med. Chem. 19, 3763–3778 (2012).
Baskin, I. & Varnek, A. in Chemoinformatics Approaches to Virtual Screening Ch. 1, 1–43 (Royal Society of Chemistry, 2008).
Kuntz, I. D. Structure-based strategies for drug design and discovery. Science 257, 1078–1082 (1992).
Anderson, A. C. The process of structure-based drug design. Chem. Biol. 10, 787–797 (2003).
Choung, O.-H., Vianello, R., Segler, M., Stiefl, N. & Jiménez-Luna, J. Extracting medicinal chemistry intuition via preference machine learning. Nat. Commun. 14, 6651 (2023).
Lyu, J. et al. Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019). Ultralarge-scale virtual screening of a make-on-demand library identified hits with previously unknown chemical motifs that were experimentally validated.
Sadybekov, A. A. et al. Synthon-based ligand discovery in virtual libraries of over 11 billion compounds. Nature 601, 452–459 (2022).
Gorgulla, C. et al. Virtualflow 2.0—the next generation drug discovery platform enabling adaptive screens of 69 billion molecules. Preprint at bioRxiv https://doi.org/10.1101/2023.04.25.537981 (2023).
Rombach, R., Blattmann, A., Lorenz, D., Esser, P. & Ommer, B. High-resolution image synthesis with latent diffusion models. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition 10684–10695 (IEEE, 2022).
Brown, T. et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 33, 1877–1901 (2020).
Zhavoronkov, A. et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 37, 1038–1040 (2019). One of the first studies to experimentally validate ML-generated molecules and highlighted the potential for accelerated drug discovery.
Ren, F. et al. AlphaFold accelerates artificial intelligence powered drug discovery: efficient discovery of a novel CDK20 small molecule inhibitor. Chem. Sci. 14, 1443–1452 (2023). A study performing molecular docking using an AlphaFold-generated structure on ML-generated molecules with experimental validation.
Wu, C.-T. et al. COT: an efficient and accurate method for detecting marker genes among many subtypes. Bioinf. Adv. 2, vbac037 (2022).
Méndez-Lucio, O., Baillif, B., Clevert, D.-A., Rouquié, D. & Wichard, J. De novo generation of hit-like molecules from gene expression signatures using artificial intelligence. Nat. Commun. 11, 10 (2020).
Sanchez-Fernandez, A., Rumetshofer, E., Hochreiter, S. & Klambauer, G. CLOOME: contrastive learning unlocks bioimaging databases for queries with chemical structures. Nat. Commun. 14, 7339 (2023).
Nguyen, C. Q., Pertusi, D. & Branson, K. M. Molecule-morphology contrastive pretraining for transferable molecular representation. Preprint at https://arxiv.org/abs/2305.09790 (2023).
Schaller, D. et al. Next generation 3D pharmacophore modeling. Wiley Interdiscip. Rev. Comput. Mol. Sci. 10, e1468 (2020).
Imrie, F., Hadfield, T. E., Bradley, A. R. & Deane, C. M. Deep generative design with 3D pharmacophoric constraints. Chem. Sci. 12, 14577–14589 (2021).
Guo, J. et al. Link-INVENT: generative linker design with reinforcement learning. Digit. Discov. 2, 392–408 (2023).
Torge, J., Harris, C., Mathis, S. V. & Lio, P. DiffHopp: a graph diffusion model for novel drug design via scaffold hopping. Preprint at https://arxiv.org/abs/2308.07416 (2023).
Keiser, M. J. et al. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 25, 197–206 (2007).
Harris, C. et al. Flexible small-molecule design and optimization with equivariant diffusion models. In ICLR 2023—Machine Learning for Drug Discovery Workshop (OpenReview, 2023).
Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36 (1988).
Krenn, M., Häse, F., Nigam, A., Friederich, P. & Aspuru-Guzik, A. Self-referencing embedded strings (SELFIES): a 100% robust molecular string representation. Mach. Learn. Sci. Technol. 1, 045024 (2020).
Kingma, D. P. & Welling, M. Auto-encoding variational bayes. In Proc. 2nd International Conference on Learning Representations (eds Bengio, Y. & LeCun, Y.) (OpenReview, 2014).
Gómez-Bombarelli, R. et al. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 4, 268–276 (2018). One of the first studies to apply a variational autoencoder for molecular design and is a foundational work for many recently reported methods.
Simonovsky, M. & Komodakis, N. GraphVAE: towards generation of small graphs using variational autoencoders. In Artificial Neural Networks and Machine Learning—ICANN 2018 (eds Kůrková, V. et al.) 412–422 (Springer, 2018).
Goodfellow, I. et al. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 27, 2672–2680 (2014).
Guimaraes, G. L., Sanchez-Lengeling, B., Outeiral, C., Farias, P. L. C. & Aspuru-Guzik, A. Objective-reinforced generative adversarial networks (ORGAN) for sequence generation models. Preprint at https://arxiv.org/abs/1705.10843 (2017).
De Cao, N. & Kipf, T. MolGAN: an implicit generative model for small molecular graphs. Preprint at https://arxiv.org/abs/1805.11973 (2018).
Rezende, D. & Mohamed, S. Variational inference with normalizing flows. In Proc. 32nd International Conference on Machine Learning 1530–1538 (PMLR, 2015).
Shi, C. et al. GraphAF: a flow-based autoregressive model for molecular graph generation. In Proc. 8th International Conference on Learning Representations (OpenReview, 2020).
Lipman, Y., Chen, R. T., Ben-Hamu, H., Nickel, M. & Le, M. Flow matching for generative modeling. In Proc. 11th International Conference on Learning Representations (OpenReview, 2023).
Song, Y. et al. Equivariant flow matching with hybrid probability transport for 3D molecule generation. Adv. Neural Inf. Process. Syst. 36, 549–568 (2023).
Van Oord, A., Kalchbrenner, N. & Kavukcuoglu, K. Pixel recurrent neural networks. In Proc. 33rd International Conference on Machine Learning 1747–1756 (PMLR, 2016).
Popova, M., Shvets, M., Oliva, J. & Isayev, O. MolecularRNN: generating realistic molecular graphs with optimized properties. Preprint at https://arxiv.org/abs/1905.13372 (2019).
Gebauer, N., Gastegger, M. & Schütt, K. Symmetry-adapted generation of 3D point sets for the targeted discovery of molecules. Adv. Neural Inf. Process. Syst. 32, 7566–7578 (2019).
Ho, J., Jain, A. & Abbeel, P. Denoising diffusion probabilistic models. Adv. Neural Inf. Process. Syst. 33, 6840–6851 (2020).
Hoogeboom, E., Satorras, V. G., Vignac, C. & Welling, M. Equivariant diffusion for molecule generation in 3D. In Proc. 39th International Conference on Machine Learning 8867–8887 (PMLR, 2022).
Schneuing, A. et al. Structure-based drug design with equivariant diffusion models. In NeurIPS 2022 Machine Learning for Structural Biology (OpenReview, 2022). One of the first studies to leverage the flexibility of diffusion models to achieve a variety of types of conditional generation and molecule optimization.
Igashov, I. et al. Equivariant 3D-conditional diffusion models for molecular linker design. Nat. Mach. Intell. 6, 417–427 (2024).
Xu, M. et al. GeoDiff: a geometric diffusion model for molecular conformation generation. In Proc. 9th International Conference on Learning Representations (OpenReview, 2021).
Liu, Q., Allamanis, M., Brockschmidt, M. & Gaunt, A. Constrained graph variational autoencoders for molecule design. Adv. Neural Inf. Process. Syst. 31, 7795–7804 (2018).
Segler, M. H., Kogej, T., Tyrchan, C. & Waller, M. P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 4, 120–131 (2018).
Bengio, E., Jain, M., Korablyov, M., Precup, D. & Bengio, Y. Flow network based generative models for non-iterative diverse candidate generation. Adv. Neural Inf. Process. Syst. 34, 27381–27394 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Jensen, J. H. A graph-based genetic algorithm and generative model/Monte Carlo tree search for the exploration of chemical space. Chem. Sci. 10, 3567–3572 (2019).
Yang, X., Zhang, J., Yoshizoe, K., Terayama, K. & Tsuda, K. ChemTS: an efficient Python library for de novo molecular generation. Sci. Technol. Adv. Mat. 18, 972–976 (2017).
Olivecrona, M., Blaschke, T., Engkvist, O. & Chen, H. Molecular de-novo design through deep reinforcement learning. J. Cheminf. 9, 48 (2017). One of the first studies applying reinforcement learning to molecular design and is the first version of ‘REINVENT’, an industrially used method, which is still under active development.
Popova, M., Isayev, O. & Tropsha, A. Deep reinforcement learning for de novo drug design. Sci. Adv. 4, eaap7885 (2018).
You, J., Liu, B., Ying, Z., Pande, V. & Leskovec, J. Graph convolutional policy network for goal-directed molecular graph generation. Adv. Neural Inf. Process. Syst. 31, 6410–6421 (2018).
Fu, T., Gao, W., Coley, C. & Sun, J. Reinforced genetic algorithm for structure-based drug design. Adv. Neural Inf. Process. Syst. 35, 12325–12338 (2022).
Fu, T., Xiao, C., Li, X., Glass, L. M. & Sun, J. MIMOSA: multi-constraint molecule sampling for molecule optimization. In Proc. 35th AAAI Conference on Artificial Intelligence 125–133 (AAAI, 2021).
Fu, T. et al. Differentiable scaffolding tree for molecular optimization. In Proc. 10th International Conference on Learning Representations (OpenReview, 2022).
Jin, W., Barzilay, R. & Jaakkola, T. Junction tree variational autoencoder for molecular graph generation. In Proc. 35th International Conference on Machine Learning, Proc. Machine Learning Research Vol. 80 (eds Dy, J. & Krause, A.) 2323–2332 (PMLR, 2018).
Griffiths, R.-R. & Hernández-Lobato, J. M. Constrained Bayesian optimization for automatic chemical design using variational autoencoders. Chem. Sci. 11, 577–586 (2020).
Gao, W., Fu, T., Sun, J. & Coley, C. Sample efficiency matters: a benchmark for practical molecular optimization. Adv. Neural Inf. Process. Syst. 35, 21342–21357 (2022). One of the benchmarks for molecule optimization evaluating more than 25 methods on 20 commonly used oracle functions.
Du, Y. et al. ChemSpacE: interpretable and interactive chemical space exploration. In Transactions on Machine Learning Research (OpenReview, 2023).
Schrödinger release 2024-1 (Schrödinger, 2024).
OpenEye: Applications (Cadence Molecular Sciences, 2023); https://www.eyesopen.com/applications
Friesner, R. A. et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 (2004).
OpenEye: OEDocking (Cadence Molecular Sciences, 2023); https://www.eyesopen.com/oedocking
Arús-Pous, J. et al. Randomized SMILES strings improve the quality of molecular generative models. J. Cheminf. 11, 71 (2019).
Brown, N., Fiscato, M., Segler, M. H. & Vaucher, A. C. GuacaMol: benchmarking models for de novo molecular design. J. Chem. Inf. Model. 59, 1096–1108 (2019). One of the first proposed benchmarks for ML-based molecular design and many proposed tasks are still used in newer benchmarks.
Polykovskiy, D. et al. Molecular sets (MOSES): a benchmarking platform for molecular generation models. Front. Pharmacol. 11, 565644 (2020).
Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 23, 3–25 (1997).
Bickerton, G. R., Paolini, G. V., Besnard, J., Muresan, S. & Hopkins, A. L. Quantifying the chemical beauty of drugs. Nat. Chem. 4, 90–98 (2012).
Guo, J. et al. DockStream: a docking wrapper to enhance de novo molecular design. J. Cheminf. 13, 1–21 (2021).
Arnott, J. A. & Planey, S. L. The influence of lipophilicity in drug discovery and design. Expert Opin. Drug Discov. 7, 863–875 (2012).
Hopkins, A. L., Keserü, G. M., Leeson, P. D., Rees, D. C. & Reynolds, C. H. The role of ligand efficiency metrics in drug discovery. Nat. Rev. Drug Discov. 13, 105–121 (2014).
Luo, S., Guan, J., Ma, J. & Peng, J. A 3D generative model for structure-based drug design. Adv. Neural Inf. Process. Syst. 34, 6229–6239 (2021).
Harris, C. et al. Benchmarking generated poses: how rational is structure-based drug design with generative models? Preprint at https://arxiv.org/abs/2308.07413 (2023).
Ertl, P. & Schuffenhauer, A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminf. 1, 8 (2009).
Fukunishi, Y., Kurosawa, T., Mikami, Y. & Nakamura, H. Prediction of synthetic accessibility based on commercially available compound databases. J. Chem. Inf. Model. 54, 3259–3267 (2014).
Voršilák, M., Kolář, M., Čmelo, I. & Svozil, D. SYBA: Bayesian estimation of synthetic accessibility of organic compounds. J. Cheminf. 12, 35 (2020).
Coley, C. W., Rogers, L., Green, W. H. & Jensen, K. F. SCScore: synthetic complexity learned from a reaction corpus. J. Chem. Inf. Model. 58, 252–261 (2018).
Coley, C. W., Rogers, L., Green, W. H. & Jensen, K. F. Computer-assisted retrosynthesis based on molecular similarity. ACS Cent. Sci. 3, 1237–1245 (2017).
Schwaller, P. et al. Molecular transformer: a model for uncertainty-calibrated chemical reaction prediction. ACS Cent. Sci. 5, 1572–1583 (2019).
Schwaller, P. et al. Predicting retrosynthetic pathways using transformer-based models and a hyper-graph exploration strategy. Chem. Sci. 11, 3316–3325 (2020).
Segler, M. H., Preuss, M. & Waller, M. P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature 555, 604–610 (2018). Seminal work applying deep learning for retrosynthesis, which can be used to filter ML-generated molecules for synthesizability.
Genheden, S. et al. AiZynthFinder: a fast, robust and flexible open-source software for retrosynthetic planning. J. Cheminf. 12, 70 (2020).
Thakkar, A., Chadimová, V., Bjerrum, E. J., Engkvist, O. & Reymond, J.-L. Retrosynthetic accessibility score (RAscore)—rapid machine learned synthesizability classification from AI driven retrosynthetic planning. Chem. Sci. 12, 3339–3349 (2021).
Liu, C.-H. et al. RetroGNN: fast estimation of synthesizability for virtual screening and de novo design by learning from slow retrosynthesis software. J. Chem. Inf. Model. 62, 2293–2300 (2022).
Bradshaw, J., Paige, B., Kusner, M. J., Segler, M. & Hernández-Lobato, J. M. A model to search for synthesizable molecules. Adv. Neural Inf. Process. Syst. 32, 79377949 (2019).
Bradshaw, J., Paige, B., Kusner, M. J., Segler, M. H. S. & Hernández-Lobato, J. M. Barking up the right tree: an approach to search over molecule synthesis dags. Adv. Neural Inf. Process. Syst. 33, (2020).
Horwood, J. & Noutahi, E. Molecular design in synthetically accessible chemical space via deep reinforcement learning. ACS Omega 5, 32984–32994 (2020).
Gottipati, S. K. et al. Learning to navigate the synthetically accessible chemical space using reinforcement learning. In Proc. 37th International Conference on Machine Learning (eds Daumé H. & Singh, A.) 3668–3679 (PMLR, 2020).
Gao, W., Mercado, R. & Coley, C. W. Amortized tree generation for bottom-up synthesis planning and synthesizable molecular design. In Proc. 10th International Conference on Learning Representations (OpenReview, 2022).
Swanson, K. et al. Generative AI for designing and validating easily synthesizable and structurally novel antibiotics. Nat. Mach. Intell. 6, 338–353 (2024).
Fialková, V. et al. LibINVENT: reaction-based generative scaffold decoration for in silico library design. J. Chem. Inf. Model. 62, 2046–2063 (2021).
Hartenfeller, M. et al. DOGS: reaction-driven de novo design of bioactive compounds. PLoS Comput. Biol. 8, e1002380 (2012).
Ghiandoni, G. M. et al. RENATE: a pseudo-retrosynthetic tool for synthetically accessible de novo design. Mol. Inform. 41, 2100207 (2022).
Flam-Shepherd, D., Zhu, K. & Aspuru-Guzik, A. Language models can learn complex molecular distributions. Nat. Commun. 13, 3293 (2022).
Ballarotto, M. et al. De novo design of Nurr1 agonists via fragment-augmented generative deep learning in low-data regime. J. Med. Chem. 66, 8170–8177 (2023).
Gaulton, A. et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, D1100–D1107 (2012).
Ivanenkov, Y. A. et al. Chemistry42: an AI-driven platform for molecular design and optimization. J. Chem. Info. Model. 63, 695–701 (2023).
Zhu, W. et al. Discovery of novel and selective SIK2 inhibitors by the application of AlphaFold structures and generative models. Bioorg. Med. Chem. 91, 117414 (2023).
Grisoni, F. et al. Combining generative artificial intelligence and on-chip synthesis for de novo drug design. Sci. Adv. 7, eabg3338 (2021).
Li, Y. et al. Generative deep learning enables the discovery of a potent and selective RIPK1 inhibitor. Nat. Commun. 13, 6891 (2022). A study showing the complementarity of virtual screening and generative molecular design with experimental validation.
Yoshimori, A. et al. Design and synthesis of DDR1 inhibitors with a desired pharmacophore using deep generative models. ChemMedChem 16, 955–958 (2021).
Blaschke, T. et al. REINVENT 2.0: an AI tool for de novo drug design. J. Chem. Inf. Model. 60, 5918–5922 (2020).
Merk, D., Grisoni, F., Friedrich, L., Gelzinyte, E. & Schneider, G. Scaffold hopping from synthetic RXR modulators by virtual screening and de novo design. MedChemComm 9, 1289–1292 (2018).
Merk, D., Grisoni, F., Friedrich, L., Gelzinyte, E. & Schneider, G. Computer-assisted discovery of retinoid X receptor modulating natural products and isofunctional mimetics. J. Med. Chem. 61, 5442–5447 (2018).
Gillet, V. et al. Synthetically accessible de novo design using reaction vectors: application to PARP1 inhibitors. Mol. Inform. 43, e202300183 (2024).
Pun, F. W., Ozerov, I. V. & Zhavoronkov, A. AI-powered therapeutic target discovery. Trends Pharmacol. Sci. 44, 561–572 (2023).
First generative AI drug begins phase II trials with patients. Insilico Medicine https://insilico.com/blog/first_phase2 (2023).
Ren, F. et al. A small-molecule TNIK inhibitor targets fibrosis in preclinical and clinical models. Nat. Biotechnol. (2024).
Guo, J. et al. Improving de novo molecular design with curriculum learning. Nat. Mach. Intell. 4, 555–563 (2022).
Guo, J. & Schwaller, P. Augmented memory: capitalizing on experience replay to accelerate de novo molecular design. Preprint at https://doi.org/10.48550/arXiv.2305.16160 (2024).
Guo, J. & Schwaller, P. Beam enumeration: probabilistic explainability for sample efficient self-conditioned molecular design. In Proc. 12th International Conference on Learning Representations (OpenReview, 2024).
Dodds, M. et al. Sample efficient reinforcement learning with active learning for molecular design. Chem. Sci. 15, 4146–4160 (2024).
Buttenschoen, M., Morris, G. M. & Deane, C. M. Posebusters: AI-based docking methods fail to generate physically valid poses or generalise to novel sequences. Chem. Sci. 15, 3130–3139 (2024).
Watson, J. L. et al. De novo design of protein structure and function with RFdiffusion. Nature 620, 1089–1100 (2023).
Du, Y., Guo, X., Wang, Y., Shehu, A. & Zhao, L. Small molecule generation via disentangled representation learning. Bioinformatics 38, 3200–3208 (2022).
Jin, W., Barzilay, R. & Jaakkola, T. Multi-objective molecule generation using interpretable substructures. In Proc. 37th International Conference on Machine Learning 4849–4859 (PMLR, 2020).
Hoffman, S. C., Chenthamarakshan, V., Wadhawan, K., Chen, P.-Y. & Das, P. Optimizing molecules using efficient queries from property evaluations. Nat. Mach. Intell. 4, 21–31 (2021).
Madhawa, K., Ishiguro, K., Nakago, K. & Abe, M. GraphNVP: an invertible flow model for generating molecular graphs. Preprint at https://arxiv.org/abs/1905.11600 (2019).
Kadurin, A. et al. The cornucopia of meaningful leads: applying deep adversarial autoencoders for new molecule development in oncology. Oncotarget 8, 10883 (2017).
Imrie, F., Bradley, A. R., van der Schaar, M. & Deane, C. M. Deep generative models for 3D linker design. J. Chem. Inf. Model. 60, 1983–1995 (2020).
Liu, M., Yan, K., Oztekin, B. & Ji, S. GraphEBM: molecular graph generation with energy-based models. Preprint at https://arxiv.org/abs/2102.00546 (2021).
Vignac, C. et al. DiGress: discrete denoising diffusion for graph generation. In Proc. 11th International Conference on Learning Representations (OpenReview, 2023).
Nigam, A., Friederich, P., Krenn, M. & Aspuru-Guzik, A. Augmenting genetic algorithms with deep neural networks for exploring the chemical space. In Proc. 8th International Conference on Learning Representations (OpenReview, 2020).
Spiegel, J. O. & Durrant, J. D. AutoGrow4: an open-source genetic algorithm for de novo drug design and lead optimization. J. Cheminf. 12, 25 (2020). One of the representative works that leverage genetic algorithms for molecular design.
Simm, G. & Hernandez-Lobato, J. M. A generative model for molecular distance geometry. In Proc. 37th International Conference on Machine Learning, Proc. Machine Learning Research Vol. 119 (eds Daumé H. & Singh, A.) 8949–8958 (PMLR, 2020).
Ganea, O. et al. GeoMol: torsional geometric generation of molecular 3D conformer ensembles. Adv. Neural Inf. Process. Syst. 34, 13757–13769 (2021).
Klein, L., Krämer, A. & Noe, F. Equivariant flow matching. Adv. Neural Inf. Process. Syst. 36, 59886–59910 (2023).
Stärk, H., Ganea, O., Pattanaik, L., Barzilay, D. & Jaakkola, T. EquiBind: geometric deep learning for drug binding structure prediction. In Proc. 39th International Conference on Machine Learning, Proc. Machine Learning Research Vol. 162 (eds Chaudhuri, K. et al.) 20503–20521 (PMLR, 2022).
Jing, B., Corso, G., Chang, J., Barzilay, R. & Jaakkola, T. Torsional diffusion for molecular conformer generation. Adv. Neural Inf. Process. Syst. 35, 24240–24253 (2022).
Corso, G., Stärk, H., Jing, B., Barzilay, R. & Jaakkola, T. S. DiffDock: diffusion steps, twists, and turns for molecular docking. In Proc. 11th International Conference on Learning Representations (OpenReview, 2023).
Ragoza, M., Masuda, T. & Koes, D. R. Generating 3D molecules conditional on receptor binding sites with deep generative models. Chem. Sci. 13, 2701–2713 (2022).
Drotár, P., Jamasb, A. R., Day, B., Cangea, C. & Liò, P. Structure-aware generation of drug-like molecules. Preprint at https://arxiv.org/abs/2111.04107 (2021).
Joshi, R. P. et al. 3D-scaffold: a deep learning framework to generate 3D coordinates of drug-like molecules with desired scaffolds. J. Phys. Chem. B 125, 12166–12176 (2021).
Liu, M., Luo, Y., Uchino, K., Maruhashi, K. & Ji, S. Generating 3D molecules for target protein binding. In Proc. 39th International Conference on Machine Learning, Proc. Machine Learning Research, Vol. 162 (eds Chaudhuri, K. et al) 13912–13924 (PMLR, 2022).
Garcia Satorras, V., Hoogeboom, E., Fuchs, F., Posner, I. & Welling, M. E(n) equivariant normalizing flows. Adv. Neural Inf. Process. Syst. 34, 4181–4192 (2021).
Graves, A. et al. Hybrid computing using a neural network with dynamic external memory. Nature 538, 471–476 (2016).
Makhzani, A., Shlens, J., Jaitly, N., Goodfellow, I. & Frey, B. Adversarial autoencoders. Preprint at https://arxiv.org/abs/1511.05644 (2015).
Philippidis, A. Insilico joins scramble to treat solid tumors by targeting KIF18A; https://www.genengnews.com/topics/artificial-intelligence/insilico-joins-scramble-to-treat-solid-tumors-by-targeting-kif18a/ (2024).
Merk, D., Friedrich, L., Grisoni, F. & Schneider, G. De novo design of bioactive small molecules by artificial intelligence. Mol. Inf. 37, 1700153 (2018).
Merk, D., Grisoni, F., Friedrich, L. & Schneider, G. Tuning artificial intelligence on the de novo design of natural-product-inspired retinoid X receptor modulators. Commun. Chem. 1, 68 (2018).
Yu, Y. et al. A novel scalarized scaffold hopping algorithm with graph-based variational autoencoder for discovery of JAK1 inhibitors. ACS Omega 6, 22945–22954 (2021).
Moret, M., Helmstädter, M., Grisoni, F., Schneider, G. & Merk, D. Beam search for automated design and scoring of novel ROR ligands with machine intelligence. Angew. Chem. Int. Ed. 60, 19477–19482 (2021).
Jang, S. H. et al. PCW-A1001, AI-assisted de novo design approach to design a selective inhibitor for FLT-3 (D835Y) in acute myeloid leukemia. Front. Mol. Biosci. 9, 1072028 (2022).
Eguida, M., Schmitt-Valencia, C., Hibert, M., Villa, P. & Rognan, D. Target-focused library design by pocket-applied computer vision and fragment deep generative linking. J. Med. Chem. 65, 13771–13783 (2022).
Chen, N. et al. Recurrent neural network (RNN) model accelerates the development of antibacterial metronidazole derivatives. RSC Adv. 12, 22893–22901 (2022).
Tan, X. et al. Discovery of pyrazolo [3,4-d] pyridazinone derivatives as selective DDR1 inhibitors via deep learning based design, synthesis, and biological evaluation. J. Med. Chem. 65, 103–119 (2021).
Hua, Y. et al. Effective reaction-based de novo strategy for kinase targets: a case study on MERTK inhibitors. J. Chem. Inf. Model. 62, 1654–1668 (2022).
Moret, M. et al. Leveraging molecular structure and bioactivity with chemical language models for de novo drug design. Nat. Commun. 14, 114 (2023).
Song, S. et al. Application of deep generative model for design of pyrrolo [2,3-d] pyrimidine derivatives as new selective tank binding kinase 1 (TBK1) inhibitors. Eur. J. Med. Chem. 247, 115034 (2023).
Yu, Y. et al. Accelerated discovery of macrocyclic CDK2 inhibitor QR-6401 by generative models and structure-based drug design. ACS Med. Chem. Lett. 14, 297–304 (2023).
Atz, K. et al. Prospective de novo drug design with deep interactome learning. Nat. Commun. 15, 3408 (2024).
Putin, E. et al. Adversarial threshold neural computer for molecular de novo design. Mol. Pharmaceutics 15, 4386–4397 (2018).
Polykovskiy, D. et al. Entangled conditional adversarial autoencoder for de novo drug discovery. Mol. Pharm. 15, 4398–4405 (2018).
Korshunova, M. et al. Generative and reinforcement learning approaches for the automated de novo design of bioactive compounds. Commun. Chem. 5, 129 (2022).
Li, Y. et al. Discovery of potent, selective, and orally bioavailable small-molecule inhibitors of CDK8 for the treatment of cancer. J. Med. Chem. 66, 5439–5452 (2023).
Salas-Estrada, L. et al. De novo design of κ-opioid receptor antagonists using a generative deep-learning framework. J. Chem. Inf. Model. 63, 5056–5065 (2023).
Xu, J. et al. Discovery of novel and potent prolyl hydroxylase domain-containing protein (PHD) inhibitors for the treatment of anemia. J. Med. Chem. 67, 1393–1405 (2024).
Bo, W. et al. Local scaffold diversity-contributed generator for discovering potential NLRP3 inhibitors. J. Chem. Inf. Model. 64, 737–748 (2024).
Xia, Y. et al. Target-aware molecule generation for drug design using a chemical language model. Preprint at bioRxiv https://doi.org/10.1101/2024.01.08.574635 (2024).
Vakili, M. G. et al. Quantum computing-enhanced algorithm unveils novel inhibitors for KRAS. Preprint at https://arxiv.org/abs/2402.08210 (2024).
Hassen, A. K. et al. Generate what you can make: achieving in-house synthesizability with readily available resources in de novo drug design. Preprint at chemRxiv https://doi.org/10.26434/chemrxiv-2024-wtjt6 (2024).
Wang, Y. et al. Discovery of 3-hydroxymethyl-azetidine derivatives as potent polymerase theta inhibitors. Bioorg. Med. Chem. 103, 117662 (2024).
Zhao, Y. et al. Accelerating factor Xa inhibitor discovery with a de novo drug design pipeline. Chin. J. Chem. Eng. (2024).
Jiang, Y. et al. Pocketflow is a data-and-knowledge-driven structure-based molecular generative model. Nat. Mach. Intell. 6, 326–337 (2024).
Zhang, J. et al. ISM9682A, a novel and potent KIF18A inhibitor, shows robust antitumor effects against chromosomally unstable cancers. Cancer Res. 84, 5727–5727 (2024).
Huang, L. et al. A dual diffusion model enables 3D molecule generation and lead optimization based on target pockets. Nat. Commun. 15, 2657 (2024).
Acknowledgements
J.G. and P.S. acknowledge support from the NCCR Catalysis (grant number 180544), a National Centre of Competence in Research funded by the Swiss National Science Foundation. A.R.J. is funded by a Biotechnology and Biological Sciences Research Council (BBSRC) DTP studentship (BB/M011194/1). Y.W. acknowledges the support of Cornell Presidential Life Science Fellowship. We are grateful to K. Atz and A. Mueller for helpful feedback and discussion.
Author information
Authors and Affiliations
Contributions
Y.D., A.R.J. and J.G. led this work under the supervision of P.S. and T.L.B. and contributed equally. T.F. and C.H. also contributed equally. All authors contributed ideas and discussions to writing, reviewing and editing of the paper before submission.
Corresponding authors
Ethics declarations
Competing interests
A.R.J. declares a potential financial conflict of interest due to his role as a machine learning scientist at Prescient Design, Genentech. The other authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks J. B. Brown and Ola Engkvist for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary text and references.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Du, Y., Jamasb, A.R., Guo, J. et al. Machine learning-aided generative molecular design. Nat Mach Intell 6, 589–604 (2024). https://doi.org/10.1038/s42256-024-00843-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-024-00843-5
- Springer Nature Limited
This article is cited by
-
AI and ML for small molecule drug discovery in the big data era II
Molecular Diversity (2024)