Abstract
Human immunodeficiency virus (HIV) cure efforts are increasingly focused on harnessing CD8+ T cell functions, which requires a deeper understanding of CD8+ T cells promoting HIV control. Here we identifiy an antigen-responsive TOXhiTCF1+CD39+CD8+ T cell population with high expression of inhibitory receptors and low expression of canonical cytolytic molecules. Transcriptional analysis of simian immunodeficiency virus (SIV)-specific CD8+ T cells and proteomic analysis of purified CD8+ T cell subsets identified TOXhiTCF1+CD39+CD8+ T cells as intermediate effectors that retained stem-like features with a lineage relationship with terminal effector T cells. TOXhiTCF1+CD39+CD8+ T cells were found at higher frequency than TCF1−CD39+CD8+ T cells in follicular microenvironments and were preferentially located in proximity of SIV-RNA+ cells. Their frequency was associated with reduced plasma viremia and lower SIV reservoir size. Highly similar TOXhiTCF1+CD39+CD8+ T cells were detected in lymph nodes from antiretroviral therapy-naive and antiretroviral therapy-suppressed people living with HIV, suggesting this population of CD8+ T cells contributes to limiting SIV and HIV persistence.







Similar content being viewed by others
Data availability
Single-cell RNA-seq data have been deposited in the NCBI GEO database under accession number GSE266576. Proteomics data have been deposited in the PRIDE database under accession number PXD050498 and can be accessed at https://www.ebi.ac.uk/pride/archive/. All other data are present in the article as individual data points, Supplementary Information or from the corresponding authors upon reasonable request.
Code availability
The custom code used in this manuscript is available from the corresponding author upon reasonable request.
References
Blank, C. U. et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 19, 665–674 (2019).
Alfei, F. et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019).
Scott, A. C. et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019).
Khan, O. et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218 (2019).
Yao, C. et al. Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat. Immunol. 20, 890–901 (2019).
Seo, H. et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl Acad. Sci. USA 116, 12410–12415 (2019).
Beltra, J. C. et al. Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 52, 825–841 (2020).
Sekine, T. et al. TOX is expressed by exhausted and polyfunctional human effector memory CD8+ T cells. Sci. Immunol. 5, eaba7918 (2020).
Heim, K. et al. TOX defines the degree of CD8+ T cell dysfunction in distinct phases of chronic HBV infection. Gut 70, 1550–1560 (2020).
Im, S. J. et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016).
Hudson, W. H. et al. Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1+ stem-like CD8+ T cells during chronic infection. Immunity 51, 1043–1058 (2019).
Collins, D. R., Gaiha, G. D. & Walker, B. D. CD8+ T cells in HIV control, cure and prevention. Nat. Rev. Immunol. 20, 471–482 (2020).
Day, C. L. et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443, 350–354 (2006).
Trautmann, L. et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat. Med. 12, 1198–1202 (2006).
Petrovas, C. et al. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J. Exp. Med. 203, 2281–2292 (2006).
Hoffmann, M. et al. Exhaustion of activated CD8 T cells predicts disease progression in primary HIV-1 infection. PLoS Pathog. 12, e1005661 (2016).
Chew, G. M. et al. TIGIT marks exhausted T cells, correlates with disease progression, and serves as a target for immune restoration in HIV and SIV infection. PLoS Pathog. 12, e1005349 (2016).
Collins, D. R. et al. Functional impairment of HIV-specific CD8+ T cells precedes aborted spontaneous control of viremia. Immunity 54, 2372–2384 (2021).
Bengsch, B. et al. Epigenomic-guided mass cytometry profiling reveals disease-specific features of exhausted CD8 T cells. Immunity 48, 1029–1045 (2018).
Reuter, M. A. et al. HIV-specific CD8+ T cells exhibit reduced and differentially regulated cytolytic activity in lymphoid tissue. Cell Rep. 21, 3458–3470 (2017).
Nguyen, S. et al. Elite control of HIV is associated with distinct functional and transcriptional signatures in lymphoid tissue CD8+ T cells. Sci. Transl. Med. 11, eaax4077 (2019).
Buggert, M. et al. Identification and characterization of HIV-specific resident memory CD8+ T cells in human lymphoid tissue. Sci. Immunol. 3, eaar4526 (2018).
Collins, D. R. et al. Cytolytic CD8+ T cells infiltrate germinal centers to limit ongoing HIV replication in spontaneous controller lymph nodes. Sci. Immunol. 8, eade5872 (2023).
Ward, A. R., Mota, T. M. & Jones, R. B. Immunological approaches to HIV cure. Semin. Immunol. 51, 101412 (2021).
Deeks, S. G. et al. Research priorities for an HIV cure: International AIDS Society Global Scientific Strategy 2021. Nat. Med. 27, 2085–2098 (2021).
Huang, S. et al. Increased TOX expression concurrent with PD-1, Tim-3, and CD244 in T cells from patients with non-Hodgkin lymphoma. Asia Pac. J. Clin. Oncol. 18, 143–149 (2022).
Wang, X. et al. TOX promotes the exhaustion of antitumor CD8+ T cells by preventing PD1 degradation in hepatocellular carcinoma. J. Hepatol. 71, 731–741 (2019).
Gupta, P. K. et al. CD39 expression identifies terminally exhausted CD8+ T cells. PLoS Pathog. 11, e1005177 (2015).
Sade-Feldman, M. et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013 (2018).
Canale, F. P. et al. CD39 expression defines cell exhaustion in tumor-infiltrating CD8+ T cells. Cancer Res. 78, 115–128 (2018).
Abbas, H. A. et al. Single cell T cell landscape and T cell receptor repertoire profiling of AML in context of PD-1 blockade therapy. Nat. Commun. 12, 6071 (2021).
Jonsson, A. H. et al. Granzyme K+ CD8 T cells form a core population in inflamed human tissue. Sci. Transl. Med. 14, eabo0686 (2022).
Arbones, M. L. et al. Lymphocyte homing and leukocyte rolling and migration are impaired in l-selectin-deficient mice. Immunity 1, 247–260 (1994).
Takeuchi, A. et al. CRTAM confers late-stage activation of CD8+ T cells to regulate retention within lymph node. J. Immunol. 183, 4220–4228 (2009).
Guma, M. et al. The CD94/NKG2C killer lectin-like receptor constitutes an alternative activation pathway for a subset of CD8+ T cells. Eur. J. Immunol. 35, 2071–2080 (2005).
Trkola, A. et al. Genetic subtype-independent inhibition of human immunodeficiency virus type 1 replication by CC and CXC chemokines. J. Virol. 72, 396–404 (1998).
Cocchi, F. et al. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270, 1811–1815 (1995).
Ichii, H. et al. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat. Immunol. 3, 558–563 (2002).
Sun, Q. et al. BCL6 promotes a stem-like CD8+ T cell program in cancer via antagonizing BLIMP1. Sci. Immunol. 8, eadh1306 (2023).
Eberhardt, C. S. et al. Functional HPV-specific PD-1+ stem-like CD8 T cells in head and neck cancer. Nature 597, 279–284 (2021).
Sekaly, R. et al. Dual blockade of IL-10 and PD-1 leads to control of SIV viral rebound following analytical treatment interruption. Preprint at Res. Sq. https://doi.org/10.21203/rs.3.rs-3175716/v1 (2023).
Petrovas, C. et al. Follicular CD8 T cells accumulate in HIV infection and can kill infected cells in vitro via bispecific antibodies. Sci. Transl. Med. 9, eaag2285 (2017).
Yates, K. B. et al. Epigenetic scars of CD8+ T cell exhaustion persist after cure of chronic infection in humans. Nat. Immunol. 22, 1020–1029 (2021).
Bruner, K. M. et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566, 120–125 (2019).
Utzschneider, D. T. et al. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity 45, 415–427 (2016).
Kiniry, B. E. et al. Differential expression of CD8+ T cell cytotoxic effector molecules in blood and gastrointestinal mucosa in HIV-1 infection. J. Immunol. 200, 1876–1888 (2018).
Passaes, C. et al. Optimal maturation of the SIV-specific CD8+ T cell response after primary infection is associated with natural control of SIV: ANRS SIC study. Cell Rep. 32, 108174 (2020).
Rutishauser, R. L. et al. TCF-1 regulates HIV-specific CD8+ T cell expansion capacity. JCI Insight 6, e136648 (2021).
Mvaya, L. et al. Differential localization and limited cytotoxic potential of duodenal CD8+ T cells. JCI Insight 7, e154195 (2022).
Fardoos, R. et al. HIV specific CD8+ TRM-like cells in tonsils express exhaustive signatures in the absence of natural HIV control. Front. Immunol. 13, 912038 (2022).
McGary, C. S. et al. CTLA-4+PD-1− memory CD4+ T cells critically contribute to viral persistence in antiretroviral therapy-suppressed, SIV-infected rhesus macaques. Immunity 47, 776–788 (2017).
Li, H. et al. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc. Natl Acad. Sci. USA 113, E3413–E3422 (2016).
Langner, C. A. & Brenchley, J. M. FRugally optimized DNA cctomer (FRODO) qPCR measurement of HIV and SIV in human and nonhuman primate samples. Curr. Protoc. 1, e93 (2021).
Strongin, Z. et al. The role of CD101-expressing CD4 T cells in HIV/SIV pathogenesis and persistence. PLoS Pathog. 18, e1010723 (2022).
Hoang, T. N. et al. Baricitinib treatment resolves lower-airway macrophage inflammation and neutrophil recruitment in SARS-CoV-2-infected rhesus macaques. Cell 184, 460–475 (2021).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
DeTomaso, D. & Yosef, N. FastProject: a tool for low-dimensional analysis of single-cell RNA-seq data. BMC Bioinf. 17, 315 (2016).
Bolotin, D. A. et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381 (2015).
Black, S. et al. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat. Protoc. 16, 3802–3835 (2021).
Deleage, C. et al. Defining HIV and SIV reservoirs in lymphoid tissues. Pathog. Immun. 1, 68–106 (2016).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Skowronek, P. et al. Rapid and in-depth coverage of the (phospho-)proteome with deep libraries and optimal window design for dia-PASEF. Mol. Cell Proteom. 21, 100279 (2022).
Bruderer, R. et al. Optimization of experimental parameters in data-independent mass spectrometry significantly increases depth and reproducibility of results. Mol. Cell Proteom. 16, 2296–2309 (2017).
Lazar, C., Gatto, L., Ferro, M., Bruley, C. & Burger, T. Accounting for the multiple natures of missing values in label-free quantitative proteomics data sets to compare imputation strategies. J. Proteome Res. 15, 1116–1125 (2016).
Acknowledgements
This manuscript is dedicated in loving memory of our colleague and friend T. Hoang. We thank S. Jean, S. Ehnert, C. Wallace, J. Wood and all veterinary and animal care staff at the Emory National Primate Research Center. We also thank the Emory Flow Cytometry core (K. Gill) for flow cytometry support, the Emory National Primate Research Center Genomics Core for RNA-seq support, the NIH Tetramer Core facility at Emory University for providing tetramer, K. Easley for providing statistical advice and Accelevir Diagnostics for intact proviral DNA measurements. Peptide pools of SIVmac239 Gag were provided by the HIV Reagent Program, which were contributed by the Division of AIDS at NIAID. This work was supported by UM1AI164562 (M.P.), cofunded by NIH/NHLBI/NIDDK/NINDS/NIDA/NIAID and by the NIAID, NIH under award numbers R37AI141258 (R.S.), R01AI116379 (M.P.) and R56AI150401 (M.P.). Additional support was provided by: NIH OD, ORIP, P51 OD011132 and U42 OD011023. This work was supported by the Center for AIDS Research at Emory University (Division of Intramural Research, NIAID, P30AI050409). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. 75N91019D00024; Divisions of Intramural Research of the National Institute of Allergy and Infectious Diseases, NIH 1ZIAAI001029 (J.M.B.); B03-16 project was supported by funds from the Mexican Government (Programa Presupuestal P016, Anexo 13 del Decreto del Presupuesto de Egresos de la Federación). Sequencing data were acquired on an Illumina NovaSeq6000 funded by NIH S10 OD026799 (S.E.B.). C.M.B. was supported by Swedish Research Council (grant 2023-00510). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products or organizations imply endorsement by the US government. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Z.S. and M.P. conceptualized the study. Z.S., L.R.M., C.D., M.B.P., M.A.C., C.M.B., T.N.H., E.A.U., M.G., K.N., S.L., A.R.R., J.H. and H.T.K. performed the investigations. Z.S., L.R.M., C.D., M.B.P., C.M.B., M.A.C., E.A.U., M.G., G.K.T., A.R.R. and H.T.K. did the formal analysis. P.M.R.E., M.G.-N., Y.A.L., S.A.-R. and G.R.-T. provided resources. Z.S., L.R.M., C.D., M.A.C., C.M.B. and H.T.K. implemented data visualization. C.D., R.S., D.A.K., G.S., J.B., S.E.B., D.E.G., M.R.B. and M.P. acquired funding to support the work. A.S.-C., J.B., S.E.B., D.E.G., M.R.B., H.T.K. and M.P. supervised the work. Z.S., L.R.M. and M.P. wrote the original draft. Z.S., C.D., L.R.M., H.T.K., M.R.B. and M.P. reviewed and edited the final manuscript.
Corresponding author
Ethics declarations
Competing interests
Z.S. is employed by and/or has financial interests in Merck and Co.. All other authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Ioana Staicu, in collaboration with the Nature Immunology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
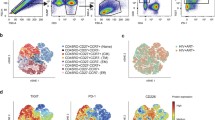
Extended Data Fig. 1 LN TOXhiTCF1+CD39+CD8+ TM cells express high levels of inhibitory receptors after SIV infection.
a) Expression of PD-1, TIGIT, CD101 and Ki-67 on TOX+ and TOX− LN CD8+ TM cells at day 42 p.i. (n = 28 macaques). b) UMAP visualization of Phenograph clusters as in a. c) Heatmap of expression of TOX, TCF1, CD39, PD-1, TIGIT, CD101, Ki-67, GzmB and EOMES across all Phenograph clusters as in a. d) Proportion of total CD8+ TM cell population made up by individual Phenograph clusters (n = 28) as in a. e) Frequency of TCF1+CD39+ cells within LN CD8+ TM cells in 5 unique cohorts of macaques: uninfected (n = 10), day 21 p.i. (n = 7), day 35 p.i. (n = 20), day 42 p.i. (n = 28), late chronic infection (mean 17 months p.i. (n = 10). f-g) Expression of TOX, PD-1, TIGIT, CD101, Ki-67 and GzmB in TCF1+CD39−, TCF1+CD39+ and TCF1−CD39+ subsets of CD8+ TM cells at day 35 p.i. (n = 20) (f) and late chronic infection (n = 10) (g). P values determined by Wilcoxon matched-pairs signed rank test, Kruskal-Wallis one way ANOVA with Dunn’s multiple comparison correction (e) or two-way ANOVA with Tukey’s multiple comparisons test (f-g). Box and whiskers are displayed via Tukey method. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 2 TCF1+CD39+CD8+ TM cells maintain a dual effector and stem-like proteomic profile.
a) Representative gating strategy for sorting of TCF1+CD39−, TCF1+CD39+ and TCF1−CD39+ subsets of CD8+ TM at day 42 p.i. b-c) Volcano plots displaying significantly differently expressed proteins of interest between sorted LN TCF1+CD39− and TCF1+CD39+ CD8+ TM cells (b) and between sorted LN TCF1+CD39− and TCF1+CD39+ CD8+ TM cells (c) at day 42 p.i. d) Selected proteins with significantly different expression are listed (n = 4 macaques). All statistical comparisons were performed based on two-tailed Student’s t-tests using Spectronaut and custom R scripts.
Extended Data Fig. 3 Sorting of LN SIV-specific CD8+ TM cells.
a) Sorting strategy for LN SIV-specific CM9+CD8+ TM cells for downstream scRNA-seq at day 42 p.i. b) Plots and frequencies of sorted LN SIV-specific CM9+CD8+ TM cells for all 5 macaques as in a.
Extended Data Fig. 4 Transcriptional clustering of LN SIV-specific CD8+ TM cells overlaps with established human CD8 T cell signatures.
a) Heatmap of top 20 differentially expressed genes across the 5 clusters of LN SIV-specific CM9+CD8+ TM cells at day 42 p.i. b) UMAP projection of cells matching established human CD8+ T cell signatures (Supplementary Data File 1) of cell cycling, type 1 interferon, human stem-like CD8 and human terminally differentiated (TD) CD8. c) Distribution of LN SIV-specific CM9+CD8+ TM cells across the 5 clusters for all macaques as in a. d) Scatter plot of expression levels and thresholds (dotted lines) for identifying TCF7+ENTPD1+ CD8+ TM cells as in a.
Extended Data Fig. 5 TCR analysis of expanded TCF7+ENTPD1+CD8+ TM cells demonstrates lineage relationship with terminally differentiated CD8+ TEFF cells.
a-e) UMAP projection of the top 3 clonotypes within TCF7+ENTPD1+ CD8+ TM cells and the top 5 clonotypes of TCF7+ENTPD1+ CD8+ TM cells population from each macaque and the cluster distribution frequency of those clones across all SIV-specific CM9+CD8+ TM cells at day 42 p.i. Number of total cells expressing each TCR clonotype is shown below each bar.
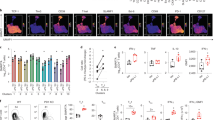
Extended Data Fig. 6 Response of TCF1+CD39+CD8+ TM cells to dual IL-10 + PD-1 mAb blockade.
a) Study schematic demonstrating dosing regimen and timing of lymph node biopsies (week -1 and week 12 post-treatment) for flow cytometry analysis placebo-treated control (n = 6) and treated (n = 9) macaques. b) Fold change of the frequency of TCF1+CD39−, TCF1+CD39+ and TCF1−CD39+ subsets of LN CD8+ TM cells, calculated using frequencies at week 12 post-treatment compared to baseline frequencies at week -1 (control (n = 6) and treated (n = 9)). c) Fold change of the frequency of LN Ki-67+ cells within TCF1+CD39−, TCF1+CD39+ and TCF1−CD39+ subsets of CD8+ TM cells (control (n = 6) and treated (n = 9)). P values determined by multiple Mann-Whitney tests with Holm-Sidak correction for multiple comparison. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 7 TCF1+CD39+CD8+ TM cells are associated with reduced viral burden.
a-b) Association of the frequency of TOX+ (a) and TCF1+ CD39+ (b, red) cells within LN CD8+ TM cells with cell-associated SIV RNA levels in LN at day 42 p.i. (n = 26). c-h) Association of the frequency of TCF1+ CD39+ (c-e, red) and TOX+ (f-h) cells within LN CD8+ TM cells with plasma viral load (c,f), total SIV DNA (d,g) and cell associated RNA (e,h) at day 35 p.i. (n = 20). All correlations were determined using two-tailed Spearman analysis.
Extended Data Fig. 8 Expression of CXCR5 by TOX+TCF1+CD39+CD8+ TM cells is associated with viral burden.
a-c) Association of the frequency of CXCR5+ cells within LN CD8+ TM cells with plasma viral load (a), total SIV DNA (b) and cell associated RNA (c) at day 42 p.i.(a n = 27 macaques, b-c n = 25). d-f) Association of frequency of CXCR5+ cells within TCF1+CD39−(d, burgundy), TCF1+CD39+ (e, red) and TCF1−CD39+ (f, pink) subsets of LN CD8+ TM cells with cell-associated SIV RNA levels in sorted LN memory CD4 T cells as in a (n = 25). g) Frequency of CXCR5+ cells within LN GzmB−GzmK+, GzmB+GzmK+ and GzmB+GzmK− CD8+ TM cells LN memory CD8 T cells as in a (n = 27). h) Representative image of immunofluorescence staining of CD39 (green) TCF1 (blue) and TOX (red). i) Representative image of immunofluorescence analysis identifying TCF1+ and TOX+ CD8+ T cells in the B cell follicle and T cell zone. Arrows designate examples of TCF1+ TOX+ CD8+ T cells. j-k) Representative image of immunofluorescence analysis identifying SIV−infected CD4+ T cells and TCF1+TOX+CD8+ T cells in the T cell zone (j) and the dark zone of the b cell follicle (k). The perimeter of analysis is defined by the red circle. Blue arrows indicate uninfected CD4+ T cells not surrounded by TCF1+TOX+CD8+ T cells. The dotted line delimits the edge of the b cell follicle. Number of events analyzed per LN area: B cell follicle SIV+ = 175, B cell follicle SIV− = 178, T cell zone SIV+ = 501, T cell zone SIV− = 475, dark zone B cell follicle SIV+ = 217, dark zone bell cell follicle SIV- = 153. All analyses were performed on samples from animals at early chronic infection (D42 p.i.). Correlations represent Spearman analysis. P values were determined using Friedman test one-way ANOVA with Dunn’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 9 TCF1+CD39+CD8+ TM cells express high levels of inhibitory receptors and low cytolytic markers in ART-suppressed PLWH.
a-d) Expression frequency of TOX, PD-1, TIGIT (a), Ki-67 (c), GzmB and GzmK (c-d) in TCF1+CD39−, TCF1+CD39+ and TCF1−CD39+ subsets of LN CD8+ TM cells in ART-suppressed PLWH (n = 10). P values determined by two-way ANOVA with Tukey’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 10 TCF1+CD39+CD8+ TM cells are not associated with viral burden in ART-naïve PLWH.
a-c) Associations of the frequency of TCF1+CD39− (burgundy), TCF1+CD39+ (red), and TCF1−CD39+ (pink) subsets within LN CD8+ TM cells from ART-naïve PLWH with plasma viral load (n = 18) (a), intact reservoir size as measured by IPDA (n = 10) (b) and total HIV DNA (n = 10) (c). d-e) Associations of the frequency of TCF1+CD39− (burgundy), TCF1+CD39+ (red), and TCF1−CD39+ (pink) subsets within LN CD8+ TM cells from ART-Suppressed PLWH with intact reservoir size as measured by IPDA (n = 10) (d) and total HIV DNA (n = 10) (e). Correlation evaluated by two-tailed Spearman analysis.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Tables 1–3.
Supplementary Data 1
scRNA-seq differentially expressed genes and reference human gene signature sets.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Strongin, Z., Raymond Marchand, L., Deleage, C. et al. Distinct SIV-specific CD8+ T cells in the lymph node exhibit simultaneous effector and stem-like profiles and are associated with limited SIV persistence. Nat Immunol 25, 1245–1256 (2024). https://doi.org/10.1038/s41590-024-01875-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-024-01875-0
- Springer Nature America, Inc.