Abstract
The lateral hypothalamic area (LHA) is essential for ingestive behavior but it remains unclear how LHA neurons coordinate feeding vs. drinking. Most LHA populations promote food and water consumption but LHA neurotensin (Nts) neurons preferentially induce water intake while suppressing feeding. We identified two molecularly and projection-specified subpopulations of LHA Nts neurons that are positioned to coordinate either feeding or drinking. One subpopulation co-expresses the long form of the leptin receptor (LepRb) and is activated by the anorectic hormone leptin (NtsLepRb neurons). A separate subpopulation lacks LepRb and is activated by dehydration (NtsDehy neurons). These molecularly distinct LHA Nts subpopulations also differ in connectivity: NtsLepRb neurons project to the ventral tegmental area and substantia nigra compacta but NtsDehy neurons do not. Intriguingly, the LHA Nts subpopulations cannot be discriminated via their classical neurotransmitter content, as we found that all LHA Nts neurons are GABAergic. Collectively, our data identify two molecularly- and projection-specified subpopulations of LHA Nts neurons that intercept either leptin or dehydration cues, and which conceivably could regulate feeding vs. drinking behavior. Selective regulation of these LHA Nts subpopulations might be useful to specialize treatment for ingestive disorders such as polydipsia or obesity.
Similar content being viewed by others
Introduction
The lateral hypothalamic area (LHA) of the brain receives inputs from osmotic and energy-sensing sites and projects to centers coordinating goal-directed ingestive behavior to maintain homeostasis1,2,3,4,5,6. Early studies defined the LHA as a “feeding center” because animals with lesion of LHA cell bodies lost all motivation to eat7,8. Less emphasized, but equally important, is that animals with LHA lesions also lost the motivation to drink water and their resulting dehydration causes death well before starvation9,10. Intriguingly, destroying passing dopaminergic fibers within the LHA similarly produces aphagia and adipsia, revealing that the LHA acts in concert with the dopamine system11. Thus, the LHA modifies both types of ingestive behavior necessary for survival, but via incompletely understood mechanisms.
The discovery of molecularly- and projection-specified populations of neurons within the LHA suggested that some of them might be specialized to coordinate drinking vs. feeding. Yet, most of the LHA populations studied to date indiscriminately promote intake of food and water. LHA neurons expressing melanin concentrating hormone promote intake of both substances and do not specifically organize feeding vs. drinking12,13. A separate population of orexin/hypocretin-expressing LHA neurons regulate arousal-dependent behaviors, including feeding, drinking and locomotor activity, but do not specify a particular ingestive behavior14,15,16. LHA neurons have also been distinguished by their expression of the classical neurotransmitters glutamate or GABA. Inhibiting LHA glutamate neurons increases intake of a palatable “meal replacement” drink17, but it is unclear if this is an effort to obtain fluid, calories or if both ingestive behaviors are modulated by these neurons. Activation of all LHA GABA neurons increases behaviors to obtain food and liquids, but also invokes gnawing at non-caloric objects such as wood or the cage floor18,19,20; thus, bulk activation of LHA GABA neurons cannot be considered to direct any specific ingestive behavior. While en masse activation of LHA GABA neurons is unlikely to occur in nature, there are subpopulations of LHA GABAergic neurons18 that may be activated by different physiologic cues to control intake. For example, LHA GABA neurons co-expressing the neuropeptide galanin mediate food seeking21. Conversely, the subset of LHA GABA neurons co-expressing the long form of the leptin receptor (LepRb) limit feeding with no effect on water intake22,23. It is therefore possible that subsets of LHA GABA neurons might be activated by distinct physiologic cues, and hence differentially control food vs. water intake. However, the studies of LHA populations to date do not explain how the LHA specifically coordinates feeding or osmolality cues to direct the appropriate ingestive behavior.
We recently characterized a large population of LHA neurons that express the neuropeptide Neurotensin (Nts) and are separate from MCH or orexin/hypocretin neurons22,24. Unlike other LHA populations that promote both food and water consumption, experimental activation of LHA Nts neurons promotes voracious drinking but restrains feeding24. Since LHA Nts neurons have been reported to contain glutamate25 or GABA26,27, we hypothesized that there might be neurochemically, molecularly, and functionally heterogeneous subsets of LHA Nts neurons to coordinate drinking vs. feeding. Indeed, some (but not all) LHA Nts neurons co-express the long form of the leptin receptor (LepRb) and GABA and are activated by the anorectic hormone leptin21,26; we refer to these as NtsLepRb neurons. This NtsLepRb population comprises a small, but essential subset of LHA Nts neurons necessary to mediate the anorectic response to leptin and proper regulation of energy balance22. Yet, mice lacking LepRb in LHA NtsLepRb neurons do not exhibit any disruptions in drinking or bodily fluid content, suggesting that LHA Nts-mediated drinking might be mediated via different LHA Nts neurons22. Some LHA Nts neurons are responsive to physiologic changes in serum osmolality, as dehydration increases expression of Nts mRNA within the LHA28; we refer to these as NtsDehy neurons. Exogenous Nts treatment also promotes drinking29, although the endogenous sources of Nts mediating this effect remained unknown. Given that experimental activation of LHA Nts neurons promotes Nts release24,27 and drinking24,27,30,31, the dehydration-induced upregulation of LHA Nts could serve as a physiologic signal to drive water seeking and intake once water becomes available32. Taken together, these data suggest that some LHA Nts neurons can be activated by cues of energy or osmolality status and might comprise separate populations to coordinate feeding or drinking behavior. We therefore assessed whether LHA NtsLepRb neurons and NtsDehy neurons are the same, or whether they are separate populations that are distinguishable via molecular, circuit and neurotransmitter criteria.
Results
Methods to Identify LHA Nts Neurons
In situ hybridization (ISH) identifies many Nts-expressing cells within the LHA (Fig. 1A,B)28 but does not easily permit determination of co-expressed transcripts or circuit tracing. Nts immunofluorescence (Nts-IF) permits such analyses, but only labels fibers, not cell bodies (Fig. 1C) unless mice were pre-treated with the axonal transport inhibitor, colchicine (Fig. 1D)33,34. Since colchicine induces neuronal dysfunction and lethality its use prohibits study of Nts contributions to normal physiology. Nonetheless, Nts-ISH and -IF confirm a large population of LHA neurons that actively express Nts (Fig. 1B,D). We reasoned that NtsCre mice would be useful to examine the molecular expression, projections, activation responses and neurochemistry of this large population of LHA Nts neurons without physiology-disrupting colchicine treatment. First, we verified the fidelity of this model for identifying Nts neurons by colchicine-treating NtsCre mice on a Cre-inducible GFP reporter background35 (NtsCre; GFP mice). We examined two brain regions that have been shown via Nts-ISH to contain numerous Nts neurons: the subthalamic nucleus (STN) and the LHA36. Similarly, NtsCre; GFP mice have dense populations of GFP-labeled neurons within the STN and LHA that co-label with Nts-IF (Fig. 1E–K). Thus, NtsCre; GFP mice reliably identify Nts neurons and can be used to characterize features of LHA Nts neurons.
Visualization of LHA Nts Neurons. (A) Coronal schematic from mouse brain atlas. The shaded square identifies the portion of the LHA shown in subsequent microscopy panels. (B) Nts-expressing cell bodies detected via ISH (courtesy of the Allen Brain Atlas36). (C) Nts-Immunofluoresence (IF) only identifies fibers within the LHA unless (D) mice were pretreated with ICV colchicine, which inhibits axonal transport and permits detection of Nts-IF within cell bodies (white outline arrows). (E) Nts-IF (red) in colchicine-treated NtsCre; GFP mouse that expresses GFP in Nts neurons (green). (F–H) Insets show the STN from E. (F) Many GFP-labeled Nts cell bodies are found within the STN consistent with Nts-ISH36 and (G) the Nts-IF cell bodies in this region (H) entirely overlap with the GFP (Nts) cells (yellow outline arrows). (I–K) Insets show the LHA from (E), where (I) the GFP-labeled cell bodies and (J) Nts-IF cell bodies (K) overlap (yellow outline arrows). Together these data confirm that NtsCre; GFP mice correctly identify Nts-expressing cells, and can be used to visualize them.
Nts Cre; GFP Mice Confirm that Some LHA Nts Neurons Project to the Midbrain
LHA Nts neurons project to two midbrain regions, the ventral tegmental area (VTA) and the substantia nigra compacta (SNc)26,37, which regulate motivated behaviors and motor function, respectively38,39. We therefore hypothesized that different subsets of LHA Nts neurons might project to the VTA or SNc. First, we identified LHA Nts projecting neurons by injecting NtsCre; GFP mice with FluoroGold (FG) in the VTA (Fig. 2A–D) or the SNc (Fig. 2E–H), which is taken up by terminals and transported retrogradely to the soma of origin. Examination of VTA-injected mice revealed many LHA cell bodies that accumulated FG, some of which also contained GFP and hence identify LHA Nts neurons that project to the VTA (Fig. 2B–D, yellow arrows). Yet, many LHA Nts neurons did not accumulate FG from the VTA (Fig. 2B–D, green arrows). This could be because some LHA Nts neurons do not project to the VTA, or if there was insufficient FG throughout the VTA subregions containing LHA Nts terminals. However, we also found adjacent FG-labeled cells lacking GFP, indicating non-Nts containing LHA cells that provide input to the VTA and that FG coverage was sufficient to label various LHA soma (Fig. 2B–D, red arrows). Analysis of SNc-injected mice revealed similar groups of labeled neurons (Fig. 2F–H). Together, these data verify that that some LHA Nts neurons project to the VTA and SNc, but nearly twice as many project to the VTA vs. the SNc. (Fig. 2I). Our methodology prevented determination of whether any LHA Nts neurons provide collateral projections to these areas. However, the differing number of projections hint that there may be some separate subpopulations of LHA Nts neurons that project to the VTA, SNc, or sites other than the midbrain.
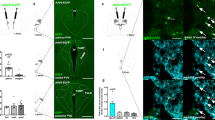
LHA Nts Neurons Project to the VTA and SNc (A) NtsCre; GFP mice were injected in the VTA with the retrograde tract tracer FluoroGold (FG). (B–D) Representative insets from the LHA showing (B) GFP-labeled Nts cell bodies (green) and (C) cell bodies that have accumulated FG (red) and from the VTA. (D) Some GFP-labeled Nts cells contain FG, indicating LHA Nts neurons that project to the VTA (yellow arrows) while GFP-labeled Nts cells lacking FG do not project to the VTA (green arrows). Some non-Nts cells also project to the VTA (red arrows). (E) NtsCre; GFP mice received FG into the SNc. F-H) Insets from the LHA show (F) GFP-labeled Nts cell bodies (green) and (G) cell bodies that have accumulated FG from the SNc (red). (H) The LHA contains cells co-labeled with GFP and FG indicating that they project to the SNc (yellow arrows), as well as GFP-labeled Nts neurons that lack FG and do not project to the SNc (green arrows). There are also non-Nts neurons that project to the SNc (red arrows). For (A–H), VTA-injected n = 12, SNc-injected n = 11. (I) Quantitation of the number of LHA Nts cells co-labeled with GFP and FG after FG injection into the VTA (n = 6) or the SNc (n = 5), **p < 0.01 via Student’s t-test. Together, these data demonstrate that some LHA Nts neurons project to the VTA and the SNc, with more projections targeting the VTA. Abbreviations: mt = mammillothalamic tract; f = fornix; LHA = lateral hypothalamic area.
NtsLepRb Neurons Are a Subset of LHA Nts Neurons that Project to the Midbrain
Some LHA Nts neurons can be molecularly distinguished via expression of LepRb (NtsLepRb neurons) and we reasoned that they might project to either the VTA or SNc. To identify NtsLepRb neurons we treated NtsCre; GFP mice with vehicle (Fig. 3A–C) or leptin (5 mg/kg i.p. 2–4 hr), which induces phosphorylation of STAT3 (pSTAT3) specifically in LepRb-expressing neurons. Some GFP-labeled LHA Nts neurons contained leptin-induced pSTAT3 and these are NtsLepRb neurons (Fig. 3D–F, cyan-outlined magenta arrows). Yet, many GFP-labeled LHA Nts neurons did not contain pSTAT3 (Fig. 3D–F, cyan arrows). These data confirmed that there are at least two molecularly distinct populations of LHA Nts neurons: one population expresses LepRb and can be revealed by leptin-induced pSTAT3 (the NtsLepRb neurons), but another LHA Nts population lacks LepRb.
LHA NtsLepRb Neurons Project to the Midbrain NtsCre; GFP mice were treated with (A–C) vehicle or (D–F) leptin (5 mg/kg, IP, 2–4 hr) to permit detection of (A) GFP-labeled Nts neurons (green) and (B) phosphorylated STAT3 (pSTAT3), a marker for leptin-activated LepRb neurons (blue). Cyan arrows label Nts neurons without pSTAT3. Magenta arrows identify pSTAT3 that does not co-label with GFP (e.g. LepRb neurons that do not express Nts). Cyan-outlined magenta arrows identify GFP-labeled Nts neurons that co-localize with pSTAT3 and are NtsLepRb neurons. (G–N) NtsCre; GFP mice received FG in the VTA or SNc and were treated with vehicle or leptin to permit identification of LepRb neurons via induction of pSTAT3. Examination of the LHA from VTA-injected mice revealed (G) GFP-labeled Nts neurons, (H) FG-labeled neurons that project to the VTA and (I) pSTAT3 neurons. (J) Merged panels identify some neurons containing GFP, FG and pSTAT3 that are NtsLepRb neurons that project to the VTA (white arrows). (K–N) Examination of the LHA from SNc-injected mice reveals (K) GFP-labeled Nts neurons, (L) FG-labeled neurons that project to the SNc and M) pSTAT3 neurons. (N) Merged panels identify some neurons containing GFP, FG and pSTAT3 that are NtsLepRb neurons that project to the SNc (magenta-outlined yellow arrows). Key for other arrows: cyan arrows = Nts-GFP only neurons; magenta arrows = pSTAT3-only (LepRb) neurons; cyan-outlined magenta arrows = NtsLepRb neurons that do not project to the VTA/SNc; yellow arrows = FG-only neurons that project to the VTA/SNc; cyan-outlined yellow arrows = Nts neurons that project to the VTA/SNc but do not contain LepRb; magenta-outlined yellow arrows = VTA/SNc projecting LepRb neurons that do no express Nts; white arrows = VTA/SNc-projecting NtsLepRb neurons. These data demonstrate that at least some NtsLepRb neurons project to the VTA and the SNc. VTA-injected vehicle-treated n = 6; female VTA-injected leptin-treated n = 10; SNc-injected vehicle-treated n = 5; SNc-injected leptin-treated n = 9.
Next, we asked whether the molecularly-specified NtsLepRb neurons are also projection-specified. NtsCre; GFP mice were injected with FG to label LHA Nts neurons that project to the VTA or SNc, and also treated with leptin to permit pSTAT3-mediated identification of NtsLepRb neurons. This paradigm identified NtsLepRb neurons that accumulated modest amounts of FG from the VTA (Fig. 3G–J, white arrows) and from the SNc (Fig. 3K–N, white arrows). In each case we also observed NtsLepRb neurons that did not accumulate FG; these may be NtsLepRb neurons that project to the midbrain region that was not injected with FG or to a yet undetermined site outside of the midbrain (Fig. 3G–N, cyan-outlined magenta arrows). As expected, we also observed LHA Nts neurons lacking LepRb, some of which project to the midbrain (Fig. 3G–P, cyan-outlined yellow arrows), but other LHA Nts neurons did not accumulate FG and hence do not project to either midbrain region (Fig. 3G–P, cyan arrows). Together, these data signify that NtsLepRb neurons are a molecularly-specified subset of LHA Nts neurons, some of which project to the VTA and SNc.
NtsDehy Neurons Are a Subset of LHA Nts Neurons that Do Not Project to the Midbrain
Since dehydration disrupts bodily osmolality and upregulates Nts mRNA in the LHA28, we postulated that it might also modify the activity of LHA Nts neurons. To test this, we provided NtsCre; GFP mice with ad lib water (euhydration) or removed water overnight (dehydration), then examined the LHA for GFP and cFos (a marker of recent neuronal depolarization). During euhydration we observed few LHA Nts neurons with cFos, suggesting that LHA Nts neurons are not activated during normal fluid balance (Fig. 4A–C). In contrast, overnight dehydration increased cFos within some (but not all) LHA Nts neurons (Fig. 4D–F, cyan outlined magenta arrows). Thus, dehydration activates a subset of LHA Nts neurons, termed NtsDehy neurons. We then assessed midbrain projections of NtsDehy neurons in dehydrated NtsCre; GFP mice previously injected with FG in the VTA or SNc. Although we observed NtsDehy neurons in the LHA (Fig. 4G–N, cyan-outlined magenta arrows), none accumulated FG from the VTA or SNc. FG accumulation within other LHA neurons confirmed successful retrograde labeling (Fig. 4G–N, yellow and cyan-outlined yellow arrows). Together with Fig. 3, these data indicate that some NtsLepRb neurons project to the midbrain but NtsDehy neurons do not. Given that NtsDehy neurons and NtsLepRb neurons have different projection targets, they must comprise distinct subsets of LHA Nts neurons.
LHA NtsDehy Neurons Do Not Project to the Midbrain. (A–F) NtsCre; GFP mice were given ad libitum water (Euhydrated) or were dehydrated overnight. Brains were assessed for GFP-labeled Nts neurons (green) and cFos, a marker of recent neuronal depolarization (blue). Cyan arrows label Nts-only neurons and magenta arrows identify dehydration activated neurons that do not express Nts. Cyan-outlined magenta arrows identify Nts-GFP neurons that co-express cFos (NtsDehy neurons). (G–N) NtsCre; GFP mice were injected with FG into the VTA or SNc (to identify midbrain projecting neurons) and dehydrated overnight (to identify dehydration-activated neurons via cFos). Assessment of the LHA revealed some NtsDehy neurons (Cyan-outlined magenta arrows) that did not accumulate FG. Similarly, (K-N) NtsDehy neurons were found within the LHA of SNc-injected mice but none of these contained FG. Key for arrows: cyan arrows = Nts-only neurons; magenta arrows = cFos-only (dehydration-activated) neurons; cyan-outlined magenta arrows = NtsDehy neurons that do not project to the VTA/SNc; yellow arrows = FG-only neurons that project to the VTA/SNc; cyan-outlined yellow arrows = Nts neurons that project to the VTA/SNc but are not activated by dehydration; magenta-outlined yellow arrows = dehydration-activated VTA/SNc projecting neurons that do no express Nts. No arrows are present to label VTA/SNc-projecting NtsDehy neurons because no such neurons were found. VTA-injected, euhydrated n = 5; VTA-injected, dehydrated n = 9; SNc-injected, euhydrated n = 5, SNc-injected, dehydrated n = 9.
NtsLepRb and NtsDehy Neurons Are Separate Subpopulations of LHA Nts Neurons
We reasoned that if NtsLepRb neurons are distinct from NtsDehy neurons they would not be activated by dehydration. The requirement for leptin or dehydration to functionally identify NtsLepRb and NtsDehy neurons meant that we could not simultaneously label these subsets in NtsCre; GFP mice. Instead, to test our hypothesis we assessed cFos in brains from dehydrated NtsCre; GFP mice (where GFP identifies all LHA Nts neurons) and LepRbCre; GFP mice (where GFP identifies all LepRb neurons). Since the majority of LHA LepRb neurons co-express Nts, the LepRbCre; GFP mice identify the NtsLepRb neurons as well as some non-Nts expressing neurons26. While dehydration significantly increased the percentage of LHA Nts neurons containing cFos (Fig. 5A,C), it did not alter the proportion of LHA LepRb neurons containing cFos compared to the euhydrated state (Fig. 5B,C). Because LHA LepRb neurons contain the subset of NtsLepRb neurons, the absence of induced LepRb-cFos indicates that NtsLepRb neurons are not activated by dehydration. Conversely, other Nts neurons lacking LepRb can be activated by dehydration. These data indicate that NtsLepRb neurons are functionally distinct from NtsDehy neurons, as distinguished via their response to dehydration. Furthermore, these subpopulations of LHA Nts neurons are molecularly distinct, such that LepRb expression can be used to distinguish NtsLepRb neurons from NtsDehy neurons. Taken together, data from Figs 3–5 reveal that LHA Nts neurons are not a homogeneous population but contain molecularly and projection-specified subsets of NtsLepRb neurons and NtsDehy neurons that are differentially activated in response to changes in energy or fluid balance, respectively.
LHA NtsLepRb Neurons are Distinct from NtsDehy Neurons. Mice that express GFP in all LepRb neurons (LepRbCre; GFP mice) and mice expressing GFP in all Nts neurons (NtsCre; GFP mice) were euhydrated or dehydrated overnight. Brains were immunostained for GFP and cFos, a marker of recent neuronal activation. (A-A’) In NtsCre; GFP mice dehydration induced cFos within LHA Nts neurons. (B-B’) In LepRbCre; GFP mice dehydration induced cFos in the LHA, but there is no colocalization of cFos with LepRb neurons. (C) Percentage of NtsGFP and LepRbGFP in the LHA that contain cFos (e.g. activated neurons) in NtsCre; GFP mice and LepRbCre; GFP mice. Dehydration specifically increases the percentage of GFP + cFos neurons in the LHA, indicating that some LHANts neurons are activated by dehydration. Dehydration did not increase the percentage of GFP + cFos in LepRbGFP neurons, which encompass the population of NtsLepRb neurons. These data signify that dehydration does not activate LepRb neurons and therefor the LepRb expressing subpopulation of LHA NtsLepRb neurons is necessarily distinct from LHA NtsDehy neurons.
Verification of Cre-Dependent Reagents to Determine Classical Neurotransmitter Content
Given the heterogeneity of LHA Nts neurons at the molecular and circuit level, we hypothesized that they might differ in other ways, such as in classical neurotransmitter content. Neuropeptide-expressing neurons may contain GABA or glutamate, release of which determines whether synaptic targets are inhibited or activated, respectively25. LHA Nts neurons have been reported as GABAergic24 or glutamatergic40, thus, we sought to define whether subpopulations might be neurochemically distinguishable. Since GABA and glutamate cell bodies cannot be labeled using immunoreagents, we crossed vGatCre and vGlut2Cre mice with Cre-inducible GFP reporter mice to label these cells with GFP (vGatCre; GFP and vGlut2Cre; GFP mice). A limitation of this model is that neurons can alter neurotransmitter expression over lifespan41, but recombination during development produces permanent GFP-labeling that may not reflect the neurochemistry of the mature neuron. Thus, we also injected adult vGatCre and vGlut2Cre mice with an AAV Cre-Lox-red fluorescent protein (RFP) so that only actively-expressing GABA and glutamate-expressing neurons undergo recombination to express RFP. Finally, we compared these reporter models with ISH data in brain regions known to primarily contain glutamatergic neurons (the STN) or GABAergic neurons (the zona incerta, ZI) to verify the fidelity of each method for identifying neurotransmitter-expressing neurons in the adult brain. We observed similar distributions of vgat-ISH, and GFP- or AAV- Lox-RFP-labeled vGAT neurons in the ZI, but no labeled neurons in the adjacent STN (Fig. 6A–F). In contrast, ISH, GFP- and AAV-Lox-RFP labeling of vGlut2 cells all confirm the absence of glutamatergic neurons in the ZI but each method identified glutamatergic neurons in the STN (Fig. 6G–L). Hence, these genetic and viral methods accurately identify GABA and glutamate neurons and can be used to examine the neurotransmitter content of the LHA.
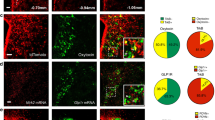
Recombianse Methods to Visualize GABA, Glutamate and Nts Neurons (A) ISH for vGat from the Allen Brain Atlas36. (B,B’) Digital magnifications of the STN and Zona Incerta from (A) show that the STN lacks vGat expression while the ZI contains many vGat-expressing cell bodies. (C,D,D’) vGatCre; GFP mice show a similar distribution of vGat-GFP labeled cells as vGat-ISH (A), including few vGat-GFP cells in the STN and many within the ZI. (E,F,F’) vGatCre mice injected with AAV-Lox-RFP have no RFP labeling in the STN but many ZI-labeled RFP cells, and mirror the distribution observed via vGat-ISH and from vGatCre; GFP mice. (G) ISH for vGlut2 from the Allen Brain Atlas. Boxed regions from (G) are digitally magnified in (H,H’) and identify many vGlut2 cell bodies in the STN but none within the ZI. (I,J,J’) vGlut2Cre; GFP mice identify a similar distribution of GFP-labeled vGlut2 cells as the vGlut2-ISH, including many GFP vGlut2-GFP cells in the STN but none in the ZI. (K,L,L’). Likewise, vGlut2Cre mice injected in the LHA with AAV-Lox-RFP have some viral spread and vGlut2-RFP labeled cell bodies in the STN but none in the ZI. Collectively these data demonstrate that vGatCre and vGlut2Cre mice can be used with Cre-inducible reporter mice or AAVs to reliably identify vGat and vGlut neurons. (M–N”) Nts-ISH identifies many Nts-containing cell bodies within the LHA and some within the ZI and STN, courtesy of Allen Brain atlas36. (O–P”) WT mouse injected with AAV-Frt-GFP in the LHA shows no induced GFP expression within the LHA, ZI or STN. (Q–R”) NtsFlpO mouse injected with AAV-Frt-GFP in the LHA shows many GFP-labeled cell bodies around the injection site and within the ZI and STN similar to the distribution of Nts-ISH (M). These data confirm the specificity of NtsFlpO model and AAV-Frt-GFP to identify Nts neurons in a Cre-independent manner. Abbreviations: mt = mammillothalamic tract; f = fornix; LHA = lateral hypothalamic area, STN = Sub-thalamic Nucleus, ZI = Zona Incerta.
Nts FlpO Mice Identify LHA Nts Neurons Without the Use of Cre
Defining the neurotransmitter content of LHA Nts neurons requires simultaneous labeling of neurons expressing Nts and vGat or vGlut2. Problematically, the use of vGatCre and vGlut2Cre mice necessary for detection of GABA and glutamate neurons precludes simultaneous Cre-dependent detection of Nts using NtsCre mice. To overcome this limitation, we generated NtsFlpO mice to visualize Nts neurons via a non-Cre, FlpO-dependent mechanism. To verify the specificity of the system we injected NtsFlpO mice and control mice lacking FlpO (WT) with an AAV that drives FlpO-inducible expression of GFP (AAV-Frt-GFP) (Fig. 6M–R”). We observed no GFP-labeled cells in the LHA, ZI and STN when WT mice were injected with AAV-Frt-GFP, despite the many Nts-expressing cells in these regions demonstrated via Nts-ISH (Fig. 6M–P”); these data confirm the FlpO-dependence for GFP expression. By contrast, injection of AAV-Frt-GFP into the LHA of NtsFlpO mice resulted in GFP-labeled cell bodies within the LHA, ZI and STN (Fig. 6Q,R”). The distribution of GFP cells in NtsFlpO mice is similar to that of Nts-ISH (Fig. 6M) and NtsCre; GFP mice, although fewer cells are reported; these results are consistent with the limited recombination efficiency of FlpO as compared to Cre42.
Determination of Classical Neurotransmitter Content Within LHA Nts Neurons Using the Dual Recombinase System
We generated dual recombinase mice to simultaneously label Nts cells and vGat or vGlut2-expressing cells by crossing NtsFlpO mice to vGatCre or vGlut2Cre mice, producing NtsFlpO; vGatCre and NtsFlpO; vGlutCre mice respectively. To test the fidelity of these dual recombinase mice, we injected them in the ZI with AAV-Frt-GFP (to permit FlpO-mediated expression of GFP that identifies Nts neurons) and AAV-Lox-RFP (to permit Cre-mediated expression of RFP for detection of vGat or vGlut2 neurons). Since the ZI contains primarily GABAergic but not glutamatergic neurons as well as some Nts neurons (Figs 1 and 6), the dual recombinase system should only result in GFP and RFP co-labeling of ZI cells in NtsFlpO; vGatCre mice, but not in NtsFlpO; vGlutCre mice. As anticipated, dual AAV injection into the ZI of NtsFlpO; vGlutCre mice yielded many GFP-labeled Nts neurons, none of which contain RFP-vGlut2 (Fig. 7A,B”, cyan arrows). The GABAergic ZI is in fact devoid of RFP-vGlut2, despite the robust induction of RFP in surrounding regions known to contain glutamate (Fig. 7A). By contrast, in dual AAV-injected NtsFlpO; vGatCre mice, we observed that all of the GFP-labeled Nts cells co-label with RFP-vGat, indicating that ZI Nts neurons are GABAergic (Fig. 7C,D”; white arrows indicate co-labeled cells, magenta arrows identify RFP-vGat cells that do not contain GFP-Nts). Taken together, these data confirm that the ZI Nts cells are GABAergic, but not glutamatergic, as would be expected of this primarily GABAergic brain region. Our findings also confirmed the reliability of the dual recombinase system to distinguish classical neurotransmitter content, so we next used it to determine whether subsets of LHA Nts neurons can be discriminated via their classical neurotransmitter expression. Dual AAV injection into the LHA of NtsFlpO; vGlutCre mice identified GFP-Nts neurons confined within the perifornical LHA (Fig. 7E,F”, cyan arrows) and RFP-vGlut2 neurons (Fig. 7E,F”, magenta arrows), but we did not observe any LHA cells that co-expressed both labels. By contrast, dual AAV injection into the LHA of NtsFlpO; vGatCre mice identified GFP-Nts neurons within the perifornical LHA, most of which co-labeled with RFP-vGat (Fig. 7G,H”, white arrows). We also observed many RFP-vGAT neurons that did not co-label with GFP-Nts (Fig. 7G,H” magenta arrows). Together these data suggest that LHA Nts neurons are predominantly GABAergic, and they comprise a subset within the larger population of LHA GABA neurons.
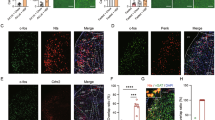
Neurotransmitter Content of LHA Nts Neurons. (A,B”) NtsFlpO; vGlut2Cre mice were injected in the ZI with AAV-Frt-GFP (to identify Nts neurons, green) and AAV Cre-RFP (to identify glutamate neurons, red). Digital magnification of the boxed area in A revealed many GFP-Nts neurons (cyan arrows) but no vGlut2 neurons in the ZI, consistent with the GABAergic nature of this brain region. (C,D”) The same dual AAV injection into the ZI of NtsFlpO; vGatCre mice identified vGat/GABA neurons in the ZI (magenta arrows), and all observed GFP-Nts neurons contain vGat2 (white arrows). These data confirm that the dual recombinase method discerns GABA vs glutamate-containing areas of the brain while also permitting identification of Nts neurons. (E,F”) Dual AAV injection into the LHA of NtsFlpO; vGlut2Cre mice identifies RFP-vGlut2 neurons (magenta arrows) and GFP-Nts neurons (cyan arrows). No overlapping RFP and GFP neurons were observed, indicating that LHA Nts neurons do not contain glutamate. (G,H”) Dual AAV injection into the LHA of NtsFlpO; vGatCre mice revealed many RFP-vGat neurons (magenta arrows), and also GFP-Nts neurons that overlapped with RFP-vGat cells (white arrows). Together, these data indicate that LHA Nts neurons express GABA but not glutamate. NtsFlpO; vGlut2cre n = 5, NtsFlpO; vGatCre n = 6). (I,N”) vGlut2Cre; GFP mice and vGatCre; GFP mice were treated with colchicine to permit detection of Nts-IF (red) and vGat- or vGlut2-GFP (green). (J,J”) In vGlut2Cre; GFP mice, many GFP-vGlut2 cell bodies are found in the LHA (magenta arrows) along with Nts-IF cell bodies (cyan arrows), but no overlapping cells were found. (K–K”) The ZI from vGlut2Cre; GFP mice contained a few Nts-IF neurons (cyan arrows) but no vGlut2-GFP cells, consistent with the GABAergic neurochemistry of the ZI. (M–M”) The LHA of vGatCre; GFP mice contained many GFP-vGat cell bodies (magenta arrows) and Nts-IF cells that all co-labeled with GFP-vGat (white arrows). (N-N”) Similarly, co-labeling was observed in the GABAergic ZI, (N-N”) but not in vGlut2Cre; GFP mice. These data confirm that LHA Nts neurons contain vGat and are GABAergic, but do not contain vGlut/glutamate. vGlut2Cre; GFP mice n = 4; vGatCre; GFP mice n = 5. Abbreviations: mt = mammillothalamic tract; f = fornix; LHA = lateral hypothalamic area, STN = Sub-thalamic Nucleus, ZI = Zona Incerta.
Determination of Classical Neurotransmitter Content of LHA Nts Neurons Using Colchicine-Mediated Nts-IF
The dual recombinase method suggests that LHA Nts neurons do not contain glutamate based on the absence of neurons co-expressing both Nts-GFP and RFP-vGlut2. This negative result could also be an artifact, perhaps if there were inefficient AAV-Lox-RFP infection within NtsFlpO; vGlutCre mice that resulted in under-detection of LHA glutamate neurons. We therefore sought to validate the classical neurotransmitter content of LHA Nts neurons using an independent strategy that did not depend on AAV-mediated recombination. We attempted to generate dual-reporter mice, but commercially available FlpO reporter lines proved ineffective for simultaneous labeling of Nts and vGat or vGlut2 neurons. This also raised concern that limited efficiency of FlpO-mediated recombination in NtsFlpO mice might under-report LHA Nts neurons and diminish the likelihood of detecting a small population of glutamatergic LHA Nts neurons.
Alternately, we treated vGatCre; GFP and vGlut2IRESCre; GFP mice (validated in Fig. 6) with colchicine, allowing for simultaneous visualization of GFP-labeled vGat and vGlut2 and Nts-IF (as in Fig. 1). Similar to findings using the dual recombinase strategy, we observed numerous Nts-IF cell bodies within the LHA and ZI of colchicine-treated mice (Fig. 7I–N”). While the ZI from vGlut2Cre; GFP mice was devoid of GFP-vGlut2 neurons (Fig. 7I,K–K”), vGatCre; GFP mice had numerous GFP-vGat neurons in the ZI, many of which also contained Nts-IF (Fig. 7L,N–N”, white arrows). These data are consistent with the GABAergic phenotype of the ZI and our findings using the dual recombinase system (Fig. 7G,H) that ZI Nts neurons are GABAergic but not glutamatergic. We observed many Nts-IF labeled cell bodies within the LHA, as well as GFP-labeled LHA glutamate neurons (Figure I’) and GFP-labeled GABA neurons (Fig. 7L). Despite the robust GFP-labeling induced in both lines, we did not observe any Nts-IF cell bodies within the LHA that co-localized with GFP-vGlut2 (Fig. 7J–J”). By contrast, essentially all of the LHA Nts-IF cell bodies overlapped with GFP-vGat (Fig. 7M–M”, white arrows), but many GFP-vGat neurons did not contain Nts-IR (magenta arrows). In sum, colchicine-mediated Nts-IR recapitulated our findings using the dual-recombinase system: that LHA Nts neurons are GABAergic but comprise a subset of the larger population of LHA GABA neurons.
Discussion
The LHA is essential for the motivation to eat and drink but the neural mediators of these behaviors have yet to be fully understood. While most LHA populations promote food and liquid intake, LHA Nts neurons divergently regulate ingestive behavior by suppressing feeding and promoting drinking22,24. We therefore hypothesized that separate subpopulations of LHA Nts neurons might exist to coordinate opposing feeding and drinking behavior. Here we characterized two separate subpopulations of LHA Nts neurons that are differentially activated by leptin (NtsLepRb neurons) or dehydration (NtsDehy neurons). While all LHA Nts neurons are GABAergic, the NtsLepRb and NtsDehy subpopulations differ in molecular expression of LepRb and at the circuit level, and hence can be distinguished via these criteria (Fig. 8). These data demonstrate the heterogeneity of LHA Nts neurons and their specific responsiveness to either energy or fluid balance cues suggest that they may coordinate different ingestive behaviors (feeding vs. drinking).
Model of LHA Nts Subpopulations. All LHA Nts neurons co-express the classical neurotransmitter, GABA and some project to the VTA and SNc. Of these, there are distinct subpopulations of LHA Nts neurons that respond to different anorectic cues; some are leptin-sensing (NtsLepRb neurons) and some are dehydration-activated (NtsDehy neurons). NtsLepRb neurons project the VTA and SNc, where they can access the mesolimbic DA circuit, but NtsDehy neurons do not. Collectively, these data describe functionally, molecularly and projection-distinct subpopulations of LHA Nts neurons that may contribute to adaptive energy balance.
Here we used NtsCre and NtsFlpO mice to non-invasively and reliably identify Nts neurons, which permitted their study under normal, physiologic conditions. Using NtsCre mice, we found that dehydration treatment increased activation from ~6% at baseline to ~18%, demonstrating that ~12% of the LHA Nts neurons are NtsDehy neurons (Fig. 5C). Additionally, NtsLepRb neurons make up a separate 15% of LHA Nts neurons22. While these are modestly sized populations, they can significantly influence homeostasis; for example, mice lacking leptin-regulation via NtsLepRb neurons have impaired response to energy balance cues and diminished dopamine signaling that causes overweight22,26. Characterization of the remaining 70% of LHA Nts neurons at the molecular and circuit level may provide insights about their function. For example, some LHA Nts neurons are activated by LPS-mediated inflammation and inhibit local orexin/hypocretin neurons, and these may contribute to illness-behavior43. Nts signaling is also implicated in regulation of analgesia, thermoregulation, stress and addiction44, so it will be important to determine if/how the remaining LHA Nts neurons contribute to these diverse aspects of physiology. Characterizing the heterogeneity of LHA Nts neurons may also suggest intersectional or pharmacological strategies to target specific subpopulations of LHA Nts neurons, and hence selective physiological outputs.
LHA neurons project widely throughout the brain and differentially modify behavior depending on their targets44. Our finding that NtsLepRb neurons, and not NtsDehy neurons, project to the midbrain suggests that there are distinct LHA Nts neural mechanisms for leptin-mediated suppression of feeding vs. regulation of drinking and fluid balance. Experimental activation of all LHA Nts neurons causes release of Nts to the VTA, and dopamine release into the nucleus accumbens27 that can modify motivated intake behavior27,45,46. Thus, at least some portion of anorectic leptin regulation via NtsLepRb neurons could occur via direct projections to, and modulation of, mesolimbic dopamine signaling. This is consistent with the requirement of leptin action via NtsLepRb neurons for regulating body weight and the integrity of the mesolimbic dopamine system, which are due in part to Nts signaling via VTA neurons expressing neurotensin receptor-1 (NtsR1)26,37. In contrast, NtsDehy neurons must act via other yet-to-be determined projection targets, and do not directly modulate dopamine signaling to modify physiology. While the function of NtsDehy neurons and their projection sites remains to be established, the activation of NtsDehy neurons in response to dehydration suggests that they may coordinate fluid need with the motivation to drink. The discovery of specific subsets of LHA Nts neurons also hints at why experimental activation of all LHA Nts neurons results in diverging ingestive behaviors. Such activation simultaneously induces NtsLepRb neurons that act partially via the VTA (and may be anorectic) as well as the NtsDehy neurons that regulate separate targets, and it is possible that these populations suppress feeding and promote drinking, respectively24. Since NtsLepRb and NtsDehy neurons are induced by separate physiological cues (leptin or dehydration), it remains to be determined whether there are any physiological situations in which these subpopulations are concurrently activated. In any case, our data confirm that NtsLepRb and NtsDehy neurons have distinct circuitry, thus projection-specific modulation may be a useful strategy to discern their respective contributions to ingestive behavior.
Despite the molecular and circuit heterogeneity of LHA Nts neurons, they all contain the same classical neurotransmitter, GABA. LHA Nts neurons presumably inhibit synaptic targets via release of GABA, as well as regulating postsynaptic and adjacent neurons via release of Nts. It remains to be determined if GABA and Nts are always co-released from LHA Nts neurons. Indeed, different physiological stimuli bias the release of neurotransmitter vs. neuropeptide signals in some LHA neurons, and the receipt of these messages depends on the repertoire of receptors expressed on target neurons, which can also vary47,48,49. Our finding that LHA Nts neurons are GABAergic is consistent with other reports of overlapping LHA Nts and GABAergic neurons27,50 but contrasts with a report of glutamatergic LHA Nts neurons that directly project to the VTA40. This discrepancy may be because AAV-Frt-GFP induced GFP expression was confined to the perifornical LHA (~Bregma −1.34 to 1.7051), and limited our characterization to this area only. However, the glutamatergic LHA Nts neurons were identified around the “rostral lateral hypothalamus” corresponding to Bregma −0.40, where the rostral LHA merges into the preoptic area, and well beyond the perifornical LHA. Hence, the GABAergic perifornical LHA Nts neurons studied here could be anatomically and neurochemically distinct from LHA Nts neurons of the hypothalamus-preoptic continuum.
While all LHA Nts neurons contain GABA, they are a subset of the vast population of LHA GABA neurons. This may account for the strikingly different behaviors observed after experimental activation of LHA Nts neurons (suppression of feeding, increased drinking) vs. activation of all LHA GABA neurons (increased feeding, drinking and gnawing directed at non-biological objects)19,20,24,27. Since activation of LHA Nts and LHA GABA neurons promotes drinking, LHA Nts neurons contribute to at least some of the polydipsic effect. The orexigenic effect observed with activation of all LHA GABA neurons likely masks anorectic effects mediated by the subset of LHA Nts neurons encompassed within them. Our findings agree with reports of functionally-distinct subpopulations of LHA GABA neurons18, and LHA Nts neurons are a functionally unique subset within the larger population of all GABA neurons that suppress feeding instead of promoting it. This arrangement may also explain differences in VTA regulation that have been ascribed to these populations. Some LHA GABA neurons project to the VTA, where they disinhibit VTA GABA neurons that in turn releases inhibition of DA neurons to facilitate DA release and feeding19,52. Some GABA-containing LHA Nts neurons, including NtsLepRb neurons, also project to the VTA, and their precise synaptic targets are yet to be defined.
Taken together, our data reveal the heterogeneity of LHA Nts neurons. Since LHA Nts neurons are differentially regulated by energy status (leptin) vs. fluid status (dehydration), and comprise separate subpopulations, there may be separate neural mechanisms to coordinate feeding and drinking necessary for survival. If true, then these data may suggest strategies to selectively modify the LHA Nts neurons that control feeding vs. those that modify drinking.
Materials and Methods
Animals
Adult male and female mice were used for studies. Some NtsCre; GFP and LepRbCre; GFP mice were generated and treated with euhydration or dehydration at the University of Michigan, under the supervision of the Unit for Laboratory Animal Medicine (ULAM). These procedures were approved by the University of Michigan Institutional Animal Care and Use Committee (IACUC). All other mice were generated from a breeding colony at Michigan State University. MSU mice were cared for by Campus Animal Resources (CAR) and all animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at Michigan State University. All mouse experiments were performed in accordance with Association for Assessment and Accreditation of Laboratory Animal Care and National Institutes of Health guidelines. In all cases mice were housed in a 12 h light/12 h dark cycle and had ad libitum access to water and chow diet unless otherwise noted.
Generation of Nts FlpO Knock-In Mice
We modified the targeting vector used to generate NtsCre mice26 to create an NtsFlpO targeting vector. Briefly, the IRES-Cre was replaced with an IRES-FlpO sequence, such that it is inserted between the stop codon and the polyadenylation site of the sequence encoding the 3’ end of the mouse Nts gene. An frt-flanked NEO cassette lied upstream of the IRES-FlpO for selection purposes. The linearized NtsFlpO targeting vector was electroporated into mouse R1 embryonic stem (ES) cells (129sv background) and cells were selected with G418. DNA from ES cell clones was analyzed via qPCR for loss of homozygosity using Taqman primer and probes for the genomic Nts insertion sites (Nts-IRES: Forward: TGAAAAGGCAGCTGTATGAAAATAA, Nts-IRES: Reverse: TCAAGAATTAGCTTCTCAGTAGTAGTAGGAA, Nts-IRES: Probe: CCAGAAGGCCCTACATTCTCAAGAGG. NGF was used as a copy number control. Putative positive ES clones were expanded, confirmed for homologous recombination by Southern blot and injected into mouse C57BL/6 blastocysts to generate chimeras. Chimeric males were mated with C57BL/6 females (Jackson Laboratory) and germline transmission was determined initially via progeny coat color, then confirmed via conventional PCR for FlpO (as described below).
Breeding and Genotyping
The NtsCre; GFP and LepRbCre; GFP mice were generated and genotyped as described previously23. For all other experiments we utilized Ntscre mice (Jackson stock #017525) that had been bred onto the C57/Bl6 background (Jackson #000664) mice for at least seven generations. To visualize Nts, vGat and vGlut2-expressing neurons, heterozygous Ntscre mice, homozygous Slc32a1tm2(cre)Lowl [Jackson stock #028862] and Slc17a6tm2(cre)Lowl [Jackson stock # 028863] mice were crossed with homozygous Rosa26EGFP-L10a mice35 and progeny heterozygous for both alleles were studied (Ntscre; GFP mice, vGatCre; GFP and vGlut2Cre; GFP mice respectively). To simultaneously detect Nts and vGat or vGlut2 we utilized a dual-recombinase strategy. Briefly, we interbred NtsFlpO mice (to permit FlpO-mediated recombination) and Slc32a1tm2(cre)Lowl or Slc17a6tm2(cre)Lowl mice (that enable Cre-mediated recombination) to generate progeny that were heterozygous for FlpO and Cre. These mice were injected with FlpO- and Cre-dependent reporters to visualize Nts and vGAT/vGlut-expressing neurons as described below. Genotyping was performed using standard PCR using the following primer sequences: Ntscre: common forward: 5′ ATA GGC TGC TGA ACC AGG AA, Cre reverse: 5′ CCA AAA GAC GGC AAT ATG GT and WT reverse: 5′ CAA TCA CAA TCA CAG GTC AAG AA. Rosa26EGFP-L10a: mutant forward: 5′ TCT ACA AAT GTG GTA GAT CCA GGC, WT forward: 5′ GAG GGG AGT GTT GCA ATA CC and common reverse: 5′ CAG ATG ACT ACC TAT CCT CCC. NtsFlpO: Nts-FlpO-WT: Forward: CCAGGAAGATATCCTTGATAACGTCAAT, Reverse: GCAAGAAACATCACATCCAATAAAGCA N, Nts-FlpO-M: Forward: TGACCTACCTGTGCTGGATGAT, Reverse: CCACGTTCTTGATGTCGCTGAA. vGatCre: VgatIRESCre Common Forward: CTTCGTCATCGGCGGCATCTG, VgatIRESCre -WT Reverse: CAGGGCGATGTGGAATAGAAA, VgatIRESCre -Mutant Reverse: CCAAAAGACGGCAATATGGT.
Stereotaxic Injections
Stereotaxic surgeries were performed as described previously22 using coordinates from the mouse brain atlas of Paxinos and Franklin51. To facilitate detection of Nts containing cell bodies via Nts-IF, adult Ntscre; GFP, vGatCre; GFP and vGlut2Cre; GFP mice received injections of colchicine (10 μg in a volume of 500 nL) into the lateral ventricle (A/P −0.2, M/L −1.0, D/V −2.1), and were euthanized via cardiac perfusion ~48 hours after treatment. For tract tracing studies, Ntscre; GFP mice were injected unilaterally with 75 nL of the retrograde tracer FluoroGold (FG) into the VTA (A/P: −3.2, M/L: +/−0.48, D/V: −4.65) or SNc (A/P: −3, M/L +/−1.3, D/V −4.7), and recovered for 7–10 days to allow for accumulation of FG in cell bodies of origin. Animals were only included in the study if the FG injection was targeted to and confined within the VTA or SNc. To simultaneously identify Nts and vGAT or vGlut, adult NtsFlpO; vGatCre and NtsFlpO; vGlut2Cre mice were injected in the LHA with 400 nL of AAV-Frt-GFP adenovirus (provided by David Olson, University of Michigan) followed by 400 nL of AAV-hM3Dq-mCherry purchased from the UNC Vector Core (here termed AAV-Lox-RFP); these were infused at a rate of 100 nL/minute. LHA coordinates were A/P: −1.34, M/L +/−1.5, D/V −4.9, angle: 6°. Mice recovered for 2–3 weeks to permit sufficient time for recombinase-mediated reporter expression.
Leptin or Dehydration Treatment
Some FG-injected Ntscre; GFP mice were treated with PBS or recombinant mouse leptin (5 mg/kg, i.p.) purchased from the National Hormone and Peptide Program (Los Angeles Biomedical Research Institute, Los Angeles, CA) then were perfused 2–4 hours later to enable detection of leptin-induced pSTAT3. In our hands, both of these leptin treatment times produce comparable pSTAT3. Males and females were studied but no notable differences between sexes were observed so they were pooled for analysis: VTA-injected vehicle-treated n = 6; female VTA-injected leptin-treated n = 10; SNc-injected vehicle-treated n = 5; SNc-injected leptin-treated n = 9.
Other FG-injected Ntscre; GFP mice underwent a dehydration-activation paradigm in which they were either had ad libitum access to water (euhydration) or the water bottle was removed for 12 hr during the dark cycle (when mice drink most of their daily water). Mice were perfused the following morning and brain sections were stained for cFos, GFP and FG (see below). Due to the lack of observable differences between sexes, males and females were pooled for analysis: VTA-injected, euhydrated n = 5; VTA-injected, dehydrated n = 9; SNc-injected, euhydrated n = 5, SNc-injected, dehydrated n = 9.
Adult male 8–12 wk old NtsCre; GFP and LepRbCre; GFP mice were also treated via euhydration or had water bottles removed for 24 hr (including during the dark cycle) to induce dehydration then were perfused (Euhydrated NtsCre; GFP n = 7, LepRbCre; GFP n = 5; Dehydrated NtsCre; GFP n = 4, LepRbCre; GFP n = 3). Brains were analyzed for cFos and GFP, and 3 representative LHA sections spanning the same Bregma sections from each mouse were analyzed using Photoshop to count the number of GFP-only labeled neurons and GFP neurons containing cFos. Graphed data represent the average percentage of GFP neurons containing cFos out of the total number of GFP-labeled neurons ±SEM. Significant differences between genotypes and treatments were determined via 2-way ANOVA with Bonferroni posttests.
Immunohistochemistry and Immunofluorescence (IF)
Mice were treated with a lethal dose of i.p. pentobarbital and perfused transcardially with either 10% formalin or 4% paraformaldehyde (Sigma-Aldrich 158127) containing 0.4% picric acid (Sigma-Aldrich 197378). Brains were removed, post-fixed in the same fixative overnight at 4 °C, dehydrated with 30% sucrose/PBS for 2–3 days, then sectioned into 30 µm slices using a sliding microtome (Leica). Brain sections were analyzed by immunohistochemistry and/or IF as previously described22. For activation studies, brain sections first were exposed to either rabbit-anti pSTAT3 (1:500, Cell Signaling) or goat-anti cFos (1:500, Santa Cruz) followed by incubation with species specific Alexa-488 conjugated (Jackson ImmunoResearch, 1:200) or Alexa-568 conjugated antibodies (LifeTech, 1:200) and visualization with DAB (Sigma). IF labeling was performed exposing sections to primary antibodies, including chicken anti-GFP (1:2000, Abcam), rabbit anti-FG (1:500, Fluorochrome), rabbit-anti Nts (1:500, Phoenix) and/or anti-dsRed (1:1000, Clontech), followed by incubation with species-specific secondary antibodies conjugated to AlexaFluor 488 or 568 fluorophores (1:200, Life Technologies or Jackson ImmunoResearch). Immunolabeled brain sections were analyzed using an Olympus BX53 fluorescence microscope outfitted with transmitted light to analyze DAB-labeling as well as FITC and Texas Red filters for IF. Microscope images were collected using Cell Sens software and a Qi-Click 12 Bit cooled camera, and images were analyzed using Photoshop software (Adobe). We collected images from all mice within an experiment and compared them side-by-side to select the representative images included in the manuscript.
Quantification of Nts Neuronal LHA –> VTA and LHA –> SNc Projections
FG-injected Ntscre; GFP mice were perfused and brains were stained as described above. An intact section between Bregma −1.3 and −1.5 was taken from each sample for quantification. The area medial and dorsal to the Fornix and ventral to the MT was counted to determine the number of FG (VTA/SNc-projecting) and GFP (Nts) labelled neurons in each. Males and females were studied but no notable differences between sexes were observed so they were pooled for analysis: VTA-injected n = 6; female; SNc-injected vehicle-treated n = 5. Graphed data represent the average number of GFP neurons containing FG. Significant differences between brain areas were determined via Student’s t-test and were calculated using GraphPad Prism (GraphPad Software Inc., San Diego, CA). Error bars depict ± standard error of the mean (SEM). **p < 0.01.
In Situ Hybridization (ISH) Data
Some figures include Nts, vGat and vGlut2 ISH data from the coronally-sectioned adult mouse brain courtesy of the Allen Brain Atlas (Lein et al.36), which are identified within our figures as “Courtesy of Allen Brain”. The entire datasets can be referenced via the Allen Brain Atlas website: Nts: http://mouse.brain-map.org/experiment/show/73788032; vGat: http://mouse.brain-map.org/experiment/show/72081554; vGlut2: http://mouse.brain-map.org/experiment/show/73818754.
Data Availability Statement
No datasets were generated during this study. Links to the publicly available datasets of ISH data that were analyzed for this work are listed in the previous section.
References
Watts, A. G. & Boyle, C. N. The functional architecture of dehydration-anorexia. Physiology & behavior 100, 472–477, https://doi.org/10.1016/j.physbeh.2010.04.010 (2010).
Goto, M., Canteras, N. S., Burns, G. & Swanson, L. W. Projections from the subfornical region of the lateral hypothalamic area. The Journal of comparative neurology 493, 412–438, https://doi.org/10.1002/cne.20764 (2005).
Hahn, J. D. & Swanson, L. W. Distinct patterns of neuronal inputs and outputs of the juxtaparaventricular and suprafornical regions of the lateral hypothalamic area in the male rat. Brain research reviews 64, 14–103, https://doi.org/10.1016/j.brainresrev.2010.02.002 (2010).
Saper, C. B., Swanson, L. W. & Cowan, W. M. An autoradiographic study of the efferent connections of the lateral hypothalamic area in the rat. The Journal of comparative neurology 183, 689–706, https://doi.org/10.1002/cne.901830402 (1979).
Hahn, J. D. & Swanson, L. W. Connections of the lateral hypothalamic area juxtadorsomedial region in the male rat. The Journal of comparative neurology 520, 1831–1890, https://doi.org/10.1002/cne.23064 (2012).
Hahn, J. D. & Swanson, L. W. Connections of the juxtaventromedial region of the lateral hypothalamic area in the male rat. Front Syst Neurosci 9, 66, https://doi.org/10.3389/fnsys.2015.00066 (2015).
Anand, B. K. & Brobeck, J. R. Localization of a “feeding center” in the hypothalamus of the rat. Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine 77, 323–324 (1951).
Anand, B. K. & Brobeck, J. R. Hypothalamic control of food intake in rats and cats. The Yale journal of biology and medicine 24, 123–140 (1951).
Morrison, S. D., Barrnett, R. J. & Mayer, J. Localization of lesions in the lateral hypothalamus of rats with induced adipsia and aphagia. The American journal of physiology 193, 230–234 (1958).
Teitelbaum, P. & Epstein, A. N. The lateral hypothalamic syndrome: recovery of feeding and drinking after lateral hypothalamic lesions. Psychological review 69, 74–90 (1962).
Ungerstedt, U. Adipsia and aphagia after 6-hydroxydopamine induced degeneration of the nigro-striatal dopamine system. Acta physiologica Scandinavica. Supplementum 367, 95–122 (1971).
Duncan, E. A., Proulx, K. & Woods, S. C. Central administration of melanin-concentrating hormone increases alcohol and sucrose/quinine intake in rats. Alcoholism, clinical and experimental research 29, 958–964 (2005).
Clegg, D. J. et al. Intraventricular melanin-concentrating hormone stimulates water intake independent of food intake. American journal of physiology. Regulatory, integrative and comparative physiology 284, R494–499 (2003).
Inutsuka, A. et al. Concurrent and robust regulation of feeding behaviors and metabolism by orexin neurons. Neuropharmacology 85C, 451–460, https://doi.org/10.1016/j.neuropharm.2014.06.015 (2014).
Tabuchi, S. et al. Conditional ablation of orexin/hypocretin neurons: a new mouse model for the study of narcolepsy and orexin system function. The Journal of neuroscience: the official journal of the Society for Neuroscience 34, 6495–6509, https://doi.org/10.1523/JNEUROSCI.0073-14.2014 (2014).
McGregor, R., Wu, M. F., Barber, G., Ramanathan, L. & Siegel, J. M. Highly specific role of hypocretin (orexin) neurons: differential activation as a function of diurnal phase, operant reinforcement versus operant avoidance and light level. The Journal of neuroscience: the official journal of the Society for Neuroscience 31, 15455–15467, https://doi.org/10.1523/JNEUROSCI.4017-11.2011 (2011).
Stamatakis, A. M. et al. Lateral Hypothalamic Area Glutamatergic Neurons and Their Projections to the Lateral Habenula Regulate Feeding and Reward. The Journal of neuroscience: the official journal of the Society for Neuroscience 36, 302–311, https://doi.org/10.1523/JNEUROSCI.1202-15.2016 (2016).
Jennings, J. H. et al. Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell 160, 516–527, https://doi.org/10.1016/j.cell.2014.12.026 (2015).
Nieh, E. H. et al. Decoding neural circuits that control compulsive sucrose seeking. Cell 160, 528–541, https://doi.org/10.1016/j.cell.2015.01.003 (2015).
Navarro, M. et al. Lateral Hypothalamus GABAergic Neurons Modulate Consummatory Behaviors Regardless of the Caloric Content or Biological Relevance of the Consumed Stimuli. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology 41, 1505–1512, https://doi.org/10.1038/npp.2015.304 (2016).
Qualls-Creekmore, E. et al. Galanin-Expressing GABA Neurons in the Lateral Hypothalamus Modulate Food Reward and Noncompulsive Locomotion. The Journal of neuroscience: the official journal of the Society for Neuroscience 37, 6053–6065, https://doi.org/10.1523/JNEUROSCI.0155-17.2017 (2017).
Brown, J. A. et al. Loss of Action via Neurotensin-Leptin Receptor Neurons Disrupts Leptin and Ghrelin-Mediated Control of Energy Balance. Endocrinology 158, 1271–1288, https://doi.org/10.1210/en.2017-00122 (2017).
Leinninger, G. M. et al. Leptin acts via leptin receptor-expressing lateral hypothalamic neurons to modulate the mesolimbic dopamine system and suppress feeding. Cell metabolism 10, 89–98, https://doi.org/10.1016/j.cmet.2009.06.011 (2009).
Woodworth, H. L. et al. Neurotensin Receptor-1 Identifies a Subset of Ventral Tegmental Dopamine Neurons that Coordinates Energy Balance. Cell Rep 20, 1881–1892, https://doi.org/10.1016/j.celrep.2017.08.001 (2017).
van den Pol, A. N. Neuropeptide transmission in brain circuits. Neuron 76, 98–115, https://doi.org/10.1016/j.neuron.2012.09.014 (2012).
Leinninger, G. M. et al. Leptin action via neurotensin neurons controls orexin, the mesolimbic dopamine system and energy balance. Cell metabolism 14, 313–323, https://doi.org/10.1016/j.cmet.2011.06.016 (2011).
Patterson, C. M. et al. Ventral tegmental area neurotensin signaling links the lateral hypothalamus to locomotor activity and striatal dopamine efflux in male mice. Endocrinology, en20141986, https://doi.org/10.1210/en.2014-1986 (2015).
Watts, A. G., Kelly, A. B. & Sanchez-Watts, G. Neuropeptides and thirst: the temporal response of corticotropin-releasing hormone and neurotensin/neuromedin N gene expression in rat limbic forebrain neurons to drinking hypertonic saline. Behavioral neuroscience 109, 1146–1157 (1995).
Hawkins, M. F., Baker, J. D. & Baumeister, A. A. Neurotensin-induced polydipsia: a structure-activity study. Brain research 487, 188–191 (1989).
Watts, A. G., Sanchez-Watts, G. & Kelly, A. B. Distinct patterns of neuropeptide gene expression in the lateral hypothalamic area and arcuate nucleus are associated with dehydration-induced anorexia. The Journal of neuroscience: the official journal of the Society for Neuroscience 19, 6111–6121 (1999).
Watts, A. G. & Sanchez-Watts, G. Rapid and preferential activation of Fos protein in hypocretin/orexin neurons following the reversal of dehydration-anorexia. The Journal of comparative neurology 502, 768–782 (2007).
Watts, A. G. Osmotic stimulation differentially affects cellular levels of corticotropin-releasing hormone and neurotensin/neuromedin N mRNAs in the lateral hypothalamic area and central nucleus of the amygdala. Brain research 581, 208-216, 0006-8993(92)90710-Q [pii] (1992).
Kahn, D., Abrams, G. M., Zimmerman, E. A., Carraway, R. & Leeman, S. E. Neurotensin neurons in the rat hypothalamus: an immunocytochemical study. Endocrinology 107, 47–54, https://doi.org/10.1210/endo-107-1-47 (1980).
Woodworth, H. L., Brown, J. A., Batchelor, H. M., Bugescu, R. & Leinninger, G. M. Determination of neurotensin projections to the ventral tegmental area in mice. Neuropeptides, https://doi.org/10.1016/j.npep.2018.02.003 (2018).
Krashes, M. J. et al. An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature 507, 238–242, https://doi.org/10.1038/nature12956 (2014).
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176, https://doi.org/10.1038/nature05453 (2007).
Opland, D. et al. Loss of neurotensin receptor-1 disrupts the control of the mesolimbic dopamine system by leptin and promotes hedonic feeding and obesity. Molecular metabolism 2, 423–434, https://doi.org/10.1016/j.molmet.2013.07.008 (2013).
Wise, R. A. & McDevitt, R. A. Drive and Reinforcement Circuitry in the Brain: Origins, Neurotransmitters, and Projection Fields. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology, https://doi.org/10.1038/npp.2017.228 (2017).
Bissonette, G. B. & Roesch, M. R. Development and function of the midbrain dopamine system: what we know and what we need to. Genes Brain Behav 15, 62–73, https://doi.org/10.1111/gbb.12257 (2016).
Kempadoo, K. A. et al. Hypothalamic Neurotensin Projections Promote Reward by Enhancing Glutamate Transmission in the VTA. The Journal of neuroscience: the official journal of the Society for Neuroscience 33, 7618–7626, https://doi.org/10.1523/JNEUROSCI.2588-12.2013 (2013).
Dulcis, D. et al. Neurotransmitter Switching Regulated by miRNAs Controls Changes in Social Preference. Neuron 95, 1319–1333 e1315, https://doi.org/10.1016/j.neuron.2017.08.023 (2017).
Kranz, A. et al. An improved Flp deleter mouse in C57Bl/6 based on Flpo recombinase. Genesis 48, 512–520, https://doi.org/10.1002/dvg.20641 (2010).
Grossberg, A. J. et al. Inflammation-induced lethargy is mediated by suppression of orexin neuron activity. The Journal of neuroscience: the official journal of the Society for Neuroscience 31, 11376–11386, https://doi.org/10.1523/JNEUROSCI.2311-11.2011 (2011).
Schroeder, L. E. & Leinninger, G. M. Role of central neurotensin in regulating feeding: Implications for the development and treatment of body weight disorders. Biochim Biophys Acta 1864, 900–916, https://doi.org/10.1016/j.bbadis.2017.12.036 (2018).
Ferrario, C. R. et al. Homeostasis Meets Motivation in the Battle to Control Food Intake. The Journal of neuroscience: the official journal of the Society for Neuroscience 36, 11469–11481, https://doi.org/10.1523/JNEUROSCI.2338-16.2016 (2016).
Berridge, K. C. & Kringelbach, M. L. Pleasure systems in the brain. Neuron 86, 646–664, https://doi.org/10.1016/j.neuron.2015.02.018 (2015).
Barbano, M. F., Wang, H. L., Morales, M. & Wise, R. A. Feeding and Reward Are Differentially Induced by Activating GABAergic Lateral Hypothalamic Projections to VTA. The Journal of neuroscience: the official journal of the Society for Neuroscience 36, 2975–2985, https://doi.org/10.1523/JNEUROSCI.3799-15.2016 (2016).
Baimel, C., Lau, B. K., Qiao, M. & Borgland, S. L. Projection-Target-Defined Effects of Orexin and Dynorphin on VTA Dopamine Neurons. Cell Rep 18, 1346–1355, https://doi.org/10.1016/j.celrep.2017.01.030 (2017).
Muschamp, J. W. et al. Hypocretin (orexin) facilitates reward by attenuating the antireward effects of its cotransmitter dynorphin in ventral tegmental area. Proceedings of the National Academy of Sciences of the United States of America 111, E1648–1655, https://doi.org/10.1073/pnas.1315542111 (2014).
Leinninger, G. M. Location, location, location: the CNS sites of leptin action dictate its regulation of homeostatic and hedonic pathways. International journal of obesity 33(2), S14–17, https://doi.org/10.1038/ijo.2009.66 (2009).
Paxinos, G. & Franklin, B. The Mouse Brain in Stereotaxic Coordinates. Second Edition edn, (Academic Press, 2001).
Nieh, E. H. et al. Inhibitory Input from the Lateral Hypothalamus to the Ventral Tegmental Area Disinhibits Dopamine Neurons and Promotes Behavioral Activation. Neuron, https://doi.org/10.1016/j.neuron.2016.04.035 (2016).
Acknowledgements
We thank co-author David P. Olson (University of Michigan) for graciously sharing the Cre-inducible RosaeGFP-L10a reporter line and AAV-Frt-GFP used in these studies and for guidance in designing NtsFlpO mice. Thanks to Martin G. Myers, Jr. for generously providing LepRbCre; GFP mice for experiments. We also thank the transgenic cores at the University of Michigan and Van Andel Research Institute for assistance generating NtsR1FlpO mice. This research was supported by grants from the NIH to JAB (T32-ES00725527, F31-DK107081) and GML (R01-DK103808), and Anna Wright was supported by an MSU College of Natural Science Undergraduate Research Award.
Author information
Authors and Affiliations
Contributions
J.B. conducted the research, analyzed the data and wrote the manuscript. A.W. assisted in data collection for the dual recombinase model and assisted in preparation of Figs 6 and 7. R.B. bred and genotyped mice used for the studies. G.L. performed the experiment described in Fig. 5 and L.C. analyzed the data under the direction of G.L., who then prepared the figure. D.P. provided the AAV-Frt-GFP and guidance on its use for experiments presented in Fig. 7. G.L. oversaw the research and edited the manuscript. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Brown, J.A., Wright, A., Bugescu, R. et al. Distinct Subsets of Lateral Hypothalamic Neurotensin Neurons are Activated by Leptin or Dehydration. Sci Rep 9, 1873 (2019). https://doi.org/10.1038/s41598-018-38143-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-38143-9
- Springer Nature Limited
This article is cited by
-
Circuit coordination of opposing neuropeptide and neurotransmitter signals
Nature (2023)
-
Ontogenetic rules for the molecular diversification of hypothalamic neurons
Nature Reviews Neuroscience (2022)