Abstract
Mumps is a common childhood infection caused by the mumps virus (MuV). Aseptic meningitis and encephalitis are usual symptoms of mumps together with orchitis and oophoritis that can arise in males and females, respectively. We have used computational tools: RNA22, miRanda and psRNATarget to predict the microRNA-mRNA binding sites to find the putative microRNAs playing role in the host response to mumps virus infection. Our computational studies indicate that hsa-mir-3155a is most likely involved in mumps infection. This was further investigated by the prediction of binding sites of hsa-mir-3155a to the MuV genome. Additionally, structure prediction using MC-Fold and MC-Sym, respectively has been applied to predict the 3D structures of miRNA and mRNA. The miRNA-mRNA interaction profile between has been confirmed through molecular docking simulation studies. Taken together, the putative miRNA (hsa_miR_6794_5p) has been found to be most likely involved in the regulation of transcriptional activity in the MuV infection.
Similar content being viewed by others
Introduction
Mumps is a painful, systemic and contagious disease characterized by inflammation of one or both of the parotid glands, which is often accompanied by more serious complications, such as meningitis, pancreatitis or orchitis1. Mumps virus (MuV) is a non-segmented negative-strand RNA virus belonging to the family Paramyxoviridae2,3. The viral genome is composed of a single, linear, negatively stranded RNA chain, composed of 15,384 nucleotides (bp), which encodes six structural proteins and at least two non-structural proteins4.
MicroRNAs (miRNAs) are a novel class of non-coding RNAs, which are encoded in the genomes of invertebrates, and vertebrates5. Functionally, miRNA regulate the translation and stability of its target mRNA on the basis of the complementarity of partial sequences6. Although the number of newly identified miRNAs is still increasing, the target mRNAs of human miRNAs are yet to be determined. Multiple computational approaches have been employed to predict the involvement of various miRNAs in different cellular functions7,8. During mumps virus replication, human miRNAs bind with viral mRNA with complementarity sequences and interfere in the viral protein synthesis and viral replication processes9. Multiple nucleoproteins such as N, V/P/I (V/phospho‐/I proteins), matrix (M), fusion (F), small hydrophobic (SH), haemagglutinin‐neuraminidase (HN) and large (L) ones play an important role in cell signaling and functioning of genomic virus10. There are multiple bioinformatics tools for miRNA analyses, which includes miRNA target prediction (TargetScan and DIANA-microT), viral genome analysis (Geneious Prime and DNASTAR Lasergene) and gene expression analysis (DESeq2 and EdgeR). These tools analyze miRNA sequences and predict their potential target genes. By identifying human genes targeted by miRNAs that are also known to be involved in viral replication we can use miRNAs to regulate the transcriptional activity during virus infection. Here, we introduced a computational approach which predicts human miRNAs that bind to the mumps virus gene segments and might lead to the prevention of MuV infection. A set of miRNAs was retrieved from miRNA databases and has been examined towards the interaction with the mumps virus genome through in silico experiments using three specific miRNA prediction algorithms (RNA22, miRanda, and psRNATarget). Predicted miRNAs were subjected to computational molecular docking studies to check the mRNA nucleotides that bind to miRNAs and take part in the initiation of viral replication. Furthermore, MD simulations were performed to check the stability of the docked mRNA-miRNA complexes. In future studies such miRNAs could be used as lead compounds for the design of RNA-based vaccines against mumps viral infection.
Computational methodology
MuV genome retrieval and annotation
The computational methodology applied in the present work results from our preceding research in which we explored the miRNAs as suitable targets for the treatment of influenza11. The complete genome sequence of MuV with the accession number NC_002200.1 has been downloaded from National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/). Moreover, the CLC Genome Workbench application (v9.5.2)12 has been used to view and analyze sequences to obtain all necessary information.
Retrieval of human mature miRNAs
The 264 human miRNAs were retrieved from online miRbase database (http://www.mirbase.org/), a collection of miRNAs from different species13,14. miRBase is a comprehensive database that provides information on miRNAs, which are short non-coding RNA molecules involved in the regulation of gene expression. miRBase serves as the central repository for miRNA sequences, annotations, and related information. Each entry in the miRBase database displays information about the location and sequence of a precursor and mature miRNAs sequences (Supplementary, Table S1).
miRNA target prediction in MuV genome
Three online tools: RNA22 (http://cm.jefferson.edu/rna22v1.0/), miRanda15, and psRNATarget (http://plantgrn.noble.org/psRNATarget/) were employed to predict targets of human miRNAs in the mump virus genome, to locate miRNA targeting regions. The sequences of human miRNAs and the genome of MuV, each in FASTA format, were processed through those algorithms using specific parameters.
RNA22 v2.0 interactive predictions
The RNA22 (https://cm.jefferson.edu/rna22/Interactive/) algorithm predicts target patterns which can be statistically significant for miRNA motifs of mature miRNAs16. The RNA22 is one of the most advanced miRNA prediction algorithms17. We followed practical strategies of using RNA22 discussed by Witkos and Koscianska18. Sensitivity and specificity values were kept at 63% and 61%, respectively and the seed size of 7 was selected with 1 unpaired base allowed in the seed region and no limit was set to the maximum number of G:U wobbles in the seed region. Minimum number of paired-up bases was set to 12 while the maximum folding energy was kept at -14 kcal/mol.
miRanda
miRanda is an miRNA target scanner to predict mRNA targets for microRNAs using dynamic-programming alignment and thermodynamics15. The miRanda algorithm compares the complementarity of miRNAs with 3'UTR regions. The outcome is a weighted sum of match and mismatch scores for base pairs and gap penalties. These values are derived from the binding energy of the duplex structure, the evolutionary conservation of the entire target site, and its location within the 3-unit thermal resistance. The basic principle parameters are sequence complementarity, free energy of RNA-RNA duplex and cross-species conservation of the target site15. miRanda was employed after defining the settings (gap open penalty = −9.0, gap extend penalty = − 4.0, score threshold = 140, energy threshold = −20 kcal/mol and scaling parameter = 4.0), respectively.
psRNATarget
psRNATarget analysis server19 contains two important analytical functions: (1) retrieving the corresponding comparisons between miRNA and target transcript using a scheme allowed scoring goals, and (2) the accessibility test by calculating the inactive energy (UPE) required to “open” a second structure around the target site of miRNA in mRNA. psRNATarget incorporates the latest findings in miRNA target detection20. The sequences of virus mature miRNAs and target mRNA in FASTA format were submitted to psRNATarget server as input files. psRNATarget uses varying 'seed' region that is sensitive; in particular, the region containing nucleotides 2–7 was initially described in vertebrate miRNA studies19.
Rstudio and Ggplot2
RStudio (https://rstudio.com/)21 and ggplot2 package22 (https://cran.r-project.org/web/packages/ggplot2/index.html) were employed for graphical representations of miRNA prediction. R is a statistical programming language, and ggplot2 is an open-source software for data visualization. They greatly improve the quality and aesthetics of graphics and efficiency of creating plots.
miRNA and mRNA retrieval
MC-Fold (https://major.iric.ca/MC-Fold/) and MC-Sym (https://major.iric.ca/MC-Sym/) were used to predict three dimensional (3D) models of miRNAs. MC-Fold is a program that takes as input an RNA sequence and produces as output a list of sub-optimal secondary structures, whereas MC-Sym is a program that takes as input a tuple of sequence and it’s secondary structure and produces as output a list of tertiary structures23. However, for mRNA structure prediction, RNAComposer web server (https://rnacomposer.cs.put.poznan.pl/) was used to predict 3D models24. The RNAComposer system offers a new user-friendly approach for the fully automated prediction of large 3D RNA structures. The method is based on the machine translation principle and operates on the RNA FRABASE database acting as the dictionary relating RNA secondary structure and tertiary structure elements. Moreover, UCSF Chimera molecular graphics software, was used to visualize the miRNA and mRNA models25.
miRNA and mRNA docking
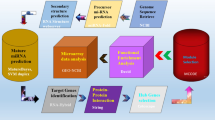
The PatchDock algorithm was employed to check miRNA-mRNA interactive profiles based on flexibility and scorings of docking algorithms26. PatchDock is a geometry-based molecular docking algorithm that aims to find docking transformations that result in good molecular shape complementarity. Docking results were further evaluated through using Discovery Studio (4.1) and UCSF Chimera 1.10.1 visualizing tools. The proposed methodology has been shown in Fig. 1.
Results and discussion
Organization of the MuV genome
The MuV is a single-stranded, non-segmented, and negative-strand RNA virus, having a shape that varies among spherical and pleiomorphic with a diameter of about 200 nm, composed of 15,384 nucleotides 27. The MuV genome is contained in a helical nucleocapsid that is enclosed in a tri-layered envelope which contains two types of spikes, including the HN protein and the F protein. The L protein combines with the P protein to form the RNA-dependent RNA polymerase (RdRp), which transcribes and replicates the genome28. It contains seven transcription units with seven genes and two noncoding regions at the 5′ and 3′ ends of the genome. Different protein segments (N, V/P, M, F, SH, HN and L) have been observed in sequential order in MuV genome. The major protein is L as compared to other proteins which may have significant importance in the MuV virulence (see Fig. 2).
Prediction of target sites in MuV genome for miRNAs binding
miRNAs regulate many mRNAs by binding in parallel sequences in 3’-untranslated regions causing RNA disruption29. In our computational studies, mRNA target sites for 264 miRNAs from miRbase were predicted and analyzed (hairpin and genomic sites) based on miRNA-mRNA compatibility analysis (Supplementary Table S1). All above mentioned tools were used in the combination to maximize the accuracy of miRNA target predictions. RNA22 predicts miRNAs based on pattern-recognition in the target sequence of MuV genome. Sensitivity and specificity values were kept at 63% and 61%. Figure 3 shows the positions of most targeted sites (within the nucleotides range from 0 to 15,000 bp) with the folding energy values ranging from -17 to -27 kcal/mol. Additionally, predicted sites in the genome corresponded to genes that were encoding proteins F, HN, L, M, NP, SH, V/P and NA, respectively.
miRanda implements various parameters, including conservation of target sites to predict miRNAs target sites. The miRanda results were analyzed based on miRNA-mRNA sequence complementarity, minimum free energy (ranging from −20 to −35 kcal/mol) and scoring values (from 145 to 169), respectively, within the nucleotide range 0–15,000 bp. The algorithm was run with the following settings: (gap open penalty = −9.0, gap extend penalty = −4.0, score threshold = 140, energy threshold = −20 kcal/mol and scaling parameter = 4.0). Our results show that genes encoding proteins F, HN, L, M, NP, SH, V/P and NA, respectively have been observed, that might be involved in metabolic pathways during MuV infections (Fig. 4).
psRNATarget predicts numerous hybridization sites in MuV genome that bind miRNAs to MuV. Figure 5 shows the positions of most targeted sites within the nucleotide sequence (ranging from 0 to 15,000 bp) with expect values ranging from 3.5 to 5.0. Genes encoding proteins F, HN, L, M, NP, SH, V/P and NA, respectively have been observed.
It has been observed that there are some common miRNAs predicted by all three different tools which shows the efficacy and credibility of our results.
The comparative results show that a total of 265, 284 and 28 miRNAs have been predicted by RNA22, miRanda and psRNATarget, respectively (Fig. 6). The comparison of all predictions shows that hsa_mir_6794_5p is a common miRNA predicted by all methods that exhibits very good binding with targeted sites in the MuV genome (see Table 1). Eighteen miRNAs are common for RNA22 and miRanda predictions (Table 2), whereas, the single miRNA (has-miR-6794-5p) is common for RNA22 and psRNATarget predictions. Furthermore, 16 miRNAs are common for psRNATarget and miRanda predictions (Table 2). Moreover, by analyzing proteins encoded by the MuV genome, we have identified multiple proteins: nucleoproteins (N), V/P/I (V/phospho‐/I proteins), matrix (M), fusion (F), small hydrophobic (SH), haemagglutinin‐neuraminidase (HN) and large (L) proteins that may have a significant role in cellular functionality of MuV10. It has been observed that HN is directly involved in the mumps virus replication cycle30. Therefore, hsa_mir_6794_5p interacting with mRNA of HN may stop the replication cycle of MuV and as the result suppress the MuV viral infection.
Prediction of hsa-mir-6794-5p structure and its cellular functionality
The predicted structural models of both premature and mature hsa-mir-6794-5p and their sequences are shown in Fig. 7. The premature and mature miRNA (hsa-mir-6794-5p) consists of 68 and 20 base pairs, respectively. The coiled and twisted models of both premature and mature hsa-mir-6794-5p have been depicted in Fig. 7.
The matching and mismatching of miRNA-mRNA nucleotides is good computational approach to predict the possible binding of miRNAs to the MuV genome sequence. Our results show that hsa_miR_6794_5p targets multiple gene encoded proteins at multiple loci in the MuV genome. During replication, has-mir_6794_5p can bind to the mRNA virus, with incomplete complementarity, leading to the suppression of synthesis of protein and viral replication. The cellular has_mir_6794_5p significantly inhibits the replication of MuV by binding to the viral genome31. The miRNA of has_mir_6794_5p cells have been reported to target the major genes (the L protein) and the phosphoprotein (the P protein). These findings strongly suggest that binding of the cellular miRNAs to the MuV genome could be another way of preventing the virus attack.
Conservation analysis of target sites
The miRNAs target sites have a tendency to be enriched at 5′ end of protein-coding genes and 3′ untranslated regions (UTRs) which affects proteins and their associated signaling pathways32. We analyzed the conserved nucleotide sequences in different mumps virus strains to explore the accuracy and reliability of predicted hsa_miR_6794_5p MuV genome interactions throughout the opposite strain sequences (see Fig. 8).
Proteins encoded by the MuV genome and their involvement in mumps infection
Our results show that multiple proteins are involved in binding of miRNAs to the MuV genome. The predicted frequencies of occurrence of proteins F, V/P, NP, L, M, SH, and HN in our miRNAs studies are shown in Fig. 9.
Nucleoprotein (NP)
The MuV NP is composed of 549 amino acids and has N-terminal base and an unstructured C-terminal tail. It has been observed that N-terminal domain is a major component responsible for the inhibition of RNA, binding of P proteins, and nucleocapsid formation33. The primary function of NP is involved in the formation of nucleocapsid in non-strand virus (NSRV). The structural analysis showed that both NP and P are present in the form of soluble complexes with RNA (NP-RNA and P-RNA complexes). Furthermore, under a transmission electron microscope, the NP-RNA complex appears as a nucleocapsid with a diameter of about 20 nm and 13 subunits. The NP-RNA ring is composed of single-stranded RNA fragment with 78 nucleotides. The MuV NP may also provide weaker protection of the RNA than the NP of a few other negative-strand RNA viruses, indicating the level of NP association34.
virus/phosphoproteins (V/P)
The V-protein of MuV plays a critical role in pathogenesis35. The V-protein is composed of 224 amino acids and particularly cysteine is abundantly present at C terminus in all paramyxoviruses35. The prior research showed that V-protein disrupts the interferon signaling pathway (IFN) by degradation of STAT1, a critical component of IFN-activated gene expression36. Moreover, V-protein has also been shown to be associated with receptor-activated C kinase (RACK1), which contains Trp-Asp replication and mediates interactions between IFN receptor and STAT135. Our results show that hsa_miR_6794_5p may stop the V protein synthesis by disturbing the cellular transcriptional and translational activities.
Matrix protein (M)
Matrix (M) protein (composed of 89 residues) acts as an adapter that links viruses to ribonucleoprotein complexes and glycoprotein viruses to cell plasma components37. The M protein expression is sufficient for the production of effective virus-like-particles, without the need for assistance with any other components of the viral structure, such as viral glycoproteins or NP proteins38. The M protein multimerizes and creates grids which interact with lipid membranes and cytoplasmic tails of HN and F proteins. At the same time, the M dimer associates with nucleocapsids through positively charged regions39. Moreover, it has been observed that budding and viral particle release is triggered by MuV M–N interaction40.
Fusion proteins
The MuV fusion protein (F) plays a crucial role for the entry process and spread of infection by mediating fusion between viral and cellular membranes, as well as, between infected and neighboring cells, respectively41. The crystal structure of the F core of MuV has been determined at 2.2 Å resolution42. The F protein has significant role in the pathogenesis of mump virus infection. The structural study of the F protein reveals that it contains 538 amino acids and the three amino acids at positions 221, 323, and 373 in the F protein of the mumps virus are directly involved in epitopes fusion43. The interactions between the virus and cellular layers involve two inactive heptad repeat (HR) domains in the ectodomain of coated glycoproteins. This complex takes shape of a helix in which three HR1 peptides form a very central coil and three HR2 peptides pack against hydrophobic grooves on the middle coil-coil surface in the form of oblique antiparallel strands 42. Our results show that miRNAs that bind to the MuV gene could inhibit the mumps infections by disturbing the F protein biosynthesis. Moreover, proteolysis and fusogenicity (process of facilitating fusion in cells) during the mumps infection could also be controled by inhibiting the F protein formation.
Small hydrophobic
The small hydrophobic (SH) protein is composed of 57 amino acids and has a molecular weight of 6,719 kDa44,45. The mump SH is a type I membrane protein expressed in infected cells46 and is involved in the disruption of immune system by inhibiting tumor necrosis factor alpha (TNF-α) mediated apoptosis and NF-κB activation47. The above literature data suggest that SH is involved in the mump infection. Therefore, the inhibition of the synthesis of SH protein could be used to prevent mumps infection.
Haemagglutinin‐neuraminidase
The Haemagglutinin‐neuraminidase (HN) is composed of 582 amino acids48. HN glycoproteins, found in the virus envelope are responsible for adhesion and mixing with targeted cells49. In the paramyxoviruses, HN plays a key role in membrane fusion process during viral infiltration, viral release and dissemination49. The neuraminidase activity of MuV-HN has a significant effect on viral budding, as it inhibits viral inflammation and facilitates the release of the virus over a host cell surface. The prior literature data suggest the significant role HN in mumps infection. Therefore, HN could be used as a target molecule in the prevention of mumps infection by inhibiting the HN involved cellular mRNA transcriptional process.
Large protein
The large protein is the largest protein with molecular weight 250 kDa in comparison with other viral structural units50. The prior results showed that viral membrane fuses with the host cell membrane and M–N proteins contacts disrupt which leads to the release of the nucleocapsid, P and L proteins into the cytoplasm where all processes of viral gene expression occur50,51. During the transcription process, L transcribes the small segment of 40–60 bp in assistance with P and releases an uncapped leader RNA and starts mRNA synthesis of the first protein gene. Furthermore, the L protein also takes part in multiple modifications such as 5′ end capping or 3′ end poly-A tailing50. Therefore, the L protein may be used as a target molecule to cure the mump infections by controlling the cellular transcriptional regulatory events through the interaction with miRNA (hsa_miR_6794_5p).
Molecular docking analysis
Based on our predictions and literature data, miRNA and the mRNA encoding the L protein was employed in docking procedure to check the binding conformation of hsa_miR_6794_5p with mRNA of L. The 3D structures of hsa_miR_6794_5p and mRNA of L were analyzed in detail to determine the significance of various nucleotides in the bound conformation. The best docking scoring value is −568.71 for the docking complex which demonstrates the best geometric shape complementarity. The detailed docking scoring values have been shown in Fig. 10.
The interactions between the non-coding RNAs (e.g. lncRNA; miRNA) and receptor molecules plays an important regulatory role in various biological processes52,53. Therefore, multiple computational methods have been used to predict their interactive profiles54,55,56,57. The interactive behavior of hsa_mir-6794-5p and mRNA of L was thoroughly examined, and our results show that multiple nucleotides of miRNA_6794 interact closely with mRNA of L. The nucleotides guanine and adenine (GA) at positions 7 and 8 of miRNA_6794 formed close interaction with mRNA of L. Similarly, uracil, guanine and guanine (UGG) at positions 10, 11 and 12 also showed close interactions with mRNA L. Similarly, different regions in mRNA-L such as CG, CU, C and U have been predicted as target receptors having potential to binds with hsa_mir_6794-5p (see Fig. 11). The docking results showed that binding of hsa_mir-6794-5p to mRNA-L may stop the translation process of the protein and can possibly prevent mumps virus infection. Of course, our computational predictions require future, detailed clinical studies.
Mechanistic pathways of miRNAs
miRNAs are small, highly conserved non-coding RNA molecules (containing about 22 nucleotides) involved in the regulation of different gene expression (Fig. 12). miRNAs are transcribed by RNA polymerases II and III, generating precursors that undergo a series of cleavage events to form mature miRNAs58. The pri-miRNA (primary transcript) is converted into pre-miRNA (stem-loop structure about 70 base pairs long) using DROSA enzyme, core nuclease that executes the initiation step of miRNA processing in the nucleus 59,60.
miRNAs and immune response
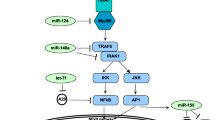
miRNAs regulate the immune responses (including both acquired and innate immunity) against mumps infection by activating various protein signaling cascades in humans61. During infection, a variety of intracellular signaling pathways are activated at different receptors sites such as TNF receptor-related substances (TRAFs), and interferon-regulatory features (IRFs) through different proteins SH, HN, M, L, F and V/P, respectively. The HN is involved in the mumps infection; therefore hsa_miR_6794_5p that binds with HN may disrupt its downstream signaling pathway (Fig. 13). Moreover, L and P are also key players in the activated signaling pathways after incorporation of pathogen within the host cells62. Our computational results indicate that hsa_miR_6794_5p has potential to bind to complementarity sites of mRNA of NH and stop the further transcriptional process. As a result, hsa_miR_6794_5p may play an important role in the prevention of mumps63 .
In the signaling pathway, the replication of viral genome is a two-step process modulated by the viral polymerase complex (composed of the phosphoprotein (P) and the large protein (L)) in which antigenome mediators are produced by genomic templates prior to production of negative sense genomes. Helical nucleocapsid is synthesized by a combination of newly formed nucleoprotein (NP) through the nascent genomic RNA. After that, the RNA-dependent RNA polymerase complex (RNP) interacts with newly formed structure and is transported to plasma membranes by M protein where it interacts with the surface glycoprotein before membrane removal and release of viral particles.
Conclusion
miRNA‐based therapy is a new promising approach including computational modeling against different diseases such as cardiovascular and neurological disorders, tumorigenesis, and viral infections64,65,66,67. The current research represents an in-silico method for detecting miRNAs aimed at silencing the MuV genome. Our results show that hsa_miR_6794-5p exhibits very good binding complementarity with the mumps genome. Furthermore, several proteins have been predicted as useful targets for the mumps therapeutics. Overall, the L protein was repeatedly predicted as having a significant role in the mumps infection. In addition, the docking results showed a binding pattern of hsa_mir_6794_5p against the mRNA of the L protein. Our results explore the nucleotide interaction profiles which may play an important role in the inhibition activity of the L gene transcription. The present work proposes a systematic approach to find the miRNAs by using miRNA target prediction algorithms and aims to silence the virus that affects humans by RNA interference. The identification of therapeutic miRNAs may be ranked among the most revolutionary and exciting therapeutic discoveries for pharmaceutical industry, after successful completion of several phases of clinical trials. Additionally, a better understanding of miRNA-lncRNA interactions54 will lead in the future to the design of miRNAs based chemical scaffolds and novel vaccines.
Data availability
The data used or analyzed in the current study are available at different online resources including https://www.ncbi.nlm.nih.gov/; http://www.mirbase.org/; http://cm.jefferson.edu/rna22v1.0/; https://tools4mirs.org/software/target_prediction/miranda/; http://plantgrn.noble.org/psRNATarget/; https://major.iric.ca/MC-Fold/; https://major.iric.ca/MC-Sym/; and https://rnacomposer.cs.put.poznan.pl/, respectively. All the relevant data has been mentioned in the manuscript whereas, rest of supporting information has been mentioned in the supplementary file.
Abbreviations
- MuV:
-
Mumps virus
- miRNAs:
-
MicroRNAs
- NP:
-
Nucleoprotein
- V/P:
-
Virus/phosphoproteins
- M:
-
Matrix
- F:
-
Fusion
- SH:
-
Small hydrophobic
- HN:
-
Haemagglutinin‐neuraminidase
- L:
-
Large
- UTRs:
-
Untranslated regions
- HR:
-
Heptad repeat
- TRAFs:
-
TNF receptor-related substances
- IRFs:
-
Interferon-regulatory features
References
Zamir, C. S., Schroeder, H., Shoob, H., Abramson, N. & Zentner, G. Characteristics of a large mumps outbreak: Clinical severity, complications and association with vaccination status of mumps outbreak cases. Hum. Vaccin. Immunother. 11, 1413–1417 (2015).
Wu, H. et al. Mumps virus infection disrupts blood-testis barrier through the induction of TNF-α in Sertoli cells. FASEB J. 33, 12528–12540 (2019).
Rubin, S. A. et al. Recent mumps outbreaks in vaccinated populations: no evidence of immune escape. J. Virol. 86, 615–620 (2012).
Hviid, A., Rubin, S. & Mühlemann, K. Mumps. Lancet 371, 932–944 (2008).
Lagos-Quintana, M., Rauhut, R., Meyer, J., Borkhardt, A. & Tuschl, T. New microRNAs from mouse and human. RNA 9, 175–179 (2003).
Oliveto, S., Mancino, M., Manfrini, N. & Biffo, S. Role of microRNAs in translation regulation and cancer. World J. Biol. Chem. 8, 45 (2017).
Li, L., Xu, J., Yang, D., Tan, X. & Wang, H. Computational approaches for microRNA studies: A review. Mamm. Genome 21, 1–12 (2010).
Mathuria, A., Mehak & Mani, I. Advances in Bioinformatics. 113–136 (Springer, 2024).
Wilson, R. L. et al. Function of small hydrophobic proteins of paramyxovirus. J. Virol. 80, 1700–1709 (2006).
Furukawa, T. et al. Role of the CM2 protein in the influenza C virus replication cycle. J. Virol. 85, 1322–1329 (2011).
Hassan, M. et al. Prediction of site directed miRNAs as key players of transcriptional regulators against influenza C virus infection through computational approaches. Front. Mol. Biosci. 9, 866072 (2022).
Matvienko, M. CLC Genomics Workbench. Plant and Animal Genome (. Sr. Field Application Scientist, CLC Bio, 2015).
Saini, H. K., Enright, A. J. & Griffiths-Jones, S. Annotation of mammalian primary microRNAs. BMC Genomics 9, 1–19 (2008).
Kozomara, A. & Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42, D68–D73 (2014).
John, B. et al. Human microRNA targets. PLoS Biol. 2, e363 (2004).
Huynh, T. et al. A pattern-based method for the identification of microRNA-target sites and their corresponding RNA. RNA Complex. Cell 126, 1203–1217 (2006).
Loher, P. & Rigoutsos, I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics 28, 3322–3323 (2012).
Witkos, T., Koscianska, E. & Krzyzosiak, W. Practical aspects of microRNA target prediction. Curr. Mol. Med. 11, 93–109 (2011).
Dai, X., Zhuang, Z. & Zhao, P. X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46, W49–W54 (2018).
Dai, X. & Zhao, P. X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159 (2011).
Racine, J. S. RStudio: A platform-independent IDE for R and Sweave. 167-172 (2012).
Wickham, H., Chang, W. & Wickham, M. H. Package ‘ggplot2’. Create Elegant Data Visualisations Using the Grammar of Graphics. Version 2. 1–189 (2016).
Parisien, M. & Major, F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 452, 51–55 (2008).
Biesiada, M., Purzycka, K. J., Szachniuk, M., Blazewicz, J. & Adamiak, R. W. RNA Structure Determination. 199–215 (Springer, 2016).
Meng, E. C., Pettersen, E. F., Couch, G. S., Huang, C. C. & Ferrin, T. E. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinform. 7, 1–10 (2006).
Schneidman-Duhovny, D., Inbar, Y., Nussinov, R. & Wolfson, H. J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363–W367 (2005).
Vaidya, S. R., Raut, C. G., Chowdhury, D. T. & Hamde, V. S. Complete genome sequence of mumps virus isolated from Karnataka State, India. Genome Announc. https://doi.org/10.1128/genomea.01429-01416 (2017).
Maldonado, Y. A. & Shetty, A. K. Principles and Practice of Pediatric Infectious Diseases 1180–1185. e1182 (Elsevier, 2023).
Titze-de-Almeida, S. S. et al. The promise and challenges of developing miRNA-based therapeutics for Parkinson’s disease. Cells 9, 841 (2020).
Yao, Q. & Compans, R. W. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology 223, 103–112 (1996).
Wichadakul, D., Mhuantong, W., Jongkaewwattana, A. & Ingsriswang, S. BMC Genomics. 1–8 (BioMed Central).
Zhou, X., Duan, X., Qian, J. & Li, F. Abundant conserved microRNA target sites in the 5′-untranslated region and coding sequence. Genetica 137, 159–164 (2009).
Mufson, M. A., Örvell, C., Rafnar, B. & Norrby, E. Two distinct subtypes of human respiratory syncytial virus. J. Gen. Virol. 66, 2111–2124 (1985).
Cox, R. et al. Characterization of a mumps virus nucleocapsid like particle. J. Virol. 83, 11402–11406 (2009).
Xu, P. et al. The V protein of mumps virus plays a critical role in pathogenesis. J. Virol. 86, 1768–1776 (2012).
Mann, B. A. et al. Vaccinia virus blocks Stat1-dependent and Stat1-independent gene expression induced by type I and type II interferons. J. Interferon & Cytokine Res. 28, 367–380 (2008).
Pohl, C., Duprex, W. P., Krohne, G., Rima, B. K. & Schneider-Schaulies, S. Measles virus M and F proteins associate with detergent-resistant membrane fractions and promote formation of virus-like particles. J. Gen. Virol. 88, 1243–1250 (2007).
Pei, Z., Bai, Y. & Schmitt, A. P. PIV5 M protein interaction with host protein angiomotin-like 1. Virology 397, 155–166 (2010).
Battisti, A. J. et al. Structure and assembly of a paramyxovirus matrix protein. Proc. Natl. Acad. Sci. 109, 13996–14000 (2012).
Li, M. et al. Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus-like particles. J. Virol. 83, 7261–7272 (2009).
Hüttl, S., Hoffmann, M., Steinmetzer, T., Sauder, C. & Krüger, N. The amino acid at position 8 of the proteolytic cleavage site of the mumps virus fusion protein affects viral proteolysis and fusogenicity. J. Virol. 94, e01732-e11720 (2020).
Liu, Y. et al. Structural characterization of mumps virus fusion protein core. Biochem. Biophys. Res. Commun. 348, 916–922 (2006).
Šantak, M., Örvell, C. & Gulija, T. K. Identification of conformational neutralization sites on the fusion protein of mumps virus. J. Gen. Virol. 96, 982–990 (2015).
Elango, N., Kövamees, J., Varsanyi, T. & Norrby, E. mRNA sequence and deduced amino acid sequence of the mumps virus small hydrophobic protein gene. J. Virol. 63, 1413–1415 (1989).
Franz, S. et al. Mumps virus SH protein inhibits NF-κB activation by interacting with tumor necrosis factor receptor 1, interleukin-1 receptor 1, and Toll-like receptor 3 complexes. J. Virol. 91, e01037-e11017 (2017).
Takeuchi, K., Tanabayashi, K., Hishiyama, M. & Yamada, A. The mumps virus SH protein is a membrane protein and not essential for virus growth. Virology 225, 156–162 (1996).
Franz, S. et al. Mumps virus SH protein inhibits NF-κB activation by interacting with tumor necrosis factor receptor 1, interleukin-1 receptor 1, and Toll-like receptor 3 complexes. J. Virol. https://doi.org/10.1128/jvi.01037-01017 (2017).
Jin, L., Beard, S., Hale, A., Knowles, W. & Brown, D. W. The genomic sequence of a contemporary wild-type mumps virus strain. Virus Res. 70, 75–83 (2000).
Forgione, R. E. et al. Structural basis for Glycan-receptor binding by mumps virus hemagglutinin-neuraminidase. Sci. Rep. 10, 1–13 (2020).
Ivashchenko, S. Investigating the Role of Intrinsic Conformational Disorder in Mumps Virus Proteins. (Université Grenoble Alpes, 2019).
Almansour, I. Mumps vaccines: Current challenges and future prospects. Front. Microbiol. 11, 1999 (2020).
Zhang, L., Yang, P., Feng, H., Zhao, Q. & Liu, H. Using network distance analysis to predict lncRNA–miRNA interactions. Interdiscip. Sci. Comput. Life Sci. 13, 535–545 (2021).
Zhao, J., Sun, J., Shuai, S. C., Zhao, Q. & Shuai, J. Predicting potential interactions between lncRNAs and proteins via combined graph auto-encoder methods. Brief. Bioinform. 24, bbac527 (2023).
Wang, W., Zhang, L., Sun, J., Zhao, Q. & Shuai, J. Predicting the potential human lncRNA–miRNA interactions based on graph convolution network with conditional random field. Brief. Bioinform. 23, bbac463 (2022).
Sun, F., Sun, J. & Zhao, Q. A deep learning method for predicting metabolite–disease associations via graph neural network. Brief. Bioinform. 23, bbac266 (2022).
Wang, T., Sun, J. & Zhao, Q. Investigating cardiotoxicity related with hERG channel blockers using molecular fingerprints and graph attention mechanism. Comput. Biol. Med. 153, 106464 (2023).
Hu, H. et al. Gene function and cell surface protein association analysis based on single-cell multiomics data. Comput. Biol. Med. 157, 106733 (2023).
MacFarlane, L.-A. & Murphy, P. MicroRNA: Biogenesis, function and role in cancer. Curr. Genomics 11, 537–561 (2010).
Lee, Y. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003).
Nguyen, T. H. et al. Potential role of microRNAs in the regulation of antiviral responses to influenza infection. Front. Immunol. 9, 1541–1541. https://doi.org/10.3389/fimmu.2018.01541 (2018).
Zhang, Y. & Li, Y.-K. MicroRNAs in the regulation of immune response against infections. J. Zhejiang Univ. Sci. B 14, 1–7 (2013).
Ludwig, S. Disruption of virus–host cell interactions and cell signaling pathways as an anti-viral approach against influenza virus infections. Biol. Chem. 392, 837–847 (2011).
Gaur, P., Munjal, A. & Lal, S. K. Influenza virus and cell signaling pathways. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 17, 148 (2011).
Ying, H. et al. miRNAs; A novel strategy for the treatment of COVID-19. Cell Biol. Int. 45, 2045–2053 (2021).
Li, X. et al. Caspase-1 and Gasdermin D afford the optimal targets with distinct switching strategies in NLRP1b inflammasome-induced cell death. Research (2022).
Jin, J., Xu, F., Liu, Z., Shuai, J. & Li, X. Quantifying the underlying landscape, entropy production and biological path of the cell fate decision between apoptosis and pyroptosis. Chaos Solitons Fract. 178, 114328 (2024).
Jin, J. et al. Biphasic amplitude oscillator characterized by distinct dynamics of trough and crest. Phys. Rev. E 108, 064412 (2023).
Acknowledgements
A.K. acknowledges financial support from NIH Grants R01GM127701, and R01HG012117. M.H. acknowledges The Ohio State University for “President’s Postdoctoral Scholars Program (PPSP)” fellowship for financial support.
Author information
Authors and Affiliations
Contributions
Conceptualization: M.H., and M.S.I.; Methodology and Analysis: M.H., M.S.I., Z.Y., and S.S. Writing: original draft preparation, M.H., M.S.I., Z.Y., S.S.; Proof reading and final approval: A.K.
Corresponding author
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hassan, M., Shahzadi, S., Iqbal, M.S. et al. Exploration of microRNAs as transcriptional regulator in mumps virus infection through computational studies. Sci Rep 14, 18850 (2024). https://doi.org/10.1038/s41598-024-67717-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-67717-z
- Springer Nature Limited