Abstract
The aim of this study was to investigate the vertical transfer of microbiota from dams to the offspring. We studied a pair of 20 dams and its offspring. Maternal sources (colostrum, feces and vaginal secretion) and newborn fecal samples were analyzed using 16S rDNA amplicon sequencing on days 1, 3, 7, 14 and 28. Overall, newborns were maintained healthy and did not receive antimicrobial therapy. The Source Tracker analysis indicated that the newborn fecal microbiota was similar to colostrum and vaginal secretion from day 1 up to 7. However, an unknown source (probably from the environment) showed a gradual increase in its similarity with fecal samples from calves measured from day 3 to 28. The most abundant bacteria groups on meconium (day 1) and calf fecal samples on day 3 were Escherichia-Shigella and Clostridium, respectively. On day 7, the predominant genus were Bifidobacterium and Lactobacillus, while Fusobacterium was the most abundant genus on day 14, coinciding with the diarrhea peak. Faecalibacterium showed a gradual increase throughout the neonatal period. Maternal sources contribute to the neonatal microbiota, however other unknown sources (probably environment) had a strong influence on development of the gut microbiota later in the neonate period.
Similar content being viewed by others
Introduction
As in humans, the first year of a calf’s life is critical to the development of the microbiota and to the function of mucosal immunity. This period establishes a healthy or pro-inflammatory host state. This affects both short- and long-term on morbidity, growth and has an influence on future performance. The early gut microbial colonization of the GIT (gastrointestinal tract) can be influenced by the host’s physiology, the environment, the nutritional factors, including the transition from liquid to solid diets, and the use of nutraceuticals and antimicrobials agents1,2. Several studies have been conducted describing the cattle microbiome from different body sites during the transition period: gastrointestinal tract3,4, vagina5,6,7, and colostrum/milk8,9,10,11. However, vertical transfer of microbiota from dams to the offspring is poorly investigated and reported in cattle and ruminants in general12,13,14.
In humans, a so-called endogenous entero-mammary pathway has been postulated, where bacteria or bacterial components from the maternal intestine, are transported to breast milk through the bloodstream, and/or lymph by phagocytic cells. Whether this is an active part of the gut colonization process or not, there is ample evidence that human breast milk is not sterile, being an important part of the active colonization period happening after birth. Therefore, milk acts as a natural probiotic and as a source of natural inoculation in the early stages of life, with a significant role in modulating the innate immune system in newborns7,15,16,17. This hypothesis has been taken up as a paradigm of bovine neonatology.
Studies have reported three maternal sources as a possible origin of the bacterial gut colonization in newborn calves: vagina, udder skin, and colostrum12,14. The vagina microbiome appears to be the first source of vertically-transferred microbiota from dams to the offspring during parturition labor. This is supported by a large diversity similarity between the calf gut microbiome and offspring (such as methanogenic archaea), suggesting that some of the calf fecal microbiota may be derived from the birth canal during birth. Many microbes found in colostrum are also seen in the calf gastro-intestinal tract, suggesting that colostrum significantly contributes to the make-up of the calf GIT14. A very large number of OTUs (Operational Taxonomic Unit) was reported to be shared between udder skin microbiome and both luminal and mucosal-associated gut microbiota in calves from day 1 to 2 postnatal. However, this was also observed even with the immediate separation of dam and offspring before colostrum intake and/or sucking, indicating that microbes shared between the skin of the udder and the calf GIT were not maternally transferred but instead seeded directly, or indirectly, perhaps continuously, from the environment14.
The intestinal microbiota of newborns plays an important role in the development of immunity and metabolism, and metagenomic focused research aims to advance neonatal health care. Nevertheless, it is not well understood precisely how the neonatal gut bacterial microbiome is initially populated. In this study, we followed the development of the gut microbiome in calves aged 1 to 28 days, and compared the microbial composition found to that present in the dam’s colostrum, vaginal and gut microbiomes. To better understand how the maternal sources influence the acquisition of the calves GIT microbiota, we studied the relationship among the microbiota from dams to the GIT of 20 Holstein young heifers in the first 28 days of life.
Results
Number of bacterial sequences
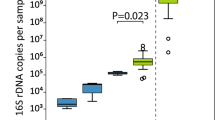
The number of bacterial sequences obtained from the V3–V4 region of 16-S sequencing profiles is shown in the Supplementary Table 1. Meconium, colostrum and vaginal secretion all showed a low bacterial DNA load, and only 1 meconium, 5 colostrum and 8 vaginal secretion samples had more than 1000 sequences. So due to this fact, meconium was removed from bioinformatic analysis. On the other hand, maternal and newborn fecal samples had bacterial loads that were much higher.
α-diversity index (species number) and richness (abundance of species)
Alpha diversity is a measure of microbiome diversity applicable to a single sample. The Chao 1 index and Shannon indices reflect the richness and diversity of species, respectively. Maternal feces and vaginal secretion had higher richness compared to colostrum and newborn fecal samples. Similar profile was observed for the Shannon index. It was observed an increase of Chao and Shannon indexes in fecal microbiome from calves over time (Fig. 1).
β-diversity index
UniFrac is a β-diversity measure that uses phylogenetic information to compare samples from different niches. Unweighted-UniFrac, considers the presence and absence of bacterial groups within clusters, while weighted-Unifrac takes into account the relative abundance of species shared between samples.
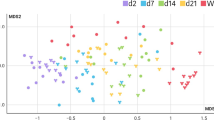
PCOA graphic represents the similarity between maternal sources and fecal microbiota in 20 Holstein calves, measured by using Unweighted UniFrac (Fig. 2a) and Weighted UniFrac (Fig. 2b).
Unweighted UniFrac reveals a degree of similarity between microbial communities in maternal feces and vaginal secretion, while colostrum samples showed less distance with calves’ fecal microbiota on days 3 and 7. Unweight and Weighted UniFrac clearly demonstrates the development of the calf’s gut microbiota, observing a decrease in distancing between feces from dams and calves on day 28.
The distance between maternal and offspring fecal microbiota are shown in Fig. 3. Calves at the end of the neonatal period have a different fecal microbiota than dams, despite the reduction in the distance over time.
Maternal microbiota as a source of bacteria transferred to the gastrointestinal system of newborn calves
Bacteria truly recovered from maternal samples and those that contaminate the collections each have an impact on the GIT microbiome in any calves raising system. These two factors were assessed using the Source Tracker model. The source bacteria for their contribution to the calves GIT colonization included all maternal and offsprings samples independently of the sequences number detected by the 16sRNA gene sequence, in addition to the control samples like compost bedding, milk replacer and drinking water from calf buckets available in calf housing.
The Source Tracker analysis indicated that the meconium microbiome from 20 newborn calves was correlated with maternal sources (vaginal secretion and colostrum), in addition to the compost bedding. The colostrum signal disappears from three days of life, however the vaginal microbiota had influence in the newborn fecal microbiota at least seven days age. On the other hand, an “unknown source of microbes” (probably from the environment) gradually increased in relation with the fecal samples from calves starting at day 3 through day 28 (Fig. 4).
Descriptive analysis of microbiota in calf samples
The description of the microbiota from the maternal niches at calving has been published in a previous paper by our team18. The quantitative estimates of bacterial phyla and families in meconium and calf fecal samples are shown in Fig. 5, respectively. Overall, the clusters generated at phylum and family level show similarity between the dam samples from day zero versus newborn fecal samples. Proteobacteria phylum was predominant in colostrum, while dams fecal samples and vaginal secretion was abundant in the Firmicutes phylum. Newborn fecal samples were abundant in Proteobacteria on day 3, followed by the Firmicutes phylum on day 7–21. Phylum Verrucomicrobia was the predominant phylum on days 7–14, while Bacteroidetes had higher abundance on day 28. Fusobacteria phylum presented a high proportion on the peak of diarrhea on day 14 (Supplementary Fig. 1).
Regarding newborn fecal samples, Proteobacteria_Enterobacteriacea and Firmicutes_Clostridiaceae were the predominant groups of bacterias on day 3. On day 7, Actinobacteria_Bifidobacteriaceae and Firmicutes Lactobacillaceae were the abundant bacterias groups. On day 14, Fusobacteria_Fusobacteriaceae was the highest abundant phylum. Finally, Bacterioidetes_Bacteriaceae, Firmicutes_Lachnospiraceae, and Firmicutes_Ruminococcaceae were the predominant groups on day 28.
Figure 5 presents a heat map depicting the abundance of bacterial groups. The major taxon (family level) displayed in the heat map were selected based on their presence in a minimum number of samples: at least one colostrum sample, two vaginal secretions, and ten fecal samples. Additionally, the taxa included must account for at least 0.1% of the global relative abundance. This selection criteria were employed to improve the clarity and quality of the figure.
The plot presented in the figure 6 shows the fit and confidence intervals for a LOESS regression applied to the selected bacterial groups in calves’ fecal samples across the newborn period (Supplementary tables 3 and 4). The most abundant bacteria group on calf fecal samples on day 3 were the genus Escherichia-Shigella and Clostridium, respectively. On day 7, the predominant genus were Bifidobacterium and Lactobacillus, while Fusobacterium was the most abundant genus on day 14. Faecalibacterium showed a gradual increase during the neonate period. Meconium samples were excluded because only one sample had more than 1000 sequences. This figure offers a secondary picture of the development of the bacterial community in calves.
The plot shows the fit and confidence intervals for a LOESS regression applied to the selected bacterial groups in calves’ fecal samples across the newborn period. The pseudo R-squared value, calculated based on the sum of squares, indicates the percentage of variance captured by the model. An empirical p-value was determined using a permutation procedure and then corrected for multiple comparisons using the False Discovery Rate (FDR) method.
Clinical evaluation
The distribution of fecal scores and rectal temperature evaluated among the 20 Holstein calves in different time points in the neonatal period are shown in the Supplementary Fig. 2. Overall, the number of calves presenting diarrhea (Fecal score 2 and 3) were very low before five days of life, in addition to the absence of neonates with fever (Rectal Temperature ≥ 39.5 °C). Cases of diarrhea had a gradual increase from day 6 up to days 12th–24th of life. Cases of fever over age were very punctual on day 13 and 14th of life.
Discussion
This study evaluated the level of vertical transfer of maternal microbes to the newborn calves and the development of GIT from birth through 28 days. The sources of seed bacteria for GIT colonization and their influence on the development of a functional microbiota have been an ongoing debate in human and veterinary neonatology and pediatrics. The understanding of the microbiome profile in newborn calves is important because of its role in the immune system maturation, a fully functional metabolism, competition to prevent disease, and its long-term effects on development of future dairy cows.
It is very important to clearly define and validate the initial state of the calf population selected for the conduction of our study. As the microbiota are modulated by factors such as: genetics, gestational issues, maternal environmental and health issues, the nutritional plan employed (timing of liquid and solid diet), the source and quality of drinking water, the age of calves studied, weaning management employed, the uncontrolled impact of antimicrobials and feed supplements, the local environment, and the health status of the calf during measurement1,2,19. The target population of our study was the newborn Holstein heifer. We collected a cluster of 20 cow-calf pairs from the same commercial farm under standardized management and a fixed long-term nutritional plan. A core feature of the experimental design was to eliminate the indirect use of antimicrobials or feed supplements in the calves to prevent effects of these products on the calves GIT microbiome development. Because of this, all calves received a liquid diet, without transition milk in the first 14 days of life, because the farm included milk from sick (and treated) postpartum cows in the waste milk stream. The intramammary antimicrobial used in the dry-cow therapy was anhydrous cephalonium. This has a withdrawal interval of 96 hours following calving. The dry period for these dams ranged from 21 up to 150 days. This opened a possibility for the indirect use of antimicrobials in the maternal colostrum given to some of the calves. The impact of the dry-cow treatment on the calf microbiome should be investigated in the future.
Overall, the alpha diversity and richness indexes were higher in the dam sources (feces and vaginal secretion) compared to the calves’ fecal samples from d3 to d28. Klein-Jöbstl et al.12 also reported that cow fecal samples had significantly higher species richness, diversity and number of observed OTUs compared with fecal samples from newborn calves in the perinatal period.
Βeta diversity analysis showed two different profiles by using Unweigh and Weight UniFrac. Unweight Unifrac (presence or absence of taxa) reveals dissimilarity between dams’ sources (feces and vagina) and calves’ fecal samples. Colostrum was the maternal source with less distance of calves’ fecal samples in the first week of life. Fecal microbiota from calves at 28 days of age appeared to be more differentiated than earlier samples, and this profile demonstrates a variance of the abundance of bacteria in the communities with increasing calf age. The dissimilarity between maternal and newborn calves’ fecal samples in the first three days of life was previously reported by Klein-Jöbstl et al.12. However, they did not follow calves until the end of the neonatal period.
Colostrum presented low load of bacterial DNA, and only five samples had more than 1,000 reads in our study. These samples had low richness and diversity, compared with other maternal sources. In addition, colostrum do not seem to be an important source of bacterias to the newborn based on Source Tracker analysis, which is not favorable for the entero-mammary hypothesis. Entero-mammary hypothesis suggest a possible vertical transfer of bacterias between dams and offsprings by exposure to free bacteria from her during initial grooming, or as carried in colostrum phagocytes, macrophages, neutrophils or dendritic cells17. Our results agree with the findings of Klein-Jöbstl et al.12. They found general dissimilarity between maternal colostrum and fecal samples from 15 Holstein-Friesian newborn calves during the immediate perinatal period. In both studies dams and offspring were separated immediately after calving. Independently of the dissimilarity between maternal colostrum and fecal samples from calves on day 3 of life, there are the description of possible regulation of the gut microbial colonization and immunity development by the bioactive factors transfer by maternal colostrum.
Source tracker procedure was other bioinformatic tools to estimate the contribution from the dam over time to the infant. The main maternal sources which contribute to the GIT colonization was the vaginal secretion. We also observed a little contribution from the bedding of compost barn on day 1 and milk replacer from day 1 to day 14. Contributions of the “unknown” sources of bacteria increased over time. These “Unknown” sources are defined as the component of the newborn microbiome that could not be associated with the investigated dam sources. This aids us to better determine if completely “new strains” were acquired at some point between the 1st and 28th days of life.
The most abundant bacteria group on meconium (day 1) and calf fecal samples on day 3 were Proteobacteria philo Enterobacter and Proteobacteria philo Escherichia-Shigella, respectively. Similar results were reported by Alipour et al.20, who reported a colonization with Escherichia/Shigella, and Clostridia of the GIT of newborn calves at the first days of life. Klein-Jönstl et al.12 also reported that the predominance of OTUs belonging to the Enterobacteriaceae family in calves during the first two days of life.
The quantification and analysis of E.coli virulence factors isolated from fecal samples from the same calves of this study was previously published by Gomes et al.21. The number of E.coli copies was minimal in meconium samples, peaking between day 3 and 7, followed by a decrease on day 14-28 of age. It was not detected enterotoxigenic E.coli (K99). Non-pathogenic E.coli can be a secondary infectious agent. It is documented that non-pathogenic strains can proliferate under a gut community dysbiosis, and cause systemic complication and death by Colisepsis.
The compost dairy barn is a widely used housing system that is adopted in tropical countries to improve the well-being of the dams due to the reduction in heat-stress. The bedded pack is a mixture of organic bedding and cattle excreta. Manure is a reservoir of a broad range of microbial organisms. These include: pathogens, which have potential to cause disease, and may pose both a public health risk and to impact future animal health. Bacteria of genus Clostridium were found in the liquid and solid phase of cattle manure22. The exposure to Clostridium spp. like Clostridium perfringens type A and type C, represent potential serious pathogens of newborn calves. They may cause newborn death through GIT inflammatory disease. DCHA23 recommends the birth of calves in clean, dry-bedding in a well-drained and well-ventilated calving environment. They also indicate that the immediate removal of calves from the maternity pen will prevent injury and illness. The compost dairy barn does not allow adequate cleaning and disinfection procedures to be achieved after each calving, or even once a week.
After seven days of age, the population of probiotic bacteria in the GIT increased over time. We observed a high abundance of Firmicutes philo_Bifidobacterium and Lactobacillus on day 7, followed by colonization with Bacteroidetes philo_Bacteroides and Firmicutes_Faecalibacterium by day 28. Fusobacteria philo_Fusobacterium was abundant on day 14. It was associated with the presence of neonatal diarrhea pathogens. Alipour et al.20 also reported an increased abundance of probiotic bacteria on day 7 compared to the first few days of life. Unfortunately, these authors do not follow calves through the whole neonatal period.
Summarizing, the similarity between maternal and calf fecal sample microbial diversity and number, with a similarity that increased with age to 28 days is probably due to the development of neonatal gastric microbial reservoirs. Unknown sources of bacteria have a clear role in the GIT microbiome from the first day to the 28th days of the calves’ life.
Methods
The procedures used for this project were approved by the Committee on Ethics in the Use of Animals from the College of Veterinary Medicine and Animal Science at University of São Paulo (CEUA number 3481060817). In this experiment, all methods were performed in accordance with relevant guidelines and regulations. Additionally, the authors complied with the ARRIVE guidelines.
Farm and dams’ management
This study was conducted in a commercial dairy farm located in São Paulo, Brazil, whose geographic coordinates are Latitude: 21° 54′ 14″ South, Longitude: 47° 37′ 12″ West, during the winter up to spring season of 2017. The cows and calves included in the study originated from the same dairy farm.
Dairy cows were screened on the farm. Those between 2nd and 5th calving were evaluated and grouped according to the expected calving date. Dry and pre-partum management followed the documented procedures of the farm. This started with dry-cow treatment with anhydrous cephalonium to prevent mastitis and was used with a commercial teat seal. Dry period ranged from 21 to 150 days. Dams were transferred from the free-stall barn to the pasture where they remained until shortly before delivery. At day -30 prior to the due date, cows were transferred to the compost barn building with a tunnel ventilation climate control system. Calving was monitored 24 hours a day by members of the research team to detect the early stages of labor. Labor was indicated by the observation of the following signals: tail twitching, restlessness, stand with tail raised and back arched, segregation, vulva and mammary gland edema, abdominal contraction and or the appearance of fetal membranes. At this time, cows were moved to the maternity pens in a separated area inside the compost barn building. In this area, they were placed on fresh wood shaving as a bedding that was changed after each calving. After birth, calves from dystocia, male, twins and/or neonates presenting congenital diseases or neonatal asphyxia were excluded from the trial. Newborn calves presented with a vitality score of ≥ 7. In addition, the included cows produced at least three litters of colostrum with minimum concentration of 50 g/L of IgG. This was estimated using a colostrometer, Brix refractometer and/or colostrum balls. All calves were evaluated for failure on the passive immune transfer (FPIT) by IgG ELISA. Any calves from this project presented FPIT. Based on this series of exclusion criteria, we obtained 20 Holstein dams and their offspring for the research.
Samples harvesting on-farm
Immediately after calving, dams were milked in the maternity pen to collect colostrum. The neonates were wiped dry using paper towels, then immediately moved into the “Cuddle Box”. This is a covered bed of good quality hay and is outside the calving pen. The calves were each presented to their own dam. The swing gate was moved against the dam to lock her into the meeting space. She was secured with a chain, to allow us to work safely under the udder. Each cow was encouraged to lick her calf.
The mammary gland of each cow was cleaned by using a soft brush to remove the dry dirt that adhered to the udder. After this the teat seal was removed. The first jets of milk were collected to evaluate the macroscopic characteristic of the colostrum. Teats were disinfected by rubbing with a sterile gauge containing an iodine solution (Riodeine®, Rioquimica), then disinfected again using alcohol 70%. Colostrum samples, collected by quarter, were harvested by manually milking in sterile 50 mL tubes (Falcon®, BD Biosciences, San Jose, CA, EUA). A blank sample (n = 2) consisting of 50 mL of ultrapure water free of DNA was collected and processed the same way of colostrum.
The perineum was initially cleaned with dry paper towels. Then the vulvar and the rectal area were disinfected separately using a sterile gauze soaked in iodine solution. Vaginal secretion was obtained, close to the cervix, using a long swab covered with a plastic sheath (Provar®). The fecal samples were harvested by using sterile dry swabs under as aseptic a method as possible. Both samples were stored frozen in DNA free microtubes. Each newborn was assessed as it entered the cuddle box. At this time, the first rectal swabs (meconium) were collected before colostrum intake. This was done as described for their dams. Unused swab (n = 2) was collected and processed the same way of vaginal and fecal samples from dams and calves.
All samples were immediately placed on dry ice (− 80 °C), after sampling except for the colostrum which was refrigerated (4 °C).
Calf management on-farm
After harvesting the preliminary samples, the dams were milked. Colostrum quality was assessed using colostrum balls (density ≥ 1.045), a refractometer (Brix Index ≥ 21%) and a commercial colostrometer (IgG ≥ 50 g/L). The neonates were each fed with fresh colostrum from their own dam, using a volume that ranged from 3.0 to 6.5 liters, depending on the quantity produced by each dam. The first feeding of each calf was offered ad libitum immediately after colostrum collection. It was done within the first hour of life for each calf. To achieve the goal of a volume of colostrum fed that was equivalent to 10% of the calf’s body weight at birth, we needed to offer a second colostrum feeding between 6 and 18 hours after birth. Colostrum was maintained under refrigeration (4 °C) until the second colostrum feeding. Colostrum for the second feeding was heated in a water bath before the second feeding. The longitudinal and descriptive presentation of the dynamic of passive immune transfer biomarkers in the calves used in this research was previously published by Gomes et al.21.
From 2nd to the 14th days of age, calves were housed in suspended individual closed pens on hay bedding. These were distributed in either covered or opened barn structures isolated from other animals. Calves were fed milk substitute (Nattimilk E Max®, Auster Animal Nutrition) from the 2nd day of life. This milk replacer contained 12.5% total solids. It was offered in two feeds of 3 liters each in bottles on the 2nd and 3rd day of life. Subsequently, the neonates were fed using individual buckets. Calves were transferred to the phase 2 calf raising system, where they were maintained from 15th to the 28th days of life. Here, they were fed with transition milk supplemented with milk substitute at 14% total solids. Starter solid feed, without probiotics or antimicrobials, was introduced in the 1st week of life ad libitum. Transition milk was not offered during the first 14 days of life to prevent the transfer of antimicrobial residues due to the post-partum treatment of cows. Since the milking system used for postpartum cows did not allow segregation between healthy and sick cows on this farm. Calves were monitored daily for disease. None of the calves in this study received oral or systemic antimicrobials. Water was offered ad libitum. Fecal samples from calves for microbiome analysis was harvested on days 3, 7, 14 and 28 of life by using sterile dry swabs, storage at − 80 °C.
Blanks and controls
A series of control samples were taken during the course of the experiment to monitor for noise and outside contamination that may confound the samples based on interaction with the environment, water or milk replacer (Supplementary Table 5). Compost bedding from maternity pen was harvested immediately after calving in the early and the end of the experiment. In addition, drinking water and diluted milk replacer used to fed calves from second day of life was sampled to evaluate a possible source of contamination over time.
Laboratory procedures
Fecal, vaginal and meconium swabs were stored in a − 80 °C freezer. Colostrum samples, harvested by quarter, were pooled by dams in the lab. To achieve this, five mL of colostrum from each quarter were mixed together in a new sterile tube in the laboratory to yield a volume of 20 mL. This pool was diluted 1:1 with 0.9% sterile saline. The mixture was centrifuged at 1500×g for 20 minutes (at 20 °C). The supernatant was removed and the pellet was suspended in the original volume of 0.9% sterile saline. This was repeated three times. The pellet containing the cellular fraction was prepared after the washes by suspending in 5 mL of sterile phosphate buffered saline (PBS). This was distributed into five aliquots of 1mL each in sterile DNAse-free microtubes and stored at − 80 °C.
Total DNA was extracted from each sample using a Power Soil 96-well DNA Isolation Kit (MoBio Laboratories Inc., Carlsbad, CA, USA) and strictly adhering to the manufacturer’s instructions. The V3–V4 region of the 16S rRNA gene was amplified using traditional PCR. The sequencing library preparation was carried out in a two-step PCR protocol, following Illumina’s recommendation. In the first PCR reaction we used the V3–V4 primers 341F–806R24 as this pair has great taxonomy coverage in bacteria and archaea25.
Bioinformatic and statistical analyses
The analysis was carried out using standard methods available within qiime 1.9.1. We used clustering at 97% sequence similarity to remove background noise and serve as taxon creation using uclust, a method still currently in use. We chose not to migrate to qiime2 for consistency with other analyses being carried out in the lab at the time. The reads were quality-filtered using Cutadapt, the forward sequences were cut at position 223 and the reverse sequences at position 98. These positions were chosen because the first quartile of the sequence quality score was greater than or equal to 20. Chimeric sequences were removed using UCHIME69 and operational taxonomic units (OTUs) using UCLUST70 with 97% similarity for bacterial sequences were considered for further analysis if they had a minimum of 5 sequences detected across all samples. Taxonomy was assigned to OTU representative sequences using the SILVA 16S database (version n132)26.
All subsequent analyses were performed with R version 4.0 and RStudio version 3.6.1 statistical programs by applying the phyloseq, vegan, gplots, ggplot2 and qiimer R library routines27,28,29. Samples were ratified to the lowest number (min. 1,000) of sequences based on the alpha diversity analysis and the number of observed OTUs. The Shannon diversity index and Chao1 richness estimator were also calculated among samples. Beta diversity was calculated using weighted and unweighted UniFrac assessment30. A heatmap was built using Ward’s hierarchical clustering method (ward.d2) and the OTUs were filtered with a minimum threshold of 1000. The Source Tracker modeling based on 16S rDNA sequencing can be used for the analysis of microbial maternal and contamination sources for meconium and fecal samples from calves in the neonatal period, and can calculate the similarities between different bacterial communities. The Source Tracker model was based on the Bayesian algorithm to predict the composition ratio of the target samples from each source sample, according to the microbial community structure distribution of the target samples and the source samples.
Calf clinical examination protocol
Fecal and Bovine Respiratory Disease (BRD) scores were assessed in accordance with the Calf Health Scoring Criteria previously published by The University of Wisconsin (Madison) by McGuirk31. The stool scores were classified on a 0–3 basis using fecal consistency. A score of 0 represented “normal” consistency, a score of 1 represented a pasty or semi-formed stool consistency, a score of 2 represented watery stool consistency, with considerable water and fecal content that adhered to the perineum and tail of the calf, and a score of 3 represented fully liquid feces that adhered to the perineum and tail. Calves with scores of 0 or 1 were considered to be free of diarrhea, but calves with scores of 2 or 3 were suffering from diarrhea. BRD was scored using a combination of the following parameters: rectal temperature, cough, nasal and ocular secretion and ear position with a score of 0–3 for each based on severity of each. Calves were assessed as having BRD when the sum of these scores was ≥ 5.0. Umbilical region was evaluated by inspection and palpation to detect inflammation.
Data availability
Raw sequencing data and metadata are available through NCBI under BioProject accession PRJNA1120923. The analyzed data and supplementary information files are available in the Zenodo repository, https://doi.org/https://doi.org/10.5281/zenodo.11114476.
References
Amin, N. & Seifert, J. Dynamic progression of the calf’s microbiome and its influence on host health. Comput. Struct. Biotechnol. J. 19, 989–1001. https://doi.org/10.1016/j.csbj.2021.01.035 (2021).
Arshad, M. A. et al. Gut microbiome colonization and development in neonatal ruminants: Strategies, prospects, and opportunities. Anim. Nutr. 7(3), 883–895. https://doi.org/10.1016/j.aninu.2021.03.004 (2021).
Muñoz-Vargas, L. et al. Fecal microbiome of periparturient dairy cattle and associations with the onset of Salmonella shedding. PLoS One 13(5), e0196171. https://doi.org/10.1371/journal.pone.0196171 (2018).
Oikonomou, G. et al. Microbiota of cow’s milk; distinguishing healthy, sub-clinically and clinically diseased quarters. PLoS One. 9(1), e85904. https://doi.org/10.1371/journal.pone.0085904 (2014).
Bicalho, M. L. S. et al. Dynamics of the microbiota found in the vaginas of dairy cows during the transition period: Associations with uterine diseases and reproductive outcome. J. Dairy Sci. 100(4), 3043–3058. https://doi.org/10.3168/jds.2016-11623 (2017).
Laguardia-Nascimento, M. et al. Vaginal microbiome characterization of Nellore cattle using metagenomic analysis. PLoS One. 10(11), e0143294. https://doi.org/10.1371/journal.pone.0143294 (2015).
Rodrigues, N. F. et al. Qualitative analysis of the vaginal microbiota of healthy cattle and cattle with genital-tract disease. Genet. Mol. Res. 14(2), 6518–6528. https://doi.org/10.4238/2015.June.12.4 (2015).
Bhatt, V. D. et al. Milk microbiome signatures of subclinical mastitis-affected cattle analysed by shotgun sequencing. J. Appl. Microbiol. 112(4), 639–650. https://doi.org/10.1111/j.1365-2672.2012.05244.x (2012).
Bonsaglia, E. C. R. et al. Milk microbiome and bacterial load following dry cow therapy without antibiotics in dairy cows with healthy mammary gland. Sci. Rep. 7, 8067. https://doi.org/10.1038/s41598-017-08790-5 (2017).
Ganda, E. K. et al. Normal milk microbiome is reestablished following experimental infection with Escherichia coli independent of intramammary antibiotic treatment with a third-generation cephalosporin in bovines. Microbiome. https://doi.org/10.1186/s40168-017-0291-5 (2017).
Oikonomou, G. et al. Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. Associations of Faecalibacterium species with health and growth. PLoS One. 8(4), e63157. https://doi.org/10.1371/journal.pone.0063157 (2013).
Klein-Jöbstl, D. et al. Microbiota of newborn calves and their mothers reveals possible transfer routes for newborn calves’ gastrointestinal microbiota. PLoS One 14(8), e0220554. https://doi.org/10.1371/journal.pone.0220554 (2019).
Li, M. et al. Comparison of changes in fecal microbiota of calves with and without dam. PeerJ. 10, e12826. https://doi.org/10.7717/peerj.12826 (2022).
Yeoman, C. J. et al. Biogeographical differences in the influence of maternal microbial sources on the early successional development of the bovine neonatal gastrointestinal tract. Sci. Rep. 8, 3197. https://doi.org/10.1038/s41598-018-21440-8 (2018).
Jost, T., Lacroix, C., Braegger, C. P., Rochat, F. & Chassard, C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 16(9), 2891–2904. https://doi.org/10.1111/1462-2920.12238 (2014).
Martín, V. et al. Sharing of bacterial strains between breast milk and infant feces. J. Hum. Lact. 28(1), 36–44. https://doi.org/10.1177/0890334411424729 (2012).
Perez, P. F. et al. Bacterial imprinting of the neonatal immune system: Lessons from maternal cells?. Pediatrics 119(3), e724–e732. https://doi.org/10.1542/peds.2006-1649 (2007).
Tardón, D. C. et al. Relationships among indicators of metabolism, mammary health and the microbiomes of periparturient Holstein cows. Animals 12(1), 3. https://doi.org/10.3390/ani12010003 (2022).
Malmuthuge, N. & Guan, L. L. Understanding the gut microbiome of dairy calves: Opportunities to improve early-life gut health. J. Dairy Sci. 100(7), 5996–6005. https://doi.org/10.3168/jds.2016-12239 (2017).
Alipour, M. J. et al. The composition of the perinatal intestinal microbiota in cattle. Sci. Rep. 8, 10437. https://doi.org/10.1038/s41598-018-28733-y (2018).
Gomes, V. et al. The role of anti-E. coli antibody from maternal colostrum on the colonization of newborn dairy calves gut with Escherichia coli and the development of clinical diarrhea. Anim. Open Space. https://doi.org/10.1016/j.anopes.2023.100037 (2023).
Pandey, P. et al. 16S rRNA analysis of diversity of manure microbial community in dairy farm environment. PLoS One 13(1), e0190126. https://doi.org/10.1371/journal.pone.0190126 (2018).
Dairy Calf and Heifer Association (DCHA). Dairy Calf and Heifer Association gold standards (2n Edition). 1–24p. (2016).
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1), 4516–4522. https://doi.org/10.1073/pnas.1000080107 (2011).
Takahashi, S., Tomita, J., Nishioka, K., Hisada, T. & Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of bacteria and Archaea using next-generation sequencing. PLoS One. 9(8), e105592. https://doi.org/10.1371/journal.pone.0105592 (2014).
Quast, C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acids Res. 41(D1), D590–D596. https://doi.org/10.1093/nar/gks1219 (2013).
McMurdie, P. J. & Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8(4), e61217. https://doi.org/10.1371/journal.pone.0061217 (2013).
Oksanen, J. F. et al. Vegan: Community Ecology Package. R Package Version 2.5–5. Available online: https://CRAN.Rproject.org/package=vegan (2019).
Wickham, H. ggplot2: Elegant graphics for data analysis (Springer-Verlag, 2016).
Anderson, M. J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. https://doi.org/10.1111/j.1442-9993.2001.01070.pp.x (2001).
McGuirk, S. M. Disease management of dairy calves and heifers. Vet. Clin. North Am. Food Anim. Pract. 24(1), 139–153. https://doi.org/10.1016/j.cvfa.2007.10.003 (2008).
Acknowledgements
The authors would like to acknowledge Roberto Jank, as well as the employers, for encouraging and allowing the conduction of our research in the Agrindus farm; the Fundação de Amparo à Pesquisa do estado de de São Paulo (FAPESP) for the financial support (Process number 2016/16748-2); and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (Cnpq) for granting a research scholarship–PQ to Prof. Viviani Gomes (Process number 249516/2013-6).
Author information
Authors and Affiliations
Contributions
V.G. and D.J.H. proposed the study and designed the experiments. V.G., F.C.R.S. and D.I.C.T collected and processed study samples. V.G. managed the project and C.H. analyzed the data. V.G. wrote the paper. C.H and D.J.H reviewed the draft paper. S.S.R. reviewed and prepared the article for submission and publication. All authors provided critical review and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gomes, V., Hoffmann, C., Castro-Tardón, D.I. et al. Vertical transfer of gut microbiota from dam to neonate calf in the early of life. Sci Rep 14, 21746 (2024). https://doi.org/10.1038/s41598-024-72296-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-72296-0
- Springer Nature Limited