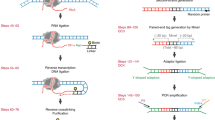
Abstract
Transcription factors (TFs) bind specific DNA sequences to regulate transcription. Apart from DNA sequences, local factors such as DNA accessibility and chromatin structure determine the affinity of a TF for any given locus. Including these factors when measuring TF–DNA affinities has proven difficult. To address this challenge, we recently developed a method called binding affinities in native chromatin by sequencing (BANC-seq). In BANC-seq, intact mammalian nuclei are incubated with a concentration range of epitope-tagged TF, followed by either chromatin immunoprecipitation or cleavage under target and release using nuclease with spike-in DNA. This allows determination of apparent dissociation constant (KdApp) values, defined by the concentration of TF at which half-maximum binding occurs, across the genome. Here we present a detailed stepwise protocol for BANC-seq, including downstream data analysis. In principle, any molecular biologist should be able to perform a BANC-seq experiment in as little as 1.5 d (excluding analysis). However, preprocessing and analysis of the sequencing data does require some experience in command-line shell and R programming.
Key points
-
BANC-seq enables the quantification of genome-wide transcription factor binding affinities in the native chromatin context. This protocol describes implementations based on chromatin immunoprecipitation or cleavage under target and release using nuclease, followed by library preparation, sequencing and data analysis.
-
Unlike traditional methods for measuring transcription factor affinities, BANC-seq measurements incorporate the effects of the chromatin landscape, DNA accessibility and the impact of cofactors.



Similar content being viewed by others
Data availability
Next-generation sequencing data used as example data for this paper can be retrieved from the Gene Expression Omnibus with accession code GSE219035.
Code availability
The workflow for pre-processing of sequencing data and to perform KdApp determination from raw count files is freely available at https://github.com/HNeikes/BANCseq_protocol.
References
Kribelbauer, J. F., Rastogi, C., Bussemaker, H. J. & Mann, R. S. Low-affinity binding sites and the transcription factor specificity paradox in eukaryotes. Ann. Rev. Cell Dev. Biol. 35, 357–379 (2019).
Weirauch, M. T. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Jolma, A. et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 20, 861–873 (2010).
Yin, Y. et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239 (2017).
Zhu, F. et al. The interaction landscape between transcription factors and the nucleosome. Nature 562, 76–81 (2018).
Neikes, H. K. et al. Quantification of absolute transcription factor binding affinities in the native chromatin context using BANC-seq. Nat. Biotechnol. 41, 1801–1809 (2023).
Gräwe, C., Makowski, M. M. & Vermeulen, M. PAQMAN: protein–nucleic acid affinity quantification by MAss spectrometry in nuclear extracts. Methods 184, 70–77 (2020).
van der Sande, M. et al. seq2science (v0.9.6). Zenodo https://doi.org/10.5281/zenodo.5948679 (2022).
Acknowledgements
The Vermeulen laboratory is part of the Oncode Institute, which is partly funded by the Dutch Cancer Society (KWF). In addition, research at the Netherlands Cancer Institute is supported by institutional grants of the Dutch Cancer Society and of the Dutch Ministry of Health, Welfare and Sport.
Author information
Authors and Affiliations
Contributions
R.G.H.L. and M.V. conceived the study. R.G.H.L. designed the methodology and analyses. H.K.N. adapted the methodology to the CUT&RUN-based protocol. R.G.H.L., H.K.N. and R.A.W. performed experiments and analysis. R.A.W., H.K.N., R.G.H.L. and M.V. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Sebastian Pott and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Neikes, H. K. et al. Nat. Biotechnol. 41, 1801–1809 (2023): https://doi.org/10.1038/s41587-023-01715-w
Supplementary information
Supplementary Information
Supplementary Table 1 and Fig. 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wester, R.A., Neikes, H.K., Lindeboom, R.G.H. et al. Quantifying genome-wide transcription factor binding affinities for chromatin using BANC-seq. Nat Protoc (2024). https://doi.org/10.1038/s41596-024-01026-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-024-01026-7
- Springer Nature Limited