Abstract
Colorectal cancer (CRC) is a wide-spread gastrointestinal cancer that is associated with augmented morbidity and mortality, and we do not yet have a deep understanding of its epidemiology and carcinogenicity. The transcriptome can reveal the complexity and heterogeneity of tumors and uncover new biomarkers or treatment options. In this study, we identified messenger RNAs (mRNAs), long non-coding RNAs (lncRNAs), round RNAs (circRNAs), and microRNAs (miRNAs) using whole-transcriptome sequencing and generated competing endogenous RNA (ceRNA) modulatory axes. We conducted whole transcriptome sequencing on 10 CRC and para-cancer (CRCP) samples and discovered 2465 differentially expressed (DE) mRNAs (DEmRNAs), 77 DE miRNAs (DEmiRNAs). 2852 DE lncRNAs (DElncRNAs) and 1477 DE circRNAs (DEcircRNAs). In addition, utilizing co-DE analysis, we generated the ceRNA axis. Subsequently, we employed the ceRNA axis to identify essential genes and corresponding associations with lncRNAs, circRNAs, and miRNAs in CRC. ceRNA regulatory network including mRNA-miRNA-lncRNA and mRNA-miRNA-circRNA. These modulatory axes potentially modulate the positive regulation of smooth muscle contraction, melanosome, plasma membrane, integral plasma membrane component and so on. Finally, the results of RNA sequencing (RNA-SEQ) were combined with the TCGA and GEO databases, and the DEGs strongly correlated with the TCGA-COAD overall survival (OS) as estimated by univariate cox and logarithmic rank analyses were cross-analyzed, and the co-upregulated DEGs were screened. Among the many DEs, KPNA2 was chosen for additional analysis. Using invitro experimentations, western blot, CCK8, EdU and other experiments were performed to verify the results. We found siRNA-based KPNA2 depletion reduces bladder cancer cells’ viability, migratory, and proliferative activities, which showed that the DEmRNA profiles were comparable to the sequencing information, confirming that the sequencing data were very reliable. These evidences highlight the ceRNA regulatory mechanisms in CRC and will aid future research into the molecular mechanisms behind colorectal cancer prevention and treatment.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) ranks third among globally prevalent cancers and is the second contributor to cancer-associated mortality1. The Global Cancer Statistics 2020 estimates over 1.9 million new CRC cases and 935,000 global deaths in 2020, thereby reaching 10% of all cancer incidences and related mortalities2. In addition, these figures continue to rise particularly in developing countries. Despite advancements in CRC screening, diagnosis, and intervention, the 5-year CRC patient survival rate remains quite unsatisfactory3. This is likely due to missed early diagnosis, timely and efficacious intervention, and the unexplored contribution of underlying signaling pathways4. More recently, the establishment of high-throughput sequencing (HTS) technology has revealed marked aberrations in non-coding RNA (ncRNA) expressions, such as, lncRNAs, transcripts with sequences > 200 nucleotides without open read frames, and circular RNAs with covalent closed-loop structures formed by reverse shear (circRNAs) in multiple stages of cancer progression5,6. ncRNAs are critical for miRNA-regulated gene expression. miRNAs partly interacts with target gene mRNA complementary sequences known as response elements (MRE)7. Endogenous RNAs carrying the same MRE competes with association to miRNAs, these RNAs are otherwise known as competitive endogenous RNA (ceRNA)8. lncRNA, circRNA, and pseudogenes often competes with miRNA using the same MRE to minimize miRNA-mediated suppression, and increase miRNA target gene (mRNA) expression8,9,10. In order to further extend the survival time of CRC patients, it is extremely important and clinically urgent to identify new biomarkers for screening, early diagnosis, prognosis, and outcome assessment for CRC treatment, and to elucidate the associated signaling networks behind CRC tumor development and progression.
Full transcriptome analyses use HTS techniques to examine the interactions between mRNAs, miRNAs, lncRNAs, and circRNAs, and to clarify the role of ncRNAs) in post-transcriptional biological processes11,12. The ceRNA hypothesis describes the interaction between coding and ncRNAs. It states that mRNA and lncRNA/circRNA interacts via MRE, and lncRNA/circRNA sequesters miRNA, thereby inhibiting its silencing effect13. At present, the pathogenesis of CRC is still unclear. The ceRNA modulatory axis enhances the current knowledge of tumor-associated networks, which are critical for diagnosis, prognostic markers, novel therapies, and predicting treatment response.
Herein, we employed whole transcriptome sequencing to determine the lncRNAs, circRNAs, mRNAs, miRNAs profiles in CRC and adjoining healthy tissues. In all, 2465 differentially expressed (DE) mRNAs (DEmRNAs) and 77 DE miRNAs (DEmiRNAs) were identified from the samples of CRC and CRCP respectively. There were 2852 DE lncRNAs (DElncRNAs) and 1477 DE circRNAs (DEcircRNAs). In addition, we generated the ceRNA network based on co-DE analysis, and using this network, we screened essential genes and corresponding associations with lncRNA, circRNA, and miRNA in colorectal cancer. ceRNA regulatory network including mRNA-miRNA-lncRNA and mRNA-miRNA-circRNA. Finally, the results of RNA sequencing (RNA-Seq) were combined with the TCGA and GEO databases, and the DE genes (DEGs) strongly correlated with the TCGA-COAD overall survival (OS) estimated using univariate cox and log-rank methods were cross-analyzed, and the co-down-regulated DEGs were screened. Western blot, CCK8, EdU and other experiments were performed to verify the results, which showed that the DE RNA profile strongly corroborated with the sequencing information, confirming the reliability of the sequencing information. The results from this study highlight novel modulatory roles of circRNA in CRC epidemiology and carcinogenicity.
Materials and methods
Patient and tissue sample collection
The excised CRC and CRCP tissues, both identified by ≥ 2 pathologists, were acquired from 10 CRC patients at Shaoxing People's Hospital between September 2022 and September 2023. No participants underwent adjuvant therapy prior to surgical resection. All CRC and CRCP samples were removed from CRC patients, prior to a flash freeze in liquid nitrogen. CRC tissue (CRC1, CRC2, CRC3, CRC4, and CRC5) and corresponding adjacent normal tissue (CRCP1, CRCP2, CRCP3, CRCP4, and CRCP5) were sampled with three bio-logical replicas, each consisting of tissue from five different patients (Fig. 1A). Colonoscopy (Fig. 1B) and radiology (Fig. 1C) findings were used to validate CRC. Cancer tissue fragments underwent fixation in 4% para-formaldehyde and sliced at 5 microns for heme and hemoglobin (HE) staining. We next assessed proliferation index and tumor differentiation (Fig. 1D). The relevant clinical data of each patient are shown in the Supplementary Table S1. All experimental protocols received ethical approval from the Academic Ethics Committee of Shaoxing People's Hospital and obtained documented informed consent from all participants. We confirm that all experiments were performed in accordance with relevant guidelines and regulations.
RNA extraction and library preparation
Total RNA isolation employed TRIzol (Invitrogen, CA, USA) and associated directions. RNA quality and quantity were assessed via NanoDrop 2000 spectrophotometer (Thermo Scientific, USA), and integrity via Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The samples with qualified purity, quantity and integrity were used for subsequent library construction. Ribo-off rRNA Depletion Kit (Vazyme, Nanjing, China) was used to remove ribosomal RNA, then the libraries were generated with the VAHTS Universal V6 RNA-seq Library Prep Kit and associated directions. Lastly, transcriptome sequencing and analysis were done via OE Biotech Co., Ltd. (Shanghai, China).
RNA sequencing analysis
RNA sequencing utilized the IIumina Novaseq 6000 platform, and generated 150 bp paired-end reads. Raw reads, generated in the fastq format, were processed using fastp14 and reads of reduced quality were eliminated to clean the reads. Thereafter, approximately 142.55 G clean reads per sample mapped to the reference genome using HISAT215. Individual gene read counts were acquired via HTSeq-count16, following which, the corresponding FPKM17 was calculated. We conducted PCA analysis in R (v 3.2.0) to assess the biological duplication of samples. DE analysis employed the DESeq218, with DEG criteria defined as Q value < 0.05 and foldchange (FC) > 2 or FC < 0.5. DEGs hierarchical cluster analysis utilized R (v 3.2.0) and demonstrated the gene expression profiles of varying groups and samples. Using R packet ggradar, we depicted the leading 30 gene radar map revealing the up- and down-regulated DEGs. Next, using the hypergeometric distribution, and functional analyses, including DEG GO19, KEGG20,21,22,23 pathway, Reactome and WikiPathways, the significant enriched term was identified in R (v 3.2.0). Using the same program, we also generated the column, chord and bubble diagrams of relevant enrichment terms. Gene Set Enrichment Analysis (GSEA) utilized the GSEA software24,25, using a predefined gene set as well as genes ranked based on the degree of DE in both samples. Lastly, we examined whether the predefined gene set was enriched at the top or bottom of the ranking list.
lncRNA analysis
DE analysis utilized DESeq218 and DElncRNA criteria was described as P-value < 0.05 and FC > 2 or FC < 0.5. Using Pearson's coefficient, we calculated the expression correlation between DELs and DEGs, the significant related lncRNA-gene pairs were screened with the criteria of p < 0.05 and | cor |> 0.8. GO, KEGG Pathway, Reactome and WikiPathways analyses were conducted for genes significantly co-expressed by lncRNAs to estimate lncRNAs physiological roles. The lncRNA and miRNA interaction was predicted by miranda (v 3.3a) software. LncRNA-RBP (RNA binding protein) pairs were obtained based on starbase database (https://www.lncrnablog.com/starbase-v2-0-for-decoding-rna-interaction-networks/). The construction of lncRNA-TF (transcription factor) -mRNA regulatory relationship depends on three conditions: (1) TF-target gene pairs obtained from GTRD database; (2) motif sequences were extracted from JASPAR database, and the motif module in Biopython predicted the lncRNA sequences associations to motif; (3) There is at least one common TF between the target gene and lncRNA. On the basis of these three conditions, the lncRNA-TF-mRNA regulatory relationship was constructed, and ggsankey (v 0.0.99999) was used to draw the Sankey diagram. Motif sequence in JASPAR database and motif module in Biopython were used to predict the motif binding region in the promoter of lncRNA, and the transcription factors regulating the lncRNAs were analyzed.
circRNA analysis
Find_circ (v 1.2) and CIRI226 software were used to identify the circRNA. The circRNA's parent gene was annotated according to its genomic position, and Circbase (http://www.circbase.org/) and CIRCpedia (http://yang-laboratory.com/circpedia/) were used to identify known circRNAs. RPB (junction reads per billion mapped reads) was used to quantify circRNA, and DEGseq27 was used to calculate the differential expression of circRNA. The P-value was calculated by NB (negative binomial distribution test). Q value < 0.05 and FC > 2 or FC < 0.5 was described as the criteria for DEcircRNA. Utilizing hypergeometric distribution, GO19, KEGG20 pathway, Reactome and WikiPathways analyses of DEcircRNA's parent gene were performed to evaluate the function of circRNA's parent genes. Pearson's coefficient was employed for calculation of the expression correlation between DECs and DEGs, the significant related circRNA-gene pairs were screened with the criteria of p < 0.05 and |cor|> 0.8. GO, KEGG Pathway, Reactome and WikiPathways enrichment analysis were conducted for genes significantly co-expressed by circRNAs to predict the function of circRNAs. The binding of circRNA and miRNA was predicted by miranda (v 3.3a) software. CircRNA-RBP (RNA binding protein) pairs were obtained based on starbase database (https://www.lncrnablog.com/starbase-v2-0-for-decoding-rna-interaction-networks/). ORF prediction of circRNA was performed using ORFfinder (v 0.4.3), and IRES28 prediction was also performed. CircRNAs containing both ORF and IRES sequence was judged to have the potential to encode proteins.
Small RNA sequencing experimental method
Total RNA isolation employed the mirVana miRNA Isolation Kit (Ambion) and associated directions. RNA quantity was assessed using Nanodrop 2000 (Thermo Fisher Scientific Inc., USA), and integrity using Agilent 2100 Bioanalyzer (Agilent Technology, USA). We utilized 1 μg total RNA from individual samples for small RNA library generation via the NEBNext Small RNA Library Prep Set for Illumina kit (Cat. No. NEB#E7330S, NEB, USA) and associated directions. In short, total RNA was ligated to adapters at both ends. Next, the adapter-ligated RNA was converted to cDNA, prior to PCR amplification. The PCR products ranged between 140–160 bp and underwent extraction and purification as small RNA libraries. Library quality was evaluated using the Agilent Bioanalyzer 2100 system, and library sequencing was done via the Illumina Novaseq 6000 platform, and 150 bp paired-end reads were created. Both small RNA sequencing and analyses were completed by OE Biotech Co., Ltd. (Shanghai, China).
Small RNA sequencing analyses
Using base calling, basic reads were transformed into sequence information. Following filtration of reduced quality reads, reads with 5’ primer contaminants and poly (A) were eliminated. Subsequently, we filtered reads lacking a 3’adapter and insert tag, as well as reads < 15 nt or > 41 nt from raw data to obtain clean reads. Clean sequence length distribution in the reference genome was obtained, then sequences underwent alignment and Bowtie29 screening against Rfam v.10.1 (http://www.sanger.ac.uk/software/Rfam)30. Thereafter, rRNA, scRNA, Cis-reg, snRNA, tRNA and other RNAs were identified and filtered. Simultaneously, we also identified cDNA and Repbase31 database of species repeat sequences using Bowtie. Mature miRNAs were recognized via alignment against miRBase v22 database (http://www.mirbase.org/)32, expression profiles of various samples were determined. Subsequently, using miRDeep233, we analyzed unannotated reads to estimate new miRNAs. Utilizing the pre-miRNA hairpin structure and miRBase database, we determined the corresponding miRNA star and miRNA mature sequences. DE miRNAs were computed, followed by filtration according to Q value < 0.05 and FC > 2 or FC < 0.5. The Q value computation employed the DEG algorithm34 in R and assessed biological replicates, whereas, the Audic Claverie statistic35 was used to analyze samples without biological replicates. The DE miRNAs targets were estimated with miranda36 in animal, using parameters: S ≥ 150, ΔG ≤ − 30 kcal/mol and strict 5' seed pairing. Additionally, using Targetfinder37, we predicted miRNA target in plants. GO and KEGG axis enrichment analyses of DEmiRNA-target-gene were respectively conducted using R according to the hypergeometric profile.
ceRNA axis evaluation
The ceRNA hypothesis is a new tool for the disease molecular pathogenesis assessment6. To identify potential interactions between DERNAs (lncRNA, miRNA, and mRNA /circRNA, miRNA, and mRNA) in CRC, two ceRNA axes were generated via whole transcription-sequenced DE circRNA and mRNA and small RNAs sequenced DE miRNA. The correlation parameters between miRNA and target mRNA were correlation ≥ 0.7 and P-value ≤ 0.05, and ceRNA was generated according to P-value ≤ 0.05. The DE circRNA-based ceRNA was further assessed by GO and KEGG analyses.
Public data set acquisition and bioinformatics assessment
TCGA-COAD, GSE33113 and GSE3958238 data sets from genome data sharing data portal (https://portal.gdc.cancer.gov/) and gene expression synthesis (GEO, https://www.ncbi.nlm.nih.gov/geo/) retrieval39. Limma R package (3.40.2 version) and GEO2R online tools (https://www.ncbi.nlm.nih.gov/geo/geo2r/) to determine each dataset CRC and DEGs between the normal group. In addition, for tumor, normal and transfer network of group differences in gene expression analysis tools TNMplot40 (https://tnmplot.com/analysis/) is used to validate KPNA2 DE. ggstatsplot R package was used to analyze the correlation between KPNA2 expression profile in TCGA-COAD data set and clinical characteristics of patients, such as, patient age, sex, tumor (T) stage, lymph node (N) stage, and metastasis (M) stage. Clinical information was acquired from TCGA and Pan-Cancer Atlas on the UCSC Xena website41 (https://xenabrowser.net/). To assess the prognostic significance of alterations in OS and disease-specific survival (DSS) levels of KPNA2 content in the TCGA-COAD dataset. Plot the Kaplan-Meyer survival curve using the R-pack survival and Survivalist.
Cell culture, treatments, and siRNA incorporation
Human CRC cancer cell lines were purchased from The World Cell Factory (iCell Bioscience Inc, Shanghai). The logarithmic growth phase cells were classified as follows: control, siKPNA2-negative control (NC), and siKPNA2 groups. Cells were combined with 5 µl siKPNA2, and Lipofectamine 3000 was prepared in Opti-MEM medium, prior to a 5-min incubation. The aforementioned were combined, followed by a 20-min incubation, then the suspension was introduced to a cell culture plate, which was incubated for 48 h, prior to use in subsequent experiments.
Western-blot analysis
Protein isolation employed RIPA buffers and quantified by BCA kits. The protein solute was extracted via 10% SDS-PAGE, prior to transfer to PVDF membrane, which was then blocked and treated with primary and secondary antibodies. KPNA2 (Proteintech, 10819-1-AP) and β-actin (Proteintech, 81115-1-RR) were imaged with enhanced chemiluminescence reagents (Merck Millipore, Billerica, MA).
Confirmation of results from in vitro experiments
Cell survivability assessment utilized the cell counting kit-8 (CCK-8) assay (CCK-8, MCE, HY-K0301), and cell migration and invasion via Transwell assay.
Statistical analysis
Plots were generated using GraphPad Prism 5.0 (GraphPad Software, San Diego, California, USA). Data analyses utilized student-T test. Significance was determined as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Results
Global response of RNAseq
Full transcriptomic sequencing of 10 samples was completed on 5 pairs of fresh colorectal and para-cancer, with each replica consisting of tissue from five different patients (Fig. 1A). Co-lonoscopy (Fig. 1B) and radiology (Fig. 1C) findings were used to validate CRC, and HE staining validated proliferation index and tumor differentiation (Fig. 1D). Each sample was bioreplicated three times using the Illumina HiSeqTM 4000 platform. Herein, we obtained complete transcriptome sequencing of 10 distinct samples, and obtained 142.65 G CleanData (Table S1). The effective data volume per sample was between 13.28 and 15.69 G, the Q30 base distribution was from 93.92 to 94.62%, and the mean GC amount was 52.04%. Reads were aligned to a reference genome, with the ratio between 96.28 and 97.68%. There was a total of 1 differential gene group, and the number of differential genes detected was 2465. 34,348 lncRNAs were screened, with 61,833,304 nt total length and mean 1800.2 nt length. There was one differential lncRNA group, and the number of detected differential lncRNAs was 2852. 13,981 new circRNAs were predicted, with 36,696,834 nt total length and 2624.76 nt mean length. There was one differential circRNA group, and the number of detected differential circRNAs was 1477. The overall workflow of the current study was depicted in Supplementary Fig. 1.
Identification of DEmRNAs
We examined 20,030 genes (Table S2) and reconstructed transcripts using Stringtie. All genes have transcription fragments read per million maps per thousand bases (FPKMs) (Table S2.1, Fig. 2A). Principal component analysis (PCA) showed that all samples were grouped along the principal component 1 (PC1) axis (Fig. 2B), CRC samples and CRCP samples were clustered respectively, and CRC4 and CRC5 were close to CRCP samples. We use all samples for subsequent analyses. We identified 2471 DEmRNAs using group comparison (Table S2.2), and 940 and 1531 genes with highly- and scarcely expressed in CRC and CRCP, respectively. The DEmRNA is depicted in a volcano map (Fig. 2C). A cluster assessment of DEmRNAs in various samples is shown in the heat map (Fig. 2D). Volcanic maps and heat maps revealed different mRNAs expression characteristics between CRC and CRCP.
mRNAs screening and assessment in CRC and CRCP. (A) Sample expression value (FPKM) was separated into various regions, and quantity of genes expressed within samples in various expression regions was computed. (B) PCA of mRNAs expression profiles within samples. (C) Volcano maps showing DE mRNAs in CRC and CRCP. Red and green dots indicate DEmRNAs. (D) Heat maps represent mean centers and normalized data relative to DE mRNAs in CRC and CRCP, p < 0.05, log2FC > 1. (E) KEGG pathway analysis of all mRNAs with parental transcripts. The figure shows only the first 20 condensed KEGG paths. (F) GO annotation of all mRNAs with parental mRNAs. The leading 30 GO terms are presented.
Next, we performed KEGG and GO evaluations to explore the possible function of DE mRNAs (Tables S2.3 and S2.4, Fig. 2E,F). Using KEGG analysis, we demonstrated that these DEmRNAs were strongly enhanced in 20 KEGG metabolic axes (p < 0.05) (Fig. 2E). The concentration pathways include Neuroactive ligand-receptor interaction; Bile secretion; Cell cycle; Nicotine addiction; ECM-receptor interaction; ABC transporters; Salivary secretion; Protein di-gestion and absorption; Mineral absorption; Pancreatic secretion, etc. GO enrichment analysis showed that there were 409 significantly enriched GO items (p < 0.05) (Table S5). There are 247 distinct biological_process (BP) classes, with most DEmRNAs annotated in the BP class as the following GO terms: digestion, collagen catabolic process, chemical synaptic transmission, transport, extracellular matrix organization, nervous system development, chromosome segregation, neuropeptide axis, voltage-gated sodium channels group, microtubule-based movement, etc. In cellular component (CC) term, a majority of the DEmRNAs were enriched in the plasma membrane, extracellular space, plasma membrane integral component, proteinaceous extracellular matrix, extracellular region, apical plasma membrane, cell surface, anchored membrane component, cell junction, kinesin The complex part. In molecular function (MF) term, most DEmRNAs were enriched in the microtubule motor activity, protein heterodimerization activity, chloride channel activity, aryl sulfotransferase activity, sulfotransferase activity, adenylate cyclase interaction, extracellular matrix structural constituent, transmembranal receptor activity, retinol binding.
Identification of DElncRNAs
We identified 24,543 lncRNAs (Table S3), and counted lncRNA types from four levels based on the location association between lncRNAs and already established protein-coding mRNAs. They are respectively direction, type, location and subtype. The statistics of the first three levels of lncRNA (direction, type and location) are shown in Fig. 3A. In all, 3876 DElncRNAs were recognized (Table S3.1). Of these, 1900 up-regulated DElncRNAs and 1976 down-regulated DElncRNAs are shown on a volcanic map (Fig. 3B). Pearson correlation computed the expression association between varying lncRNA (length < 6000nt) and different mRNAs profile, and selected the association pairs with the correlation coefficient ≥ 0.8 and P-value ≤ 0.05. The number of DE lncRNAs exceeded 500. During the analysis, top500 was selected according to the absolute value of log2_FC, and the DElncRNAs and mRNAs of the same DE comparison group were depicted in circos diagram (Fig. 3C).
lncRNAs identification and assessment in CRC. (A) Statistics of the first three levels of lncRNAs. (B) Volcano plots depicting the DElncRNAs in CRC and CRCP. Red and green dots indicate the DElncRNAs. (C) circos the DE lncRNA and gene in the same DE comparison group were presented. (D) The gene GO enrichment results of the total lncRNA coexpression differences corresponded to the top 30 lncRNA coexpression differences. (E) KEGG axis assessment of all lncRNAs with parental mRNAs. Leading 20 condensed KEGG axes provided. (F) Top20 cis regulation results chart, *p < 0.05, **p < 0.01; Left and right sides of the Y-axis represent mRNAs and lncRNAs respectively; the X-axis is the distance between mRNAs and lncRNAs, with negative and positive values indicate upstream and downstream, respectively; the same lncRNA is represented as a bar chart of the same color. (G) Trans regulatory network, red and green nodes indicate lncRNA and gene, respectively, and node size indicates quantity.
According to the co-DE results, transcripts co-expressed by each lncRNA was enriched by GO and KEGG (Table S3.2 and S3.3, Fig. 3D–E). GO terms include regulation of calcium mediated signaling in the BP category; toll-like receptor 4 axis; neural tube patterning; mitotic cytokinesis modulation; mitotic cytokinesis modulation; brain development; negative modulation of chemokine (C–C motif) ligand 5 synthesis; positive modulation of interleukin-18-based axis; cell adhesion; membrane or ganizationd, etc.; astral mi-crotubule in the CC category; clathrin-coated vesicle membrane; spindle midzone; plasma membrane; dense core granule; cell periphery; GABA-A receptor complex; cell surface; dystrophin-associated glycoprotein complex; mitotic spindle midzone, etc.; metallopeptidase activity in MF category; GABA-A receptor activity; exoribonuclease activity; sodium: proton antiporter activity; potassium: proton antiporter activity; ubiquitin-protein transferase activity; phosphatidylinosi-tol-4,5-bisphosphate binding; cystine: glutamate antiporter activity; SNARE binding; gam-ma-butyrobetaine dioxygenase activity, etc. KEGG analysis was performed, Including Proximal tubule bicarbonate reclamation, Ferroptosis, Glycosphingolipid biosynthesis-lacto and neolacto series, Mineral absorption, Renin-angiotensin system, Transcriptional misregulation in cancer, Spliceosome, Amyotrophic lateral sclerosis (ALS), etc. lncRNA cis-regulates connected upstream and downstream genes. Based on the co-DE results, FEELnc23 was utilized for screening of all coding genes 100k upstream and downstream of DElncRNA, and the intersection of DEGs with significant co-expression (Pearson correlation calculation) with this lncRNA was selected. Genes that are co-expressed and genetically adjacent are potentially modulated by this lncRNA (Fig. 3F). Using co-DE analysis and the RNA interaction software RIsearch-2.024, we next predicted the candidate co-expressed lncRNAs and mRNAs association, and the base quantity directly interacting between the 2 nucleic acids was not < 10, and the base interacting free energy was not > -100 as the screening conditions. The interacting lncRNAs and mRNAs selected may have direct regulation. top500(according to co-expression P-value) association pairs were obtained, and the lncRNA-mRNA target association network diagram was generated with network software package (Fig. 3G).
Identification of DEcircRNAs
In all, 13,981 circRNAs were screened from the RNA library using circ software (Table S4). The number of distinct circulating RNA sources is depicted in Fig. 4A; We identified 5 categories of circRNAs sources, the most prevalent being sense-overlapping circRNAs (88%). circRNAs range in length between 1 and 2000 bp (Fig. 4B), with majority circRNAs ranging in length from 201 to 300 bp (16.75%). This was followed by a length of 301 to 400 bp (14.71%) compared to public circRNA datasets in CircBase, CIRCpedia v2, circAtlas, where 685,588 circRNAs were reported in known datasets. However, 309 circRNAs were not collected as new circRNAs in the public circRNAs (Fig. 4C). The DEcircRNAs is depicted in a volcano map (Fig. 4D). Cluster assessment of DEcircRNA in various samples is shown in the heat map (Fig. 4E). Volcanic maps and heat maps revealed different circRNA expression characteristics between CRC and CRCP. Compared with CRCP tissues, there were 1477 DE-circRNAs (FC > 2, p < 0.05) in CRC tissues, with 636 highly- and 841 scarcely-expressed circRNAs (Table S4.1).
circRNAs identification and assessment in CRC. (A) Distribution profiles of various types (B) Length distribution of all identified circRNAs. (C) Total circRNAs from 10 samples relative to the CircBase, CIRCpedia v2, circAtlas. (D) Volcano plots depicting DEcircRNAs in CRC and CRCP. Red and green dots indicate DEcircRNAs. (E) Heatmaps depict the average-centered and normalized data compared to the DEcircRNAs in CRC and CRCP featuring p-value < 0.05 and log2FC > 1. (F) Total circRNAs GO annotation with parental mRNAs. Leading 30 GO terms. (G) KEGG axes assessment of total circRNAs with parental mRNAs. Leading 20 enrichment KEGG axes.
To further clarify the function of DEcircRNA, GO and KEGG assessment of DEcircRNA source genes were performed (Tables S4.2 and S4.3). The GO concentration assessment exhibited 132 strongly condensed GO terms (p < 0.05) (Fig. 4F). The biological process categories of DEcircRNAs include epithelial cell differentiation; learning or memory; positive regulation of translation; iso-prenoid biosynthetic process, telomere maintenance using recombination; negative RNA splicing modulation; osteoblast differentiation; phosphatidylethanolamine biosynthetic process; Phosphati-dylethanolamine biosynthetic process; positive synapse assembly modulation; Positive Synapse Assembly modulation; phospholipid translocation; Phospholipid translocation; And extrinsic component of endosome membrane, non-motile cilium, histone deacetylase of cellular component(CC) complex, macropinocytic cup, endoplasmic reticulum tubular network membrane, axon, ciliary basal body and other processes. In the category of molecular functior (MF), DEcircRNA is rich in SUMO binding. RNA polymerase I transcription coactivator activity; G-quadruplex RNA interaction; hydrolase activity, acting on ester bonds; poly(A) interaction; transcription factor activity, RNA polymerase I core.; D-ribitol-5-phosphate cytidylyltransferase activity; Cytidylyltransferase activity, ethanolaminephosphotransferase activity; And phospholipid-translocating ATPase activity. Using KEGG analysis, we revealed that these DEcircRNAs were strongly enriched in 18 KEGG axes (p < 0.05) (Fig. 4G). DEcircRNA derived genes are enriched in the following KEGG axes: Phosphatidylinositol signaling system, Cushing syndrome, Chronic myeloid leukemia, Leukocyte transendothelial migration, Thyroid cancer, Hepatocellular carcinoma, Prion diseases, etc.
Identification of DEmiRNAs
In order to fully understand the changes of miRNA in colorectal cancer, miRNA sequencing was performed on CRC and CRCP. We conducted 3 biological replicates per sample and sequencing utilized the Illumina HiSeqTM 2500 sequencing platform. The total number of known miRNAs in all samples was 1740 (Table S5). A difference comparison group was set up, and the number of detected differential miRNAs was 77(Table S5.1). The DEmiRNA is presented as a volcano map (Fig. 5A). DEmiRNA cluster assessment of various samples presented in a heat map (Fig. 5B). Volcanic maps and heat maps revealed different miRNA expression characteristics between CRC and CRCP.
miRNAs identification and assessment in CRC and CRCP. 1. (A) Volcano plot shows DEmiRNAs in CRC and CRCP. (B) Heat maps represent average centered and normalized data versus DEmRNAs in CRC and CRCP featuring P-value < 0.05 and log2FC > 1. (C) GO annotation of all miRNAs with parental mRNAs. Leading 30 GO terms provided. (D) KEGG axis assessment of all miRNAs with parental mRNAs. Leading 20 condensed KEGG axes provided.
The target gene of DEmiRNA was analyzed by GO (Table S5.2) and KEGG (Table S5.3) to clarify its function. The GO concentration analysis showed 2329 significantly concentrated GO items (p < 0.05) (Fig. 5C). DEmiRNA has 1565 GO BP term, namely, positive synapse maturation modulation, positive cysteine-type endopeptidase activity modulation affecting the apoptotic axis, cellular surface receptor axis; clathrin-dependent endocytosis; peripheral nervous system development; cerebral cortex radial glia directed migration; Cerebral cortex radial glia guided migration; 255 GO CC terms, namely axon juxtaparanode region, plasma membrane, proteinaceous extracellular matrix; glial limiting end-foot; BAT3 complex; paranodal junction; Schwann cell microvillus et al. And 509 GO terms in the MF category, including reduced voltage-gated calcium channel activity; G-protein coupled receptor activity; core promoter interaction; neuroligin family protein interaction; extracellular matrix interaction; double-stranded methylated DNA interaction; myosin interaction. We also conducted KEGG assessment on DEmiRNA's target genes, and identified 109 strongly enriched KEGG metabolic axes (p < 0.05) (Fig. 5D). Significant abundance of pathways include. Neurotrophin axis, Axon guidance, Focal adhesion, Cell adhesion molecules (CAMs), Type II diabetes mellitus, Renal cell carcinoma, stem cell pluripotency-regulating networks, Circadian entrainment, T cell receptor axis, Human T-cell leukemia virus 1 infection, Apoptosis, Calcium axis, Fc epsilon RI signaling pathway, Endocytosis, Salivary secretion, Chemokine axis, Human papillomavirus infection, Melanogenesis, ECM-receptor interaction, Toxoplasmosis.
Identification of competing endogenous RNAs
To clarify the global regulatory network of protein-coding RNAs and ncRNAs that are involved in CRC, ceRNA networks were constructed based on the ceRNA theory using DEmRNAs, DEmiRNAs, DElncRNAs, and DEcircRNAs. As ceRNAs are regulated by miRNAs, we predicted the target genes of DEmiRNAs as a first step. ceRNA analysis revealed 63 DEmiRNAs, 2582 DElncRNAs, 1477 DEcircRNAs, and 2465 DEmRNAs (Table S6- Table S6.3). Cytoscape software (https://cytoscape.org) was used to visualize the regulatory relationships (Fig. 6A).
ceRNA-miRNA-target gene regulatory network. (A) Network diagram was drawn for 200 mRNA-miRNA-lncRNA/circRNA relationship pairs among the leading 100 mRNA-lncRNA/circRNA pairs in the ceRNA analysis data. (B) GO assessment was performed on the mRNAs in the constructed mRNA-miRNA -lncRNA /circRNA relationship pair, and bar charts were drawn for the items in biological processes, cell components, and molecular functions in the GO enrichment analysis results. The top10 most significant GO items in the three categories of GO results were shown in the same chart. (C) KEGG analysis was performed on the mRNAs in the constructed mRNA-miRNA -lncRNA/circRNA relationship pair.2.7 Validation of RNA-seq data.
miRNAs link mRNAs and lncRNAs/circRNAs by inhibiting mRNA expression and being regulated by lncRNAs/circRNAs. According to the mRNA-miRNA-lncRNA regulatory relationships, hsa-miR-378a-3p, hsa-miR-139-3p, hsa-miR-133a-3P, hsa-miR-422a and hsa-miR-584-5P are the main nodes, affecting lncRNAs (XR-927199.2, NR-149049.1, ENST00000626251, ENST00000602290, XR-001755028.1, NR-0397904.1, NR-036480.1, ENST00000562298 and NR-015380.2) and mRNAs (LEMD1, SIM2, PRKDC, WNT5A, RIC3, etc.). According to the mRNA-miRNA-circRNA regulatory relationships, hsa-miR-19b-1-5p, hsa-miR-17-5p, hsa-miR-20a-5P, hsa-miR-96-5p, hsa-miR-21-3p and hsa-miR-335-3P are the main nodes, affecting circRNAs (circRNA-06154, circRNA-11809, circRNA-05080, circRNA-05031, circRNA-06462) and mRNAs (RYR1, IL6R, ASTN1, SIAE, etc.) (Fig. 6A, Table S6.4 and S6.5).
The mRNA in the constructed ceRNA relationship pair was analyzed by GO and KEGG, and the results were presented in Figure B-C (Table S6.6–S6.9). We determined the relevant terms and pathways. In the GO analysis of mRNA in the constructed MRNA-miRNA-lncRNA relationship pair, we found that in BP category, it was mainly related to positive cytosolic calcium ion concentration modulation. sulfur amino acid metabolic process; positive action potential modulation; formation of primary female sexual features; positive systemic arterial blood pressure modulation; Positive systemic arterial blood pressure modulation; cell division control; negative transcriptional modulation via competitive promoter binding. In the CC category, mainly with melanosome; pre-snoRNP complex; platelet dense granule lumen; neuron projection; microtubule plus-end; transcription elongation factor complex; nuclear body; presynaptic active zone and so on. In MF category, it is mainly binding with NADPH. sphingosine-1-phosphate receptor activity; acetylcholine receptor binding; NAD + kinase activity; protein phosphatase 2A interaction; protein interaction involved in protein folding; NAD interaction, etc. In the GO analysis of the mRNA in the constructed mRNA-miRNA-circRNA relationship pair, we found that the BP category was mainly related to nervous system development. cGMP catabolic process; modulation of spindle microtubular attachment to kinetochore; brain development; neuron cell–cell adhesion; bicarbonate transport; axon guidance, etc. In the CC category, mainly plasma membrane; integral plasma membrane component; cell junction; proteinaceous extracellular matrix; perikaryon; dendrite; postsynaptic density; extracellular region, etc. In MF category, it is mainly related to sulfotransferase activity. 3',5' -cyclic—GMP phosphodiesterase activity; cGMP association; chloride channel activity; delayed rectifier potassium channel activity; voltage-gated sodium channel activity; kinesin binding, etc.; In the mRNA KEGG analysis in the constructed mRNA-miRNA-lncRNA relationship pairs, AMPK and Calcium axes were the main factors. Cysteine and methionine metabolism; Sphingolipid metabolism; VEGF signaling pathway; Basal cell carcinoma is associated with basal cell carcinoma. In the mRNA KEGG analysis in the constructed mRNA-miRNA-circRNA relationship pairs, it was mainly associated with Neuroactive ligand-receptor association, Morphine addiction, and ABC transporters, Calcium axis, ECM-receptor interaction, Circadian entrainment, and Intestinal immune axis for IgA synthesis is related.
Validation of RNA-seq data
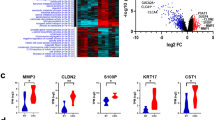
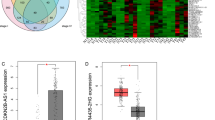
To verify RNA-seq data quality, we predicted 3932, 12,522, and 1573 upregulated DEG (CRC vs. normal sample) via bioinformatics GEO analysis (intersection of GSE33113 and GSE39582) and TCGA-COAD datasets, respectively. In addition, we used univariate Cox and logarithmic ranking methods to estimate a strong association between 727 DEGs and OS of the TCGA-COAD dataset. Lastly, cross-analysis between the aforementioned screens provides four highly prevalent up-regulated DEG, i.e. CCNA2, KPNA2, RBL1, SMOX (Fig. 7A, Table S7). Based on this, our initial investigation focuses on describing the KPNA2 contents in CRC pathogenesis. We explored the KPNA2 expression-related clinical profiles within the TCGA-COAD dataset and, as expected, KPNA2 expression was augmented in tumors versus healthy tissues (Fig. 7B) were validated in clinical tissues (Fig. 7C) and cells (Fig. 7D). CRC patients who displayed high KPNA2 contents in the primary tumor exhibited substantially worse OS and DSS (Fig. 7E).
KPNA2 up-regulated in CRC. (A) Venn chart depicting intersecting Upregulated DEG, which was evaluated by RNA-SEQ data from five of our hospital's CRC and normal tissue samples and other public datasets: TCGA-COAD, GSE33113, and GSE39582. RNAseq_DEGs_UP: Comparing the RNA-seq data of 5 CRC and normal tissue pairs from our hospital, we identified DEGs with the same expression trend in adjoining normal colonic mucosa, primary colonic adenocarcinoma, and distant liver metastasis. COAD_DEGs_UP: Based on TCGA-COAD data, DEG is up-regulated between COAD and normal group. COAD_DEGs_OS_Sig: DEG strongly correlated with TCGA-COAD data OS estimated by univariate cox and log-rank analyses. GEO_DEGs_UP: Cross-DEGs upregulation between COAD-CRC and normal group according to GSE33113 and GSE39582. (B) Comparative KPNA2 expression in normal colonic epithelial, primary, and metastatic CRC tissues identified via the TNMplot Web tool. (C) Expression of KPNA2 in normal colorectal tissue and colorectal cancer. (D) Expression of KPNA2 in normal CRCs NCM460 and CRC cell lines. (E) Kaplan–Meier analysis and control ranking testing of CRC patients in a TCGA-COAD dataset classified based on the median KPNA2 contents. Incidences with elevated KPNA2 levels were connected to substantially worse OS and DSS.
KPNA2 promotes colorectal cancer proliferation and migration
To understand the significance of KPNA2 in CRC, we further investigated the effect of elevated KPNA2 levels on colorectal cancer cell lines (HCT116 and RKO) in in vitro experiments. The technique successfully interfered with KPNA2 mRNA expression, which was confirmed by Western blotting (Fig. 8A). CCK-8 assay revealed that cell survivability was inhibited following KPNA2 depletion (Fig. 8B). In addition, EdU proliferation tests showed that inhibition of KPNA2 expression significantly reduced the positive rate of EdU in colorectal cancer cells (Fig. 8C). Scratch assay showed that the cell migration ability was inhibited after KPNA2 expression was silenced (8D).
KPNA2 promotes colorectal cancer proliferation and migration. (A) WB detection of KPNA2 relative expression in control, NC and siKPNA2 groups. (B) KPNA2 expression silencing results at different time points at CCK-8:24, 48, 72 and 96h. (C) EdU assay showed proliferating cells (HCT116 and RKO); EdU (red) and DAPI (blue) staining. Scale :50μm. (D) Scratch tests showing the ability of cells to migrate. Scale :100μm. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Discussion
Colorectal cancer is among the most prevalent cancer of the gastrointestinal tract, causing many deaths each year. The HTS data enhances our knowledge regarding the roles of various forms of RNA, particularly, ncRNAs, which are critical for life activities22. Transcriptome studies may be a valuable tool in the accurate diagnosis and prognosis of colorectal cancerAs our understanding of cancer's molecular environment grows, it becomes easier to find a valuable diagnostic, prognostic, and predictive biomarker in clinical oncology. Herein, we conducted full transcriptome sequencing analysis on 5 pairs of CRC and paracarcinoma (CRCP) samples, and identified 2465 DEmRNAs and 77 DEmiRNAs.There were 2852 DElncRNAs and 1477 DEcircRNAs.
We performed genome-wide mRNA expression profiling of colorectal cancer tissues and adjacent tissues. In addition, CRC bioinformatics analyses in GEO (GSE33113 and GSE39582 datasets) and TCGA-COAD predicted 3932, 12,522 and 1573 DEG upregulations, respectively, compared to normal samples. We used univariate Cox and logarithmic ranking techniques to estimate 727 DEGs that are intricately linked to TCGA-COAD dataset OS. Lastly, cross-analysis provided four common up-regulated DEG, namely CCNA2, KPNA2, RBL1 and SMOX (Fig. 7A, Table S7), among which KPNA2 was not reported, and we selected KPNA2 for verification. To understand the significance of KPNA2 gene in CRC, we further studied the effects of different expression levels of KPNA2 on CRC cell lines (HCT116 and RKO) in vitro experiments, and found that overexpression of KPNA2 promotes CRC proliferation and migration. Knocking down KPNA2 can inhibit CRC proliferation and migration, which is consistent with our sequencing results. The reliability of RNA-seq data was further confirmed. GO term analysis showed that The DEmRNA was mainly related to digestion, collagen catabolic process, chemical synaptic transmission, transport, plasma membrane and extracellular space, , aryl sulfotransferase activity and so on. KEGG axes analysis revealed that DEmRNA was mainly related to neuroactive ligand-receptor interactions, bile secretion, cell cycle, and nicotine addiction.
Emerging reports revealed that lncRNAs is a strong modulator of carcinogenesis, apoptosis, differentiation, proliferation, invasion, and metastasis of a variety of cancer42. lncRNAs serve as ceRNAs that associate with miRNAs to target and modulate downstream gene expressions. On the upregulated lncRNA network, miRNA levels were downregulated, however its downstream gene expression was elevated. Likewise, upon lncRNA down-regulation, miRNA expression was augmented, which diminished downstream expressions, thus impacting disease development and progression. Additionally, the same lncRNA levels can potentially regulate differing pathways in distinct diseases. Till date, there are multiple on the link between lncRNAs and CRC. We detected many lncRNAs in colorectal and paracancer, which are longer than 200 bp and do not code for proteins, but serve essential roles in modulating numerous biological processes22. Our results show that according to GO analysis, differential lncRNAs are mainly related to regulation of calcium-based cellular signaling; toll-like receptor 4 axis; neural tube patterning; astral microtubule; clathrin-coated vesicle membrane; spindle midzone; plasma membrane; dense core granule; cell periphery; metallopeptidase activity; GABA-A receptor activity; exoribonuclease activity, etc.; According to KEGG enrichment analysis, The differences of lncRNA are mainly related to Proximal tubule bicarbonate reclamation, Ferroptosis, Glycosphingolipid biosynthesis-lacto and neolacto series, Mineral absorption, Renin-angiotensin system and so on. lncRNA cis-regulates adjacent upstream and downstream gene expressions. Using co-DE data and the RIsearch-2.0 software, we predicted the association of candidate co-expressed lncRNA and mRNAs. Moreover, the threshold was set as follows: the base quantity that directly interacts between the two nucleic acid molecules was not < 10, and the base interacting free energy was not > − 100. The interacting lncRNAs and mRNAs selected maybe directly modulated, and top500(according to co-expression P-value) association pairs were acquired. Our results showed that lncRNAs with XR-923556.1, XR-001745290.1, XR-117099.1, and XR-938589.2 as the main nodes regulated multiple mRNAs.
circRNAs was once considered an insignificant by-product of pre-mRNA splicing43. Nonetheless, cyclic sequences are extremely stable in both intra- and extracellular conditions. Thus, they potentially regulate transcription, intercellular messaging, and translation44. circRNAs are ubiquitously expressed during tissue and organ development and in involved in the regulation of human diseases45. We found 309 new circRNAs. GO and KEGG assessments of decircrNA-derived genes were performed (Tables S4.2 and S4.3). The GO concentration analysis revealed 132 significantly condensed GO terms. Including epithelial cell differentiation; learning or memory; positive regulation of translation; extrinsic component of endosome membrane, non-motile cilium, histone deacetylase complex, SUMO binding; RNA polymerase I transcription coactivator activity; G-quadruplex RNA binding and other processes. Based on KEGG analysis, these DEcircRNAs were strongly enriched in 18 KEGG axes, described as follows: Phosphatidylinositol signaling system, Cushing syndrome, Chronic myeloid leukemia, Leukocyte transendothelial migration, Thyroid cancer, Hepatocellular carcinoma, Prion- diseases, etc.
miRNAs are small ncRNAs spanning approximately 22 nts that may act on cogenes or tumor suppressor genes. Trs aberrant profile causes cancer development in humans46,47. We found 77 DemiRNAs. According to GO enrichment analysis, DEmiRNA is mainly involved in positive synapse maturation regulation and positive cysteine-type endopeptidase activity modulation in apoptotic signaling pathway, juxtaparanode region of axon, plasma membrane, low voltage-gated calcium channel activity; G-protein coupled receptor activity; Equal correlation. Additionally, we conducted KEGG assessment on DEmiRNA's target genes, and identified 109 strongly enriched KEGG metabolic axes (p < 0.05) (Fig. 5D). Significant abundance of pathways include Neurotrophin axis, Axon guidance, Focal adhesion, CAMs, Type II diabetes mellitus, Renal cell carcinoma, stem cell pluripotency-regulating networks, Circadian entrainment, T cell receptor axis, Human T-cell leukemia virus 1 infection, etc.
In 2011, the concept of "ceRNA" was introduced, suggesting that ceRNA forms a large-scale transcriptome modulatory axis, which significantly enhances the human genome functional genetic knowledge. It also potentially controls cancer development and progression6。More recently, lncRNA or circRNA relevance as ceRNA in cancer has been extensively examined. The study suggests that the ceRNA axis can potentially present novel biomarkers and robust therapeutic targets for CRC48,49. Nevertheless, there are no extensive assessments of ceRNA networks in CRC. Our results showed that the target genes of DEmiRNAs as a first step. ceRNA analysis revealed 63 DEmiRNAs, 2582 DElncRNAs, 1477 DEcircRNAs, and 2465 DEmRNAs (Table S6-Table S6.3). miRNAs link mRNAs and lncRNAs/circRNAs by inhibiting mRNA expression and being regulated by lncRNAs/circRNAs. According to the mRNA-miRNA-lncRNA regulatory relationships, hsa-miR-378a-3p, hsa-miR-139-3p, hsa-miR-133a-3P, hsa-miR-422a and hsa-miR-584-5P are the main nodes, affecting lncRNAs (XR-927199.2, NR-149049.1, ENST00000626251, ENST00000602290, XR-001755028.1, NR-0397904.1, NR-036480.1, ENST00000562298 and NR-015380.2) and mRNAs (LEMD1, SIM2, PRKDC, WNT5A, RIC3, etc.). According to the mRNA-miRNA-circRNA regulatory relationships, hsa-miR-19b-1-5p, hsa-miR-17-5p, hsa-miR-20a-5P, hsa-miR-96-5p, hsa-miR-21-3p and hsa-miR-335-3P are the main nodes, affecting circRNAs (circRNA-06154, circRNA-11809, circRNA-05080, circRNA-05031, circRNA-06462) and mRNAs (RYR1, IL6R, ASTN1, SIAE, etc.) (Fig. 6A, Table S6.4 and S6.5). GO enrichment analysis, which is related to positive cytosolic calcium ion concentration regulation; sulfur amino acid metabolic process; positive action potential modulation; formation of primary female sexual features; melanosome; pre-snoRNP complex; platelet dense granule lumen; neuron projection; binding; sphingosine-1-phosphate receptor activity; acetylcholine receptor binding and other pathways are related. KEGG enrichment analysis was mainly related to AMPK and Calcium axes. Cysteine and methionine metabolism; Sphingolipid metabolism; VEGF axis and so on. Using ceRNA data, 200 mRNA-miRNA-circRNA relationship pairs in the leading 100 mRNA-circRNA sequencing results were drawn in the network diagram in the first category, the results showed that, miRNAs can affect multiple mRNAs and can affect additional miRNAs by affecting mRNAs and circRNAs. For example, has-miR-19b-1-5p can affect circRNA-06154 to affect has-miR-183-5p and has-miR-552-3p), and can also affect mRNAs (MAPK10 and ASTN1, etc.) to affect hsa-miR-17-5p and hsa-miR-20a-5p. In the GO analysis, we found that in the BP category, nervous system development was the main factor; cGMP catabolic process; modulation of spindle microtubule attachment to kinetochore; plasma membrane; integral component of plasma membrane; Integral plasma membrane component; cell junction; proteinaceous extracellular matrix; sulfotransferase activity; 3',5' -cyclic—GMP phosphodiesterase activity; cGMP interaction; chloride channel activity;; In KEGG analysis, Neuroactive ligand-receptor association, Morphine addiction, ABC transporters and Calcium axis were mainly involved network, ECM-receptor association, Circadian entrainment, Intestinal immune axis for IgA synthesis, etc.
In summary, this study sequenced and analyzed DEmRNAs, DElncRNAs, DEcircRNAs, and DEmiRNAs in colorectal and para-cancer, and provided an extensive anatomy of the ceRNA network. Using data from transcriptome sequencing and patient OS, we recognized KPNA2 as a candidate CRC bioindicator. We revealed that KPNA2 protein levels are generally upregulated in CRC tissues, and KPNA2 content stratification suggests that KPNA2 augmentation is associated with worse patient prognosis. Functional assessments showed that KPNA2 inhibited CRC proliferation and metastasis in vitro, and this data strongly corroborated with RNA-seq. A limitation of this investigation is the lack of genomic sequencing that would assess gene mutation-mediated alterations in RNA transcription, but the employed axial coding genes were relatively stable, and no hypermutated genes associated with colorectal cancer were reported. This article highlights new understanding of CRC pathogenesis involving the ceRNA axis, and these lesions may play a role in CRC occurrence.
Data availability
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI Bioproject accession numbers: PRJNA1084915 and PRJNA1082046.
References
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 73(1), 17–48. https://doi.org/10.3322/caac.21763 (2023).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71(3), 209–249. https://doi.org/10.3322/caac.21660 (2021).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 66(1), 7–30. https://doi.org/10.3322/caac.21332 (2016).
Ogunwobi, O. O., Mahmood, F. & Akingboye, A. Biomarkers in colorectal cancer: Current research and future prospects. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21155311 (2020).
Winkle, M., El-Daly, S. M., Fabbri, M. & Calin, G. A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 20(8), 629–651. https://doi.org/10.1038/s41573-021-00219-z (2021).
Salmena, L. et al. A ceRNA hypothesis: The Rosetta stone of a hidden RNA language?. Cell 146(3), 353–358. https://doi.org/10.1016/j.cell.2011.07.014 (2011).
Smolarz, B. et al. miRNAs in cancer (review of literature). Int. J. Mol. Sci. https://doi.org/10.3390/ijms23052805 (2022).
May, J. M. et al. Long and short non-coding RNA and radiation response: A review. Transl. Res. 233, 162–179. https://doi.org/10.1016/j.trsl.2021.02.005 (2021).
Liu, Y. et al. Noncoding RNAs regulate alternative splicing in cancer. J. Exp. Clin. Cancer Res. 40(1), 11. https://doi.org/10.1186/s13046-020-01798-2 (2021).
Chen, L.-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 21(8), 475–490. https://doi.org/10.1038/s41580-020-0243-y (2020).
Chan, J. J. & Tay, Y. Noncoding RNA:RNA regulatory networks in cancer. Int. J. Mol. Sci. https://doi.org/10.3390/ijms19051310 (2018).
Yuan, C. et al. PceRBase: A database of plant competing endogenous RNA. Nucleic Acids Res. 45(D1), D1009-d1014. https://doi.org/10.1093/nar/gkw916 (2017).
Thomson, D. W. & Dinger, M. E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 17(5), 272–283. https://doi.org/10.1038/nrg.2016.20 (2016).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17), i884–i890. https://doi.org/10.1093/bioinformatics/bty560 (2018).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12(4), 357–360. https://doi.org/10.1038/nmeth.3317 (2015).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2), 166–169. https://doi.org/10.1093/bioinformatics/btu638 (2015).
Roberts, A. et al. Improving RNA-seq expression estimates by correcting for fragment bias. Genome Biol. 12(3), R22. https://doi.org/10.1186/gb-2011-12-3-r22 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12), 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Gene Ontology Consortium. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47(D1), D330-d338. https://doi.org/10.1093/nar/gky1055 (2019).
Kanehisa, M. et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480-484. https://doi.org/10.1093/nar/gkm882 (2008).
Ogata, H. et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27(1), 29–34. https://doi.org/10.1093/nar/27.1.29 (1999).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28(11), 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M. et al. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51(D1), D587-d592. https://doi.org/10.1093/nar/gkac963 (2023).
Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102(43), 15545–15550. https://doi.org/10.1073/pnas.0506580102 (2005).
Mootha, V. K. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34(3), 267–273. https://doi.org/10.1038/ng1180 (2003).
Gao, Y., Wang, J. & Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 16(1), 4. https://doi.org/10.1186/s13059-014-0571-3 (2015).
Wang, L. et al. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26(1), 136–138. https://doi.org/10.1093/bioinformatics/btp612 (2010).
Zhao, J. et al. IRESfinder: Identifying RNA internal ribosome entry site in eukaryotic cell using framed k-mer features. J. Genet. Genom. 45(7), 403–406. https://doi.org/10.1016/j.jgg.2018.07.006 (2018).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3), R25. https://doi.org/10.1186/gb-2009-10-3-r25 (2009).
Griffiths-Jones, S. et al. Rfam: An RNA family database. Nucleic Acids Res. 31(1), 439–441. https://doi.org/10.1093/nar/gkg006 (2003).
Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. https://doi.org/10.1002/0471250953.bi0410s05 (2004).
Griffiths-Jones, S., Saini, H. K., van Dongen, S. & Enright, A. J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 36, D154-158. https://doi.org/10.1093/nar/gkm952 (2008).
Friedländer, M. R. et al. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40(1), 37–52. https://doi.org/10.1093/nar/gkr688 (2012).
Jin, Y. & Hammell, M. Analysis of RNA-seq data using TEtranscripts. Methods Mol. Biol. 1751, 153–167. https://doi.org/10.1007/978-1-4939-7710-9_11 (2018).
Tino, P. Basic properties and information theory of Audic-Claverie statistic for analyzing cDNA arrays. BMC Bioinform. 10, 310. https://doi.org/10.1186/1471-2105-10-310 (2009).
Enright, A. J. et al. MicroRNA targets in Drosophila. Genome Biol. 5(1), R1. https://doi.org/10.1186/gb-2003-5-1-r1 (2003).
Fahlgren, N. & Carrington, J. C. miRNA target prediction in plants. Methods Mol. Biol. 592, 51–57. https://doi.org/10.1007/978-1-60327-005-2_4 (2010).
Marisa, L. et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 10(5), e1001453. https://doi.org/10.1371/journal.pmed.1001453 (2013).
Yao, H. et al. KCTD9 inhibits the Wnt/β-catenin pathway by decreasing the level of β-catenin in colorectal cancer. Cell Death Dis. 13(9), 761. https://doi.org/10.1038/s41419-022-05200-1 (2022).
Bartha, A. & Győrffy, B. TNMplot.com: A web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int. J. Mol. Sci. 22(5), 2622. https://doi.org/10.3390/ijms22052622 (2021).
Liu, J. et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173(2), 400-416.e411. https://doi.org/10.1016/j.cell.2018.02.052 (2018).
Bin, X. et al. Research progresses in roles of LncRNA and its relationships with breast cancer. Cancer Cell Int. 18, 179. https://doi.org/10.1186/s12935-018-0674-0 (2018).
Guo, J. U., Agarwal, V., Guo, H. & Bartel, D. P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15(7), 409. https://doi.org/10.1186/s13059-014-0409-z (2014).
Chen, M., Pereira-Smith, O. M. & Tominaga, K. Loss of the chromatin regulator MRG15 limits neural stem/progenitor cell proliferation via increased expression of the p21 Cdk inhibitor. Stem Cell Res. 7(1), 75–88. https://doi.org/10.1016/j.scr.2011.04.002 (2011).
Zhang, Y. et al. Biological impact of environmental polycyclic aromatic hydrocarbons (ePAHs) as endocrine disruptors. Environ. Pollut. 213, 809–824. https://doi.org/10.1016/j.envpol.2016.03.050 (2016).
Bartel, D. P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116(2), 281–297. https://doi.org/10.1016/s0092-8674(04)00045-5 (2004).
Esquela-Kerscher, A. & Slack, F. J. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer 6(4), 259–269. https://doi.org/10.1038/nrc1840 (2006).
Abdollahzadeh, R. et al. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J. Cell. Physiol. 234(7), 10080–10100. https://doi.org/10.1002/jcp.27941 (2019).
Yao, Y. et al. Integrated analysis of co-expression and ceRNA network identifies five lncRNAs as prognostic markers for breast cancer. J. Cell. Mol. Med. 23(12), 8410–8419. https://doi.org/10.1111/jcmm.14721 (2019).
Funding
This work was funded by Zhejiang basic public welfare research program (Grant# LGF21H160011) to X.Y., the medical and health research project of Zhejiang province (Grant# 2024KY1730) to G.L. and Grant#2022RC274) to J.W., Shaoxing Health Science and Technology Project to L.Z. (Grant#2022KY004) and the university-level youth research fund project of Shaoxing People's Hospital (Grant# 2023YB21) to L.Z.
Author information
Authors and Affiliations
Contributions
Conceptualization, L.Z. and Y.L.; methodology, Z.L. and H.T.; software, C.F. and Y.W.; validation, K.W., X.Y..; formal analysis, L.Z. and G.L.; investigation; resources, J.W. and G.T.; data curation, K.W. and Y.W.; writing—original draft preparation, L.Z.; writing—review and editing, Z.L.; visualization, K.W.; supervision, Z.L.; project administration, L.Z.; funding acquisition, X.Y., L.Z. and G.L. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, L., Li, Y., Fu, C. et al. Exploration and validation of ceRNA regulatory networks in colorectal cancer based on associations whole transcriptome sequencing. Sci Rep 14, 20446 (2024). https://doi.org/10.1038/s41598-024-71465-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-71465-5
- Springer Nature Limited