Abstract
The long-term physiological consequences of respiratory viral infections, particularly in the aftermath of the COVID-19 pandemic—termed post-acute sequelae of SARS-CoV-2 (PASC)—are rapidly evolving into a major public health concern1,2,3. While the cellular and molecular aetiologies of these sequelae are poorly defined, increasing evidence implicates abnormal immune responses3,4,5,6 and/or impaired organ recovery7,8,9 after infection. However, the precise mechanisms that link these processes in the context of PASC remain unclear. Here, with insights from three cohorts of patients with respiratory PASC, we established a mouse model of post-viral lung disease and identified an aberrant immune–epithelial progenitor niche unique to fibroproliferation in respiratory PASC. Using spatial transcriptomics and imaging, we found a central role for lung-resident CD8+ T cell–macrophage interactions in impairing alveolar regeneration and driving fibrotic sequelae after acute viral pneumonia. Specifically, IFNγ and TNF derived from CD8+ T cells stimulated local macrophages to chronically release IL-1β, resulting in the long-term maintenance of dysplastic epithelial progenitors and lung fibrosis. Notably, therapeutic neutralization of IFNγ + TNF or IL-1β markedly improved alveolar regeneration and pulmonary function. In contrast to other approaches, which require early intervention10, we highlight therapeutic strategies to rescue fibrotic disease after the resolution of acute disease, addressing a current unmet need in the clinical management of PASC and post-viral disease.
Similar content being viewed by others
Main
SARS-CoV-2 infection can lead to long-term pulmonary and extrapulmonary symptoms well beyond the resolution of acute disease, a condition collectively termed PASC1. The lungs are particularly susceptible to extensive damage during primary infection, potentially resulting in dyspnoea, compromised lung function, radiological abnormalities and pulmonary fibrosis (PASC-PF)7,8,9,11,12,13, which can persist for up to 2 years after infection, in contrast to the majority of extrapulmonary sequelae14. Although less appreciated, similar long-term pulmonary deficits have also been described after influenza and SARS-CoV-1 infection, suggesting that this phenomenon may be conserved across different viral aetiologies2,3. Despite progress in identifying immune signatures that are dysregulated in respiratory PASC4,5,8,15,16,17,18, little is known regarding their role in driving pathological tissue remodelling, in part owing to the lack of animal models that recapitulate features of post-viral lung disease. Recent research characterizing alveolar regeneration after severe lung injury has identified the accumulation of undifferentiated epithelial progenitors19,20,21,22,23, the persistence of which has been pathologically associated with chronic diseases such as lung fibrosis7,9,24. Currently, the mechanisms that underlie the long-term maintenance of these dysplastic injury-associated cell states after infection and their contributions to post-viral respiratory sequelae remain largely unclear.
Aberrant immune–epithelial progenitor niche
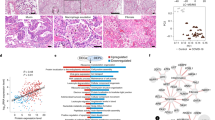
We examined diseased lungs from two cohorts of patients with PASC-PF who underwent lung transplantation at Cedars-Sinai Medical Center and University of Virginia (Supplementary Tables 1 and 2). None of the patients had any pre-existing lung disease before infection that necessitated a lung transplant. SARS-CoV-2 was absent by PCR of nasal swabs during routine clinical testing as part of the pre-transplant evaluation and was also not evident in the explanted lungs (Extended Data Fig. 1a). Lung histology showed extensive immune cell infiltration and collagen deposition in the alveolar epithelium (Fig. 1a and Extended Data Fig. 1b). Given the heterogenous distribution of tissue pathology, we performed spatial transcriptomics analysis of PASC-PF and control lungs to evaluate potentially distinct microenvironments within healthy and diseased lung areas (Extended Data Fig. 1c,d). Using an agnostic evaluation by gene set enrichment analysis, we observed robust upregulation of pathways associated with extracellular matrix deposition and fibrosis, as well as inflammation, within PASC-PF lungs compared with control lungs (Extended Data Fig. 1e). Expression of genes associated with alveolar epithelial type 1 and 2 (AT1 and AT2) cells was reduced, whereas that of genes characterizing dysplastic epithelial progenitors19,20,21,22,23,25,26 was elevated in PASC-PF lungs (Fig. 1b and Extended Data Fig. 1f). Consistent with active inflammatory processes, CD8+ T cell gene expression was enriched in PASC-PF lungs (Fig. 1b and Extended Data Fig. 1f). Furthermore, we found that large areas of PASC-PF lungs were populated by CD8+ T cells, dysplastic epithelial progenitors and CD4+ T cells (Fig. 1c and Extended Data Fig. 1f,g), whereas control lungs were primarily composed of alveolar epithelial cells (Fig. 1c and Extended Data Fig. 1f). Notably, the sporadic pockets of alveolar epithelium-rich areas in PASC-PF lungs were geographically distinct from CD8+ T cells and dysplastic repair markers (Fig. 1c). Consistent with this, a mild correlation was observed between the distribution of CD8+ T cells and dysplastic epithelial progenitors (Extended Data Fig. 1h).
a, Representative Masson’s trichrome (MT) images of control and PASC-PF lung sections. Scale bar, 2 mm. b, Quantification of the proportion of spots expressing genes characterizing various immune and epithelial populations in control (n = 2) and PASC-PF (n = 3) lungs. c, Visualization of CD8+ T cells, areas enriched for KRT5+, KRT8high and KRT17+ cells and healthy alveolar epithelium based on gene expression signatures in human control (n = 2) and PASC-PF (n = 3) lungs. d, Quantification of AT1 (AGER+) and AT2 (proSP-C+) (***P = 0.0001) cells in control (n = 5) and PASC-PF (n = 12) lung sections. e,f,h, Quantification of KRT8high transitional (e), KRT17+KRT5− aberrant basaloid (f) and CD8+ T cells (h) in control (n = 15) and PASC-PF (n = 21) lung sections. g,j, Quantification of KRT8high (*P = 0.0247) (g) and CD8+ T cells (**P = 0.0038) (j) in control (n = 3) and influenza-infected (n = 4) human lung sections. i, Unbiased analysis of the CD8+ T cell distribution in a PASC-PF lung section using QuPath. k–m, Simple linear regression of the CD8+ T cell number and KRT8high area fraction in human PASC-PF lungs (k; 5–7 random fields per lung, n = 21 lungs), human lungs after influenza infection (l; 8 random fields per lung, n = 4 lungs) and IPF lung sections (m; 5–7 random fields per lung, n = 13 lungs). Statistical analysis was performed using two-tailed unpaired t-tests (d, e–h and j); *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; NS, not significant. Data are mean ± s.e.m.
To validate these findings, we used multicolour immunofluorescence analysis in a larger cohort of samples and found reduced levels of AT1 and AT2 cells in PASC-PF lungs compared with in the control lungs27 (Fig. 1d and Extended Data Fig. 1i). Chronic persistence of ectopic KRT5+ pods, KRT8high transitional cells and KRT5−KRT17+ aberrant basaloid cells was observed in PASC-PF lungs, in proximity to areas exhibiting myofibroblast activity7,9,12 (Fig. 1e,f and Extended Data Fig. 1j–n). Thus, PASC-PF histologically resembles other fibrotic diseases such as idiopathic pulmonary fibrosis (IPF), including the sustained loss of functional alveolar epithelial cells and the persistence of metaplastic KRT5+ pods, KRT8high transitional cells and KRT17+KRT5− aberrant basaloid cells25,26,28,29 (Extended Data Fig. 1n). Notably, these features were also observed in human lungs with fibrotic remodelling after influenza viral infection (Fig. 1g and Extended Data Fig. 1q–t).
Previously, we reported that CD8+ T cell abundance in the bronchoalveolar lavage (BAL) fluid was associated with impaired lung function in convalescent individuals recovering from COVID-194,5. Similarly, CD8+ T cell numbers were elevated in PASC-PF lungs (Fig. 1h) as well as in IPF lungs compared with in the control lungs (Extended Data Fig. 1o,p). The mild correlation in localization revealed by spatial transcriptomics was corroborated by a marked spatial association between CD8+ T cells, and areas of dysplastic repair in PASC-PF lungs (Fig. 1i,k and Extended Data Fig. 2a–f). Notably, this phenomenon was conserved in lungs with fibrosis associated with influenza viral infection, which exhibited extensive CD8+ T cell infiltration (Fig. 1j and Supplementary Table 3) as well as evidence of aberrant immune–epithelial progenitor niches (Fig. 1l and Extended Data Fig. 2g). Similar niches were also observed in the lungs from two organ donors with a previous history of influenza viral infection (evidenced by high frequency of M58-66 epitope-specific CD8+ T cells) (Extended Data Fig. 2h,i). Notably, this correlation between CD8+ T cells and dysplastic epithelial progenitors was not seen in lungs from healthy donors or individuals with IPF (Fig. 1m and Extended Data Fig. 2j–n). Collectively, these data indicate that the spatiotemporal colocalization of CD8+ T cells and areas of dysplastic repair is a unique feature of post-viral pulmonary fibrosis and supports a role for immune–epithelial progenitor interactions in chronic lung sequelae.
A mouse model of post-viral lung sequelae
To investigate the role of immune–epithelial progenitor interactions, we aimed to develop a mouse model to capture the cellular and pathological features observed in PASC-PF lungs. We used a mouse-adapted (MA-10) strain of the SARS-CoV-2 virus to productively infect wild-type mice30. As ageing is associated with an increased propensity to develop lung fibrosis after alveolar injury, we included both young and aged mice in our study31,32. Aged C57BL/6 mice infected with SARS-CoV-2 MA-10 exhibited increased morbidity and mortality compared with young mice (Extended Data Fig. 3a). Yet, despite the marked acute inflammation and tissue damage (at 10 days post-infection (d.p.i.))30,33 (Extended Data Fig. 3b), only moderate pathology restricted to the subpleural regions was observed during the chronic phase (35 d.p.i.) of infection (Fig. 2a and Extended Data Fig. 3c). Moreover, KRT8high transitional cell abundance remained unchanged between aged naive and infected (35 d.p.i.) mouse lungs (Fig. 2b and Extended Data Fig. 3d,e). We next tested SARS-CoV-2 MA-10 infection in aged BALB/c mice, which was previously reported to induce more robust inflammation and fibrotic sequelae than in C57BL/6 mice31 (Extended Data Fig. 3f,g). Persistent immune cell infiltration was observed within peribronchiolar regions at 35 d.p.i., but minimal signs of collagen deposition were noted within the alveolar epithelium (Fig. 2c and Extended Data Fig. 3h). Consistent with previous observations31, BALB/c mice did not maintain pulmonary CD8+ T cells and KRT8high transitional cells (Fig. 2d and Extended Data Fig. 3i,j)—a prominent feature of human PASC-PF (Fig. 1e,h), despite the substantial alveolar damage during acute disease (Extended Data Fig. 3g). Finally, we infected transgenic mice expressing the human ACE2 receptor under the cytokeratin 18 promoter (K18-hACE2) with the wild-type SARS-CoV-2 virus (USA-WA1/2020), which is known to result in perivascular and alveolar inflammation34 (Extended Data Fig. 3k). However, alveolar injury was minimal at chronic timepoints (Fig. 2e and Extended Data Fig. 3l), with no appreciable differences in CD8+ T or KRT8high transitional cells between infected and naive mice (Fig. 2f and Extended Data Fig. 3m,n). Thus, SARS-CoV-2 infection in these three mouse models did not recapitulate key features of tissue pathology and dysplastic lung repair observed in human PASC-PF.
a,b, Evaluation of fibrotic disease (a; n = 5 (naive) and n = 5 (MA-10)) and KRT8high area (b; n = 5 (naive) and 12 (MA-10)) in aged C57BL/6 mice at 35 d.p.i. after infection with MA-10. c,d, Evaluation of fibrotic disease (c; n = 4 (naive) and n = 3 (MA-10)) and KRT8high area (d; n = 5 (naive) and n = 7 (MA-10)) in aged BALB/c mice at 35 d.p.i. after infection with MA-10. e,f, Evaluation of fibrotic disease (e; n = 3 (naive) and n = 4 (WA-1)) and KRT8high area (f; n = 4 (naive), n = 4 (WA-1)) in aged K18-hACE2 mice at 35 d.p.i. after infection with WA-1. g, Evaluation of fibrotic disease in young (Y) and aged (A) C57BL/6 mice after infection with influenza H1N1 A/PR8/34. n = 3–5 per timepoint. *P = 0.0378 (left) and 0.0412 (right), ***P = 0.0004. h,i, KRT8high transitional (h; *P = 0.0112 (left) and 0.0251 (right)) and CD8+ T cells (i; *P = 0.0190 (left), ***P = 0.0006 (right)) in young and aged lungs after IAV infection. n = 4–11 per timepoint. j, Quantification of AT1 (PDPN+) and AT2 (proSP-C+) cells after SARS-CoV-2 MA-10 and IAV infection in aged C57BL/6 mice. n = 3–4 per timepoint. k, Unbiased analysis of the CD8+ T cell distribution in an aged IAV-infected lung at 60 d.p.i. by QuPath. l, Linear regression of CD8+ T cells and KRT8high area in aged IAV-infected lungs at 60 d.p.i. 5–7 random fields per lung; n = 4. m, The experimental design for CD8+ T cell depletion. The diagram was created using BioRender. n–r, Quantification of KRT8high (n; **P = 0.0069) and KRT5+ (o; **P = 0.0085) area (n = 14–19 (control IgG), n = 7 (anti-CD8)); AT1 (PDPN+) area (p; ***P = 0.0009) and AT2 (proSP-C+) cells per mm2 (q; P = 0.0136) (n = 19 (control IgG) and n = 15 (anti-CD8)); and evaluation of lung function (r; compliance (Cst); *P = 0.0172 (left) and 0.0338 (right); resistance of the respiratory system (Rrs); *P = 0.0183 (left) and 0.0147 (right)) in aged IAV-infected mice (60 d.p.i.) treated with anti-CD8 or IgG antibody (n = 3–6 per timepoint). Data are representative of two (a–l) or three (m–r) independent experiments. Statistical analysis was performed using two-tailed unpaired t-tests (a–f and n–q), ordinary one-way analysis of variance (ANOVA) (g and r) and two-way ANOVA (h and i). Data are mean ± s.e.m.
Previously, we reported persistent lung inflammation and tissue pathology after influenza virus infection, particularly in aged C57BL/6 mice4,32,35. We therefore infected young and aged wild-type C57BL/6 mice with influenza H1N1 A/PR8/34 strain (IAV), which causes substantial viral pneumonia and alveolar damage32,35. Aged mice exhibited increased morbidity and mortality after IAV infection compared with the young mice (Extended Data Fig. 3o). In contrast to SARS-CoV-2 infection, we observed persistent immune cell infiltration and collagen deposition in the alveolar epithelium, particularly in aged mice, up to 60 d.p.i. (Fig. 2g and Extended Data Fig. 3p). Although both young19 and aged IAV-infected mice had dysplastic repair areas, aged lungs maintained significantly larger KRT5+ and KRT8high areas as well as higher levels of CD8+ T cells, similar to human PASC-PF lungs (Fig. 2h,i and Extended Data Figs. 3q and 4a,b). In addition to the quantitative differences, aged KRT8high transitional cells also expressed higher levels of PDLIM7—a marker that was recently shown to capture mouse transitional cells bearing high transcriptional similarity with aberrant basaloid cells in human patients with IPF36 (Extended Data Fig. 4c,d). Such qualitative changes were also observed in an independent dataset of post-bleomycin injury, suggesting intrinsic age-associated differences in regenerative cell states37 (Extended Data Fig. 4e). Notably, lungs exhibited considerable inflammation and collagen deposition, with high levels of CD8+ T cells and dysplastic epithelial progenitors, up to 8 months after infection (Extended Data Fig. 4f–m).
SARS-CoV-2 infection in both aged C57BL/6 and BALB/c mice induced loss of AT2 cells, whereas the AT1 cell compartment was relatively unaffected throughout the course of disease (Extended Data Fig. 5a–e). This finding contrasts with human COVID-19, in which extensive infection and loss of AT1 cells has been previously reported38,39 (Fig. 1d). However, IAV infection of both young and aged C57BL/6 mice triggered a substantial decrease in AT2 and AT1 cells during acute disease (Extended Data Fig. 5f–i), as seen in the lungs of individuals with influenza at autopsy (Extended Data Fig. 1s). Recent work demonstrating the necessity of AT1 cell loss in tissue remodelling and the emergence of the KRT8high transitional cell state40 may explain the persistence of dysplastic epithelial progenitors in IAV-infected mice but not in mice infected with SARS-CoV-2 MA-10. The differential AT1 cell death observed during acute infection across the models (Fig. 2j) is probably a reflection of the distinct tropism of the viruses in mice30,41 (Extended Data Fig. 5j).
Although dysplastic repair areas were observed in both young19,22 and aged IAV-infected mice, only aged lungs did not fully regenerate the alveolar epithelium by 30 d.p.i. (Extended Data Fig. 5h,i). The observed cellular changes were associated with sustained defects in pulmonary function in aged mice up to 60 d.p.i. after IAV pneumonia, mimicking the persistent impairment in lung function observed in patients with respiratory PASC4,12 (Extended Data Fig. 5k,l). Thus, after extensive evaluation of various mouse viral pneumonia models, IAV infection in aged mice was found to exhibit close histopathological alignment with features of post-viral disease observed in human PASC-PF and influenza-fibrosis lungs, supporting its use in studying the mechanisms underlying viral-infection-mediated lung fibrosis.
Exuberant CD8+ T cell activity impairs recovery
Similar to human PASC-PF lungs, we observed a spatial association between CD8+ T cells and dysplastic repair areas in aged IAV-infected mice, suggesting that aberrant immune–epithelial progenitor niches are conserved across different species during post-viral lung sequelae (Fig. 2k,l and Extended Data Fig. 6a–c). To understand the role of CD8+ T cells, we treated aged IAV-infected mice with CD8+ T cell depleting antibody (anti-CD8) or control antibody (IgG) starting from 21 d.p.i. (Fig. 2m). This post-acute timepoint was chosen to ensure that there was no interference with the essential antiviral activities of CD8+ T cells during infection32 (Extended Data Fig. 6d). CD8+ T cell depletion improved the histological features of disease (Extended Data Fig. 6e–g) and, importantly, reduced KRT5+ and KRT8high areas (Fig. 2n,o and Extended Data Fig. 6h). Moreover, CD8+ T cell depletion markedly increased AT1 and AT2 cells (Fig. 2p and Extended Data Fig. 6i) and improved lung function (Fig. 2r), suggesting that exuberant CD8+ T cell activity in the post-viral lung impaired alveolar regeneration, inducing chronic deficits in gas-exchange function.
Notably, depletion of CD8+ T cells resolved KRT5+ but not KRT8high areas in young mice and also did not substantially affect lung pathology (Extended Data Fig. 6j–m). To further examine whether this effect is due to intrinsic changes within the CD8+ T cell compartment with age, we transferred lung CD8+ T cells from young or aged mice after IAV infection (21 d.p.i.) into infection-matched congenic young recipients. Three weeks after transfer, AT2 cell numbers were reduced after transfer of CD8+ T cells from aged lungs compared with young lungs, while no significant differences were observed in KRT8high areas or AT1 levels (Extended Data Fig. 6n–q). These data suggest that aged CD8+ T cell transfer alone is not sufficient to cause the marked phenotype observed in virus-infected aged mice, implicating the aged microenvironment as a key mediator of chronic lung pathology. To dissect their respective contributions in impairing lung recovery, we also used low-dose and high-dose anti-CD8 treatment to deplete circulating and pulmonary CD8+ T cells, respectively32. We found that the therapeutic effects were dependent on eliminating pulmonary CD8+ T cells but not circulating CD8+ T cells32 (Extended Data Fig. 6r), suggesting that lung-resident CD8+ T cells are required for the long-term maintenance of dysplastic epithelial progenitors. Together, our results strongly implicate the activity of CD8+ T cells persisting in the lungs after viral pneumonia in driving chronic pulmonary sequelae.
Spatial profiling reveals immune–epithelial interactions
We performed spatial transcriptomics analysis of aged IAV-infected mouse lungs (60 d.p.i.) treated with IgG or anti-CD8 (Extended Data Fig. 7a–c). In addition to the histopathological similarities, dysplastic repair areas of aged IAV-infected mouse lungs were enriched with the genes and pathways that were differentially upregulated within pathological areas of human PASC-PF lungs (Extended Data Fig. 7d–f). This transcriptional congruence was dependent on CD8+ T cell activity, as anti-CD8-treated mice exhibited reduced conservation of human PASC-PF-specific genes and signalling pathways compared with the controls (Extended Data Fig. 7g–i).
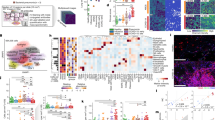
Moreover, a strong spatial association was observed between the gene expression signatures of CD8+ T cells, and KRT5+ and KRT8high areas of dysplastic repair, similar to human PASC-PF lungs (Fig. 3a). Signatures of KRT5+ and KRT8high progenitors were substantially reduced in anti-CD8-treated lungs, with a concomitant increase in healthy alveolar epithelial cells (Fig. 3b and Extended Data Fig. 7l,m). Several pathways associated with inflammatory responses were highly active within areas of dysplastic repair, whereas processes associated with growth factor activity and vascular regeneration were prominently observed in the healthy alveolar epithelium (Extended Data Fig. 7j). Previously reported transcriptional signatures of KRT8high transitional cells20,21,22 and human aberrant basaloid cells25,26 were strongly enriched within areas of dysplastic repair (Extended Data Fig. 7k). Furthermore, monocyte-derived macrophages, CD4+ T cells, interstitial macrophages, B cells and natural killer cells were enriched in dysplastic repair areas, while pro-repair tissue-resident alveolar macrophages, AT1 and AT2 cells were primarily observed within the healthy alveolar epithelium (Fig. 3b and Extended Data Fig. 7l–p). After conducting a correlation analysis, we found that CD8+ T cells and monocyte-derived macrophages physically clustered around KRT5+ and KRT8high areas of dysplastic repair and were excluded from areas enriched with alveolar macrophages and mature alveolar epithelial cells (Fig. 3c and Extended Data Fig. 7l–p). The physical distribution of these niches corresponded with the expression pattern of a gene set indicative of fibrosis, particularly enriched within KRT5+ and KRT8high areas, and was reduced in anti-CD8-treated lungs (Extended Data Fig. 7q).
a,b, Visualization (a) and quantification (b) of various immune and epithelial cell subsets as well as signalling pathways in areas enriched for KRT5+ and KRT8high cells and healthy alveolar epithelium. c, The physical distribution of monocyte-derived macrophages (MDM), alveolar macrophages (AM), KRT5+ and KRT8high areas (Krt) and healthy alveolar epithelium (AE) in aged IAV-infected (60 d.p.i.) mouse lungs. d, Unbiased analysis of the CX3CR1+ macrophage distribution in aged IAV-infected (60 d.p.i.) lungs. e, Quantification of the CX3CR1+ macrophage distribution within KRT8−/low (healthy) and KRT8high (diseased) fields in aged IAV-infected (60 d.p.i.) lungs. 3–5 random fields per lung; n = 4. *P = 0.0429. f, Quantification of CX3CR1+ macrophages in aged IAV-infected (60 d.p.i.) lungs that were treated with control IgG antibody or anti-CD8. n = 4. *P = 0.0380. g, Unbiased analysis of the distribution of CD68+ macrophages in a human PASC-PF lung using QuPath. h, Quantification of CD68+ macrophages within KRT8−/low (healthy) and KRT8high (diseased) fields in human PASC-PF lungs. 5–7 random fields per lung; n = 5. Statistical analysis was performed using two-tailed unpaired t-tests (e, f and h). Data are mean ± s.e.m.
Immunostaining identified a similar enrichment of CX3CR1+ monocyte-derived macrophages within areas of dysplastic repair (Fig. 3d,e and Extended Data Fig. 8a). Notably, CD8+ T cell depletion resulted in decreased CCL2 levels and CX3CR1+ macrophages, suggesting a potential role in macrophage recruitment and maintenance in post-viral lungs36 (Fig. 3f and Extended Data Fig. 8b–d). A similar increase in the gene signature of monocyte-derived macrophages was observed within PASC-PF lungs (Extended Data Fig. 8e), and was particularly enriched within dysplastic repair areas (Extended Data Fig. 8f–h), and corroborated by immunostaining (Fig. 3g,h and Extended Data Fig. 8i–k). Together, our data revealed a conserved finding in both mouse and human post-viral lungs, where CD8+ T cells are present in fibrotic regions, in proximity to fibroproliferative mediators such as monocyte-derived macrophages and dysplastic epithelial progenitors, constituting an aberrant niche after acute respiratory viral infections.
CD8+ T cell–macrophage axis
We postulated that the interactions between CD8+ T cells, macrophages and epithelial progenitors within this aberrant niche generated molecular cues to create a profibrotic microenvironment. IL-1R signalling and inflammasome activity were enriched in dysplastic repair areas compared with in the healthy alveolar epithelium in human PASC-PF (Fig. 4a and Extended Data Fig. 9a), and this was conserved with aged IAV-infected mouse lungs (Fig. 4b and Extended Data Fig. 9b). Notably, IL-1β has been shown to promote the expansion and persistence of transitional KRT8high cells after alveolar injury22,42. We found that CD64+ macrophages were major producers of proIL-1β in IAV-infected aged lungs (Fig. 4c and Extended Data Fig. 9c). As mature IL-1β release from cells requires caspase-1-mediated proteolytic cleavage43, we examined caspase-1 activity and observed a substantial reduction within lung macrophages after anti-CD8 treatment (Fig. 4d and Extended Data Fig. 9d,e), while inflammasome gene signatures were also attenuated (Extended Data Fig. 9f), therefore implicating CD8+ T cells in chronic inflammasome activation and IL-1β release by macrophages.
a,b, Gene expression signatures passing the cutoff (ON spots, %) for IL-1R signalling and inflammasome activity in AGERhigh (healthy) and KRT8high (diseased) areas within human PASC-PF lungs (a; n = 3) and AE (alveolar epithelium-rich; healthy) and Krt (KRT8high and KRT5+-rich; diseased) in aged IAV-infected (60 d.p.i.) mouse lungs (b; n = 2). c, Flow cytometry quantification of proIL-1β+ cells in aged naive (n = 5) and IAV-infected (42 d.p.i.; n = 9) mice. ***P = 0.0009 (left) and 0.0004 (right). d, Caspase-1 activity (FLICA) in aged IAV-infected (42 d.p.i.) lungs after control IgG antibody (n = 3) or anti-CD8 (n = 3) treatment. *P = 0.0431. e,f, Quantification of TNF+ (e; ***P = 0.0005) and IFNγ+ (f; *P = 0.0179) CD8+ T cells in human control and PASC-PF lungs. n = 5–6 per group. g, Caspase-1 activity in the lungs of aged IAV-infected mice (42 d.p.i.) after control IgG antibody (n = 5) or anti-IFNγ + anti-TNF (n = 6) treatment. **P = 0.0063. h, Schematic of the ex vivo macrophage (mac.) and CD8+ T cell coculture system. i, Caspase-1 activity. n = 4–5 wells per condition pooled from 2 mice. **P = 0.0017. j,k, Evaluation of IL-1β release by ELISA without (j; n = 4–10 wells per condition pooled from 4 mice; **P = 0.0075, ***P = 0.0008) or with (k; n = 8 wells per condition pooled from 5 mice; *P = 0.0206, **P = 0.0030) in vitro antibody blockade in the coculture. l,m, Evaluation of IL-1β levels in the BAL fluid of aged IAV-infected mice (42 d.p.i.) after IgG antibody (n = 6) or anti-CD8 (n = 9) treatment (*P = 0.0265) (l), or IgG antibody (n = 5) or anti-IFNγ + anti-TNF neutralizing antibody (n = 6) treatment (m; **P = 0.0090). n, The experimental design for the 2D AT2 culture system. o, Transitional cell marker gene expression after exposure to conditioned medium (CM). n = 2–3 wells per condition pooled from 4 mice. ***P = 0.0007. Data are representative of two (c–g, i and o) or three (j, k and m) independent experiments. Statistical analyses were performed using two-tailed unpaired t-tests (d–g and i,l,m), ordinary one-way ANOVA (j, k), and two-way ANOVA (c,o). Data are mean ± s.e.m. The diagrams in h and n were created using BioRender.
PASC-PF lungs were found to contain high levels of IFNγ+CD8+ and TNF+CD8+ T cells, similar to aged IAV-infected lungs44,45 (Fig. 4e,f and Extended Data Fig. 9g–i). Moreover, areas of dysplastic repair were enriched with IFNγ and TNF signalling in human PASC-PF lungs (Extended Data Fig. 9j). We therefore investigated whether IFNγ and TNF mediate inflammasome activation; we treated aged IAV-infected mice with neutralizing antibodies against both IFNγ and TNF and observed decreased caspase-1 activity (Fig. 4g). As this effect phenocopies anti-CD8 treatment (Fig. 4d), we directly tested whether CD8+ T cell-derived IFNγ and TNF regulated macrophage activity ex vivo (Fig. 4h) and observed that CD8+ T cells indeed augmented macrophage Il1b mRNA expression (Extended Data Fig. 9k), caspase-1 activity (Fig. 4i) and IL-1β release (Fig. 4j). The synergistic activity between macrophages and CD8+ T cells to produce IL-1β was lost when the cells were isolated from naive mice, suggesting that previous infection is necessary to prime the cells (Extended Data Fig. 9l). Although IL-1β release was significantly reduced after treatment with either IFNγ or TNF neutralizing antibody, neutralization of both cytokines was most effective, pointing to an additive effect in stimulating macrophages (Fig. 4k). Similarly, we observed decreased IL-1β levels in the BAL fluid after anti-CD8 (Fig. 4l) or anti-IFNγ + anti-TNF treatment (Fig. 4m).
We next examined whether macrophage-derived IL-1β is a negative regulator of AT2-to-AT1 transdifferentiation using a 2D primary AT2 cell culture model known to induce spontaneous differentiation into AT1 cells through the KRT8high transitional cell state46,47 (Fig. 4n). AT1 cell marker expression was reduced after exposure to conditioned medium from CD8+ T cell–macrophage coculture compared with macrophages alone, and was rescued after IL-1β neutralization (Extended Data Fig. 9m). By contrast, transitional cell marker expression exhibited the opposite pattern, with increased levels in the coculture group and a substantial reduction after anti-IL-1β treatment22 (Fig. 4o). Collectively, our results suggest that exuberant CD8+ T cell–macrophage interactions promote chronic IL-1β release to inhibit AT2 transdifferentiation by arresting the cells in the KRT8high transitional state.
Therapeutic enhancement of alveolar regeneration
Our data thus far indicate a pathological role for CD8+ T cells persisting in post-viral lungs in the development of chronic pulmonary sequelae. As depletion of CD8+ T cells is not a clinically feasible treatment strategy, we examined the therapeutic efficacy of neutralizing the cytokines that act as effectors of the profibrotic CD8+ T cell. Blocking IFNγ and TNF activity in aged IAV-infected mice ameliorated fibrotic sequelae compared with untreated controls (Fig. 5a and Extended Data Fig. 10a,b). Moreover, IFNγ and TNF neutralization diminished KRT5+ and KRT8high areas of dysplastic repair (Fig. 5b,c and Extended Data Fig. 10c) and enhanced alveolar regeneration as evidenced by the increased numbers of AT1 and AT2 cells (Fig. 5d,e and Extended Data Fig. 10c). The observed cellular changes after treatment also conferred physiological benefit by restoring pulmonary function (Fig. 5f and Extended Data Fig. 10d).
a, The experimental design for in vivo IFNγ + TNF neutralization after IAV infection. The diagram was created using BioRender. b–e, Quantification of the KRT8high area (b; **P = 0.0017), KRT5+ cell area (c; *P = 0.0404), AT1 (PDPN+) area (d; **P = 0.0073) and AT2 (proSP-C+) cells per mm2 (e; **P = 0.0046) in aged IAV-infected mice treated with anti-IFNγ + anti-TNF neutralizing antibody (n = 8) or control IgG antibody (n = 6). f, Evaluation of static compliance (*P = 0.0232 (left) and 0.0217 (right)), resistance of the respiratory system (*P = 0.0142 (left) and 0.0282 (right)) and tissue elastance (H; *P = 0.0417 (left) and 0.0355 (right)) in aged naive (n = 3) and IAV-infected mice treated with anti-IFNγ + anti-TNF neutralizing antibody (n = 9) or control IgG antibody (n = 8). g–k, Representative immunofluorescence images (g) and quantification of KRT8high cell area (h; *P = 0.0118), KRT5+ cell area (i; **P = 0.0014), AT1 (PDPN+) cell area (j; **P = 0.0011) and AT2 (proSP-C+) cells per mm2 (k; ***P = 0.0009) in aged IAV-infected lung sections treated with anti-IL-1β (n = 10) or control IgG (n = 9) antibody. l, Evaluation of static compliance (*P = 0.0361 (left) and 0.0468 (right)), resistance of the respiratory system and tissue elastance (*P = 0.0244, **P = 0.0019) in aged naive (n = 3) or IAV-infected mice treated with anti-IL-1β (n = 9) or control IgG (n = 8) antibody. m, Evaluation of the IL-1β levels in the plasma of convalescent individuals recovering from COVID-19 with or without abnormal lung function (n = 25 (normal) and n = 33 (abnormal); *P = 0.0182). LOD, limit of detection; PFT, pulmonary function test. Data are representative of three (b–f and h–l) independent experiments. Statistical analysis was performed using two-tailed unpaired t-tests (b–e, h–k and m) and one-way ANOVA (f and l). Data are mean ± s.e.m.
Next, we tested the efficacy of neutralizing IL-1β in IAV-infected aged mice and observed substantial attenuation of lung fibrosis and improved alveolar regeneration, phenocopying outcomes after CD8+ T cell depletion and neutralization of IFNγ and TNF (Fig. 5g–k and Extended Data Fig. 10e–h). Further supporting the therapeutic use of IL-1β blockade after infection, we found improved lung function in anti-IL-β-treated mice (Fig. 5l and Extended Data Fig. 10i). Notably, improvement in outcomes after IL-1β neutralization were seen only in aged mice but not young mice, which is probably due to the sustained elevation of IL-1β observed exclusively in aged mice (Extended Data Fig. 10j,k). Consistent with these data and a recent study identifying increased BAL IL-1β levels in respiratory PASC8, we found that circulating IL-1β levels were elevated in convalescent individuals recovering from COVID-19 exhibiting persistent pulmonary function deficits (Fig. 5m and Supplementary Table 4), implicating chronic IL-1β activity in the failure to restore normal lung function.
Given the efficacy of IL-6 neutralization in treating acute COVID-1948 and preclinical data suggesting rescue of fibrotic disease after bleomycin injury49, we also examined the efficacy of IL-6 blockade in our model (Extended Data Fig. 10l). In contrast to the neutralization of IFNγ + TNF or IL-1β, blocking IL-6 activity in post-viral lungs did not improve fibrotic sequelae, KRT8high and KRT5+ areas, alveolar regeneration or pulmonary function (Extended Data Fig. 10m–r). Although KRT5+ progenitors were previously shown to migrate to the distal airspace in response to IL-6 signalling49, our data indicate that their maintenance on pod formation after infection is independent of IL-6 signalling.
Collectively, these data support the neutralization of IFNγ + TNF or IL-1β in the post-acute stage of viral infection as viable therapeutic options to augment alveolar regeneration and dampen fibrotic sequelae observed after respiratory viral infections (Extended Data Fig. 10s).
Discussion
Immune-derived cues have recently been shown to influence lung repair3,22,35,42,50; however, their role in post-viral fibrosis remains unexplored. Here we describe a spatially defined aberrant immune–epithelial progenitor niche—a unique feature of lungs after viral fibrosis, but not after IPF. With insights from patient samples, we established a mouse model of post-viral lung fibrosis to demonstrate a causal role for exuberant interactions between CD8+ T cells, macrophages and epithelial progenitors in driving chronic sequelae and fibrosis after viral pneumonia. Whether the prolonged maintenance and activity of lung CD8+ T cells is a result of excessive TGF-β signalling9, chronic persistence of viral remnants51,52 or other independent mechanisms remains unclear. However, the prominent spatiotemporal association between the epithelial progenitors and CD8+ T cells in post-viral lungs strongly suggests the production of T cell recruitment and/or survival signals by the epithelium. It is vital to further extend this work and identify the contributions of other immunological factors in the post-viral lung, including interactions with the mesenchyme and the endothelium in dictating the balance between functional and pathological lung repair. Importantly, this model provides a platform to further define mechanisms of post-viral disease that is not confined to SARS-CoV-2 infection but has also been reported after viral infections such as influenza, MERS-CoV and SARS-CoV-12,3.
As PASC and other post-viral pathologies are typically diagnosed several weeks or months after the primary infection, effective treatment strategies to rescue disease in the chronic phase are critical. In contrast to anti-viral agents that require early intervention10, we show that the neutralization of IFNγ + TNF or IL-1β activity in the post-viral lung can be used as a ‘pro-repair’ strategy to augment alveolar regeneration and dampen fibrotic sequelae. Our data strongly suggest that drugs such as anakinra, an IL-1 receptor antagonist, or baricitinib, a JAK inhibitor, may be promising candidates to address this unmet need and treat respiratory PASC53,54. Considering that four respiratory viral pandemics and several more epidemics have occurred in the past two centuries, this work may have implications beyond respiratory PASC and could be foundational for potential (and probable) future viral outbreaks.
Methods
Ethics and biosafety
All aspects of this study were approved by the Institutional Review Board Committee at Cedars-Sinai Medical Center (IRB Pro00035409) and the University of Virginia (IRB 13166). Work related to SARS-CoV-2 was performed in animal biosafety level 3 (ABSL-3) facilities at the University of Virginia and influenza-related experiments were performed in animal biosafety level 2 (ABSL-2) facilities at the Mayo Clinic and the University of Virginia.
Cells, viruses and mice
The African green monkey kidney cell line Vero E6 (ATCC CRL-1587) was obtained from ATCC, tested to be free of mycoplasma contamination and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), along with 1% penicillin–streptomycin and l-glutamine at 37 °C in 5% CO2. The SARS-CoV-2 mouse-adapted strain (MA-10) was provided by B. J. Mann. The virus was passaged in Vero E6 cells, and the titre was determined by plaque assay using Vero E6 cells.
Young (aged 8–10 weeks) and aged (aged 84 weeks) female C57BL/6J mice, and 15-week-old female BALB/cJ mice (aged in-house to 10–12 months) were purchased from The Jackson Laboratory (JAX). K18-hACE2 (034860) transgenic mice as well as wild-type C57BL/6 (000664) mice were purchased from the Jackson Laboratory and bred in-house. Aged mice were received at 20 to 21 months of age from the National Institutes of Aging and all mice were maintained in the facility for at least 1 month before infection. All of the mice were housed in a specific-pathogen-free environment and used under conditions fully reviewed and approved by the institutional animal care and use committee guidelines at the Mayo Clinic (Rochester, MN) and University of Virginia (Charlottesville, VA).
For primary IAV virus infection, influenza A/PR8/34 strain (75 plaque-forming units (PFU) per mouse) was diluted in fetal bovine serum (FBS)-free Dulbecco’s modified Eagle’s medium (DMEM) (Corning) on ice and inoculated in anesthetized mice through intranasal route as described previously32. For SARS-CoV-2 infections, mice were infected with 5 × 104 PFU for C57BL/6 and 1,000 PFU for BALB/c of mouse-adapted (MA-10) virus or 200 PFU of USA-WA1/2020 for K18-hACE2 mice, intranasally under anaesthesia as described previously. Infected mice were monitored daily for weight loss and clinical signs of disease for 2 weeks, and later once a week for the duration of the experiments. The mortality rate of mice was calculated from mice that were either found dead in the cage or were euthanized when they reached 70% of their starting body weight, which is the defined humane end point in accordance with the respective institutional animal protocols. At the designated end point, the mice were humanely euthanized by ketamine–xylazine overdose and subsequent cervical dislocation. For this study, animal numbers were estimated on the basis of previous results from our infection model. Mice were assigned to treatment groups accounting for equal distribution of severity of acute disease across groups and thus investigators were not blinded to group allocation.
In vivo drug treatment
For depletion of CD8+ T cells and neutralization of cytokines, starting at 21 d.p.i., mice were intraperitoneally administered 20 μg or 500 μg of anti-CD8 antibody (53-6.7 and 2.43; BioXCell), anti-IL-1β (B122; BioXCell), anti-IFNγ (XMG1.2; BioXCell), anti-TNF (XT3.11; BioXCell) or isotype control immunoglobulin G (IgG) in 200 μl PBS once a week. Mice were euthanized 3 days after the last treatment.
Evaluation of respiratory mechanics and lung function
Lung function measurements using the forced oscillation technique and the resulting parameters have been previously described55. In brief, animals were anaesthetized with an overdose of ketamine–xylazine (100 and 10 mg per kg intraperitoneally) and paralysed with pancuronium bromide (1mg per kg intraperitoneally) to prevent respiratory efforts. Mice were then tracheostomized with a blunt 18-gauge canula (typical resistance of 0.18 cm H2O s ml−1), which was secured in place with a nylon suture. The mice were connected to the computer-controlled piston (SCIREQ flexiVent), and forced oscillation mechanics were performed under tidal breathing conditions described previously55 with a positive-end expiratory pressure of 3 cm H2O. The measurements were repeated after thorough recruitment of closed airways (two manoeuvres rapidly delivering total lung capacity of air and sustaining the required pressure for several seconds, mimicking holding of a deep breath). Each animal’s basal conditions were normalized to their own maximal capacity. Measurement of these parameters before and after lung inflation enables determination of large and small airway dysfunction under tidal (baseline) breathing conditions. Only measurements that satisfied the constant-phase model fits were used (>90% threshold determined by software). After this procedure, mice had a heart rate of around 60 beats per minute, indicating that the measurements were carried out on live individuals.
Human lung tissue specimens
Human lung samples were obtained from patients enrolled in the IRB-approved Lung Institute BioBank (LIBB) study at Cedars-Sinai Medical Center, Los Angeles, CA and at the University of Virginia, Charlottesville, VA. All of the participants or their legal representatives provided informed written consent. Lung tissues were processed within 24 h after surgical removal. Specifically, the lung tissues were cut and immediately fixed in 10% normal-buffered formalin for 24 h before tissue processing using the HistoCore PEARL–Tissue Processor (Leica) and embedding in paraffin for histological studies. The formalin-fixed paraffin-embedded cassettes were appropriately stored at room temperature until further sectioning.
For the influenza samples, lungs from deceased donors were obtained during organ acquisition for clinical transplantation through LifeLink of Georgia. As the donors were deceased at the time of collection, this study was exempt from International Review Board review. Informed consent was obtained from next of kin before explant. To ensure tissue viability, the samples were transported on ice and processed within 12 h of procurement. The sections from the human lungs were cut, embedded in OCT and then flash-frozen for immunofluorescence microscopy.
Mouse tissue processing and flow cytometry analysis
Mice were injected intravenously with 2 μg of CD45 or CD90.2 antibody labelled with various fluorochromes. Then, 2 min after injection, the mice were euthanized with an overdose of ketamine–xylazine and processed 3 min later. After euthanasia, the right ventricle of the heart was gently perfused with chilled 1× PBS (10 ml). The right lobes of the lungs were collected in 5 ml of digestion buffer (90% DMEM and 10% PBS and calcium with type 2 collagenase (180 U ml−1; Worthington) and DNase (15 μg ml−1; Sigma-Aldrich) additives). Tissues were digested at 37 °C for 1 h followed by disruption using the gentleMACS tissue dissociator (Miltenyi). Single-cell suspensions were obtained by hypotonic lysis of red blood cells in ammonium-chloride-potassium buffer and filtration through a 70 μm mesh. Cells were washed with FACS buffer (2% of FBS and 0.1% of NaN3 in PBS) and FCγ receptors were blocked with anti-CD16/32 (2.4G2). Surface staining was performed by antibody (details of which are provided in Supplementary Table 5) incubation for 30 min at 4 °C in the dark. After washing with PBS, cells were resuspended with Zombie-dye (BioLegend) and incubated at room temperature for 15 min. For IL-1β staining, cells were incubated with monesine (BioLegend) for 5 h at 37 °C and then stained with surface markers antibodies. After washing with FACS buffer, cells were fixed with fixation buffer (BioLegend) and permeabilized with intracellular staining permeabilization wash buffer (BioLegend). The cells were then stained with anti-IL-1β at room temperature for 1 h and the samples were acquired on the Attune NxT (Life Technologies) system. The data were analysed using Attune NXT software (v.5.2.0) and FlowJo (Tree Star).
Caspase-1 FLICA analysis
Caspase-1 was detected using the FAM-FLICA Caspase-1 Assay Kit (ImmunoChemistry Technologies) according to the manufacturer’s instructions. In brief, lung single cells or macrophages from in vitro culture were stained with fluorochrome-conjugated antibody cocktail for cell surface markers. After staining, cells were incubated with FLICA for 30 min at 37 °C, washed and detected by flow cytometry.
Cell isolation and ex vivo co-culture
To isolate myeloid and CD8+ T cells, single-cell suspension of the lung was generated from IAV-infected mice as described above and labelled and enriched with CD11c and CD11b or CD8 microbeads (Miltenyi Biotec) according to the manufacturer’s instructions. Purified myeloid cells were seeded in 96-well (200,000 per well) plates and incubated for 2 h at 37 °C 5% CO2 to facilitate attachment. Wells were washed with 1× PBS to select for macrophages, following which selected wells were seeded with naive or memory CD8+ T cells (40,000 per well) and further incubated for 16–18 h. Supernatant and cell pellets were collected for cytokine measurement and gene expression analyses, respectively.
AT2 cell isolation and culture
AT2 cells were isolated from naive mice as previously described46,56,57. In brief, mouse lungs were perfused with chilled PBS and intratracheally instilled with 1 ml of dispase II (15 U ml−1, Roche), tying off the trachea and cutting away the lobes from the mainstem bronchi. Lungs were incubated in 4 ml of 15 U ml−1 dispase II for 45 min while shaking at room temperature, followed by mechanical dissociation with an 18 G needle. After passage through a 100 μm filter, lungs underwent 10 min of DNase I digestion (50 μg ml−1) and were filtered through a 70 μm filter before RBC lysis. Single-cell suspensions were subjected to CD45 depletion using microbeads (Miltenyi), incubated with anti-FcgRIII/II (Fc block) and stained for CD45, EPCAM and MHC-II, and with viability die (antibody details are provided in Supplementary Table 5). Fluorescence-activated cell sorting was performed on the BD Influx cell sorter to isolate AT2 cells as described previously56 and collected in 500 μl DMEM + 40% FBS + 2% penicillin–streptomycin. Sorted AT2 cells (200,000 per well) were plated in a 96-well plate in DMEM/F12 + 10% FBS and cultured at 37 °C, 5% CO2 for 3 days before collection. For in vitro neutralization, anti-IL-1β (B122; BioXCell) was used at 10 μg ml−1 and, for stimulation, IL-1β (Peprotech) was used at 20 ng ml−1.
RNA isolation and qPCR with reverse transcription
Cells were lysed in buffer RLT and RNA was purified using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s instructions. Random primers (Invitrogen) and MMLV reverse transcriptase (Invitrogen) were used to synthesize first-strand complementary DNAs (cDNAs) from equivalent amounts of RNA from each sample. cDNA was used for quantitative PCR (qPCR) with the Fast SYBR Green PCR Master Mix (Applied Biosystems). qPCR was conducted on the QuantStudio 3 (Applied Bioscience) system. Data were generated with the comparative threshold cycle (ΔCt) method by normalizing to hypoxanthine-guanine phosphoribosyltransferase (HPRT) transcripts in each sample as reported previously58.
COVID-19 convalescent cohort
Peripheral blood specimens were obtained from patients presenting to the University of Virginia Post-COVID-19 clinic. All of the participants provided written informed consent. Pulmonary function testing (spirometry, lung volume testing, diffusing capacity of the lungs for carbon monoxide) at the time of blood draws was used to define normal and abnormal lung function.
IL-1β cytokine evaluation
Plasma from convalescent individuals recovering from COVID-19, supernatant from ex vivo culture and mouse BAL fluid after flushing of airway with 600 μl of sterile PBS were used to quantify IL-1β levels by ELISA according to the manufacturer’s instructions (R&D systems). The samples were first concentrated 5× (BAL) and 2× (cell culture supernatant) using Microcon-10kDa centrifugal filters (Millipore Sigma).
Histopathological analysis of lung tissues
Mouse lungs were routinely perfused with ice cold 1× PBS and the left lung was inflated and fixed in 10% formalin, embedded in paraffin and cut into 5-μm-thick sections. The sections were stained with haematoxylin–eosin and Masson’s trichrome. All of the sections were studied using light microscopy. The severity of fibrosis was semi-quantitatively assessed by blinded reviewers according to a modified Ashcroft scale59 and scored between 0 to 8 by examining randomly chosen fields. For histopathology, tissues were evaluated for oedema or hyaline membranes, interstitial disease and perivascular infiltrates as previously described60.
Immunofluorescence
Formalin-fixed paraffin-embedded lung tissue sections (5 μm) were deparaffinized in xylene and rehydrated. For PDLIM7 staining, tissues were fixed in 4% PFA at 4 °C for 8 h before cryoprotection in 30% sucrose. Tissues were later embedded in OCT and sectioned (5 μm). Heat-induced antigen retrieval was performed using 1× Agilent Dako target retrieval solution (pH 9) or 1× citrate solution (for frozen tissue; pH 6) in a steamer for 20 min (mouse lungs) or 45 min (human lungs), followed by blocking and surface staining. For intracellular targets, tissues were permeabilized with 0.5% Triton X-100 and 0.05% Tween-20 for 1 h at room temperature. The sections were stained with primary antibodies as listed in Supplementary Table 5 overnight at 4 °C. Subsequently, the samples were washed and incubated with fluorescent secondary antibodies as listed in Supplementary Table 5 for 2 h at room temperature. The sections were counterstained with DAPI (1:1,000, Thermo Fisher Scientific) for 3 min and mounted using ProLong Diamond Antifade mountant (Thermo Fisher Scientific). After 24 h of curing at room temperature, images were acquired using a ×10 or ×20 objective on the Olympus BX63 fluorescence microscope and pseudocolours were assigned for visualization. For all mouse samples, the left lobe of the lungs was used for imaging, for which 10–12 random fields in the distal lung were considered for quantification and the average value from each mouse was represented in respective graphs. Human lungs were similarly imaged and quantified; however, all available lobes sections were used. All images were further processed using ImageJ Fiji, cellSens, OlyVIA and/or QuPath software61. Quantitative analysis of stitched immunofluorescence images was performed using QuPath v.0.4.1, where tissue-specific expression of KRT8high was used to annotate areas of dysplastic repair and discriminate them from the remaining intact human or mouse lung tissue. CD8+ T cells and macrophages were detected using CD8α or CX3CR1 (mouse)/CD68 (human) signals, respectively. The abundance and distribution of the respective cells of interest were quantified to plot the distance from the annotation (KRT8high). Density maps were further generated to identify hotspots of the cells across the tissue with respect to the annotations. The same parameters for annotation and cell type classification were applied to all of the samples from the same experiment.
RNA in situ hybridization
RNA in situ hybridization was performed on paraffin-embedded lung sections using the RNAscope Multiplex Fluorescent Assay v2 according to the manufacturer’s instructions (Advanced Cell Diagnostics; ACD). Tissues were deparaffinized with xylene (2×, 5 min) and 100% ethanol (2×, 2 min) and incubated with hydrogen peroxide for 10 min at room temperature. Using a steamer, target retrieval was performed for 30 min followed by 30 min treatment with Protease Plus (ACD) at 40 °C. The slides were hybridized with custom probes at 40 °C for 2 h and signals were amplified according to the manufacturer’s instructions. Stained sections were captured at ×20 using the Olympus BX63 fluorescence microscope.
ST analyses
Spatial transcriptomics (ST) data (generated on the 10x Visium platform) were preprocessed using the spaceranger package (v.2.0, genome version mm10). The R package Seurat (v.4.3.0) was used for quality control and basic analyses. Only high-quality spots with sufficient gene coverage (≥2,000) were retained for downstream analysis. Similar to the single-cell RNA-sequencing data, the spatial expression data were first normalized to a log scale using the SCTransform method (v.0.3.5). The top 2,000 highly variable genes were then identified based on the variance of expression across spots for the principal component analysis input. UMAP embeddings were generated for visualization at the reduced dimensionality (top 30 principal components). Spots were clustered with a shared nearest neighbour modularity optimization-based clustering algorithm (built in Seurat). The samples were integrated and batch-effects were removed using the Harmony package (v.0.1). In addition to the spatial spot view of the expression pattern, we used UMAP to visualize the expression pattern of spots associated with the identified cell types in a lower dimension using the visualization pipeline in the Seurat package. Marker gene (differentially expressed genes (DEGs)) detection among different groups of spots was also performed with Seurat (FindMarker function), and the marker gene lists were further submitted to the gene set enrichment analysis pre-rank test for the functional annotation analysis.
We determined each spot’s potential cell type composition based on the following protocol. (1) We define alveolar epithelium (AE) spots based on the k-means clustering analysis (using knowledge-based key genes from different cell types). (2) For non-AE spots, we use the average expression of the Krt marker gene list (Supplementary Table 6) to estimate a Krt score and, similarly, average expression of Cd8 marker gene list (Supplementary Table 6) for Cd8 score. We set −0.4 as the cut-off for both the Cd8 and Krt score: spots with a Krt/Cd8 score > −0.4 were assigned as a Krt/Cd8-associated spot, and assigned blue/red colour, respectively, in the spatial map. Thus, a spot can exhibit a Krt and Cd8 signature at the same time, and we use purple to represent this category (Fig. 3a). For a given gene list, we captured the expression of relevant genes to compare the distribution of their expression index between Krt spots and AE spots. The significance of the difference was estimated using the Wilcoxon rank-sum test. The P values were then log10 transformed and displayed as bar plots. Similar to the Cd8 score and Krt score, we also defined some other cell type scores based on our knowledge-based marker gene list (Supplementary Table 6). We further used these scores to estimate the association between cell types in a specific slide or the global level (Pearson correlation coefficient). A pair of cell types with a higher Pearson correlation coefficient between their cell type score indicates a higher likelihood of their co-occurrence within the same spots.
For human ST data, most analyses were performed using methods similar to the mouse ST data. Moreover, we split spots by the expression of KRT8 and AGER using the cut-off of normalized expression index greater than 4. Spots with KRT8 > 4 and AGER ≤ 4 were defined as KRT8-only spots, whereas spots with KRT8 ≤ 4 and AGER > 4 were defined as AGER-only spots. These two spot sets were collected to compare the difference in cell type scores, key gene expression and pathway enrichment between them. In detail, we compared the percentage of ON spots (given a cell type score or gene expression index passing the cut-off) between the two groups (for example, KRT17-only spots versus AGER-only spots, or COVID spots versus control spots), and displayed the results as bar plots. A similar analysis was applied to examine the enrichment of relevant signalling pathways. Note that the cut-offs of normalized single-gene expression were set to 0.8. The cell type score (for example, CD8, KRT, AE, MDM and AM scores) was set to 1.2 for human data, as the distribution of marker gene expression varied between different species and biological samples (that is, human lung COVID samples). Detailed marker gene lists for cell types and pathways are provided in Supplementary Table 6.
To evaluate the similarity between our human and mouse models, we first detected the DEGs between all PASC-PF spots and control spots (using the human ST data). Next, we projected the COVID > control DEGs to identify mouse homologues and compared the expression of these genes in mouse spot categories (for example, IgG versus anti-CD8, or mouse Krt spots versus AE spots). Similarly, we also made this comparison for the DEGs between human KRT spots and AE spots, as well as genes from the top pathways enriched in PASC-PF spots. Wilcoxon rank-sum tests were conducted to evaluate the significance of the difference in expression. The human–mouse homologue information was downloaded from Mouse Genome Informatics (https://www.informatics.jax.org/).
Statistical analyses
Quantitative data are presented as mean ± s.e.m. Unpaired two-tailed Student’s t-tests (two-tailed, unequal variance) were used to determine statistical significance with Prism software (GraphPad) for two-group comparison. For multiple-groups comparisons, ANOVA corrected for multiple comparisons was used when appropriate (GraphPad). log-rank (Mantel–Cox) tests were used for survival curve comparison and two-way ANOVA was used to analyse differences in weight loss. P < 0.05 was considered to be significant in all statistical tests.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All relevant raw data have been deposited at the GEO under accession codes GSE267226 and GSE267228. Source data are provided with this paper.
Code availability
Relevant code used for analysis of human and mouse spatial transcriptomics data has been deposited at GitHub (https://github.com/zang-lab/covidSTanalysis).
References
Nalbandian, A. et al. Post-acute COVID-19 syndrome. Nat. Med. 27, 601–615 (2021).
Choutka, J., Jansari, V., Hornig, M. & Iwasaki, A. Unexplained post-acute infection syndromes. Nat. Med. 28, 911–923 (2022).
Narasimhan, H., Wu, Y., Goplen, N. P. & Sun, J. Immune determinants of chronic sequelae after respiratory viral infection. Sci. Immunol. 7, eabm7996 (2022).
Cheon, I. S. et al. Immune signatures underlying post-acute COVID-19 lung sequelae. Sci. Immunol. 6, eabk1741 (2021).
Vijayakumar, B. et al. Immuno-proteomic profiling reveals aberrant immune cell regulation in the airways of individuals with ongoing post-COVID-19 respiratory disease. Immunity 55, 542–556 (2022).
Phetsouphanh, C. et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat. Immunol. 23, 210–216 (2022).
Jyothula, S. S. K. et al. Fulminant lung fibrosis in non-resolvable COVID-19 requiring transplantation. eBioMedicine 86, 104351 (2022).
Gagiannis, D. et al. Clinical, imaging, and histopathological features of pulmonary sequelae after mild COVID-19. Am. J. Respir. Crit. Care Med. 208, 618–621 (2023).
Yao, C. et al. Maladaptive TGF-β signals to the alveolar epithelium drive fibrosis after COVID-19 infection. Am. J. Respir. Crit. Care Med. 208, 201–204 (2023).
Xie, Y., Choi, T. & Al-Aly, Z. Association of treatment with nirmatrelvir and the risk of post–COVID-19 condition. JAMA Intern. Med. 183, 554–564 (2023).
Al-Aly, Z., Xie, Y. & Bowe, B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature 594, 259–264 (2021).
Bharat, A. et al. Lung transplantation for patients with severe COVID-19. Sci. Transl. Med. 12, eabe4282 (2020).
Stewart, I. et al. Residual lung abnormalities after COVID-19 hospitalization: interim analysis of the UKILD Post-COVID-19 Study. Am. J. Respir. Crit. Care Med. 207, 693–703 (2022).
Bowe, B., Xie, Y. & Al-Aly, Z. Postacute sequelae of COVID-19 at 2 years. Nat. Med. 29, 2347–2357 (2023).
Merad, M., Blish, C. A., Sallusto, F. & Iwasaki, A. The immunology and immunopathology of COVID-19. Science 375, 1122–1127 (2022).
Bingham, G. C. et al. High-dimensional comparison of monocytes and T cells in post-COVID and idiopathic pulmonary fibrosis. Front. Immunol. 14, 1308594 (2024).
Arish, M., Qian, W., Narasimhan, H. & Sun, J. COVID-19 immunopathology: from acute diseases to chronic sequelae. J. Med. Virol. 95, e28122 (2023).
Klein, J. et al. Distinguishing features of long COVID identified through immune profiling. Nature 623, 139–148 (2023).
Vaughan, A. E. et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517, 621–625 (2015).
Kobayashi, Y. et al. Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat. Cell Biol. 22, 934–946 (2020).
Strunz, M. et al. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun. 11, 3559 (2020).
Choi, J. et al. Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell 27, 366–82 (2020).
Kumar, P. A. et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell 147, 525–538 (2011).
Xie, T. et al. Abnormal respiratory progenitors in fibrotic lung injury. Stem Cell Res. Ther. 13, 64 (2022).
Habermann, A. C. et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 6, eaba1972 (2020).
Adams, T. S. et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 6, eaba1983 (2020).
Rendeiro, A. F. et al. Persistent alveolar type 2 dysfunction and lung structural derangement in post-acute COVID-19. Preprint at medRxiv https://doi.org/10.1101/2022.11.28.22282811 (2022).
Jiang, P. et al. Ineffectual type 2-to-type 1 alveolar epithelial cell differentiation in idiopathic pulmonary fibrosis: persistence of the KRT8hi transitional state. Am. J. Respir. Crit. Care Med. 201, 1443–1447 (2020).
Tsukui, T. et al. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat. Commun. 11, 1920 (2020).
Leist, S. R. et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183, 1070–1085 (2020).
Dinnon, K. H. et al. SARS-CoV-2 infection produces chronic pulmonary epithelial and immune cell dysfunction with fibrosis in mice. Sci. Transl. Med. 14, eabo5070 (2022).
Goplen, N. P. et al. Tissue-resident CD8+ T cells drive age-associated chronic lung sequelae after viral pneumonia. Sci. Immunol. 5, eabc4557 (2020).
Dinnon, K. H. et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586, 560–566 (2020).
Winkler, E. S. et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 21, 1327–1335 (2020).
Wei, X., Narasimhan, H., Zhu, B. & Sun, J. Host recovery from respiratory viral infection. Ann. Rev. Immunol. 41, 277–300 (2023).
Wang, F. et al. Regulation of epithelial transitional states in murine and human pulmonary fibrosis. J. Clin. Invest. 133, e165612 (2023).
Liang, J. et al. Reciprocal interactions between alveolar progenitor dysfunction and aging promote lung fibrosis. eLife 12, e85415 (2023).
D’Agnillo, F. et al. Lung epithelial and endothelial damage, loss of tissue repair, inhibition of fibrinolysis, and cellular senescence in fatal COVID-19. Sci. Transl. Med. 13, eabj7790 (2021).
Melms, J. C. et al. A molecular single-cell lung atlas of lethal COVID-19. Nature 595, 114–119 (2021).
Konkimalla, A. et al. Transitional cell states sculpt tissue topology during lung regeneration. Cell Stem Cell 30, 1486–502 (2023).
Fiege, J. K. & Langlois, R. A. Investigating influenza A virus infection: tools to track infection and limit tropism. J. Virol. 89, 6167–6170 (2015).
Katsura, H., Kobayashi, Y., Tata, P. R. & Hogan, B. L. M. IL-1 and TNFα contribute to the inflammatory niche to enhance alveolar regeneration. Stem Cell Rep. 12, 657–666 (2019).
Kostura, M. J. et al. Identification of a monocyte specific pre-interleukin 1 beta convertase activity. Proc. Natl Acad. Sci. USA 86, 5227–5231 (1989).
Littlefield, K. M. et al. SARS-CoV-2-specific T cells associate with inflammation and reduced lung function in pulmonary post-acute sequalae of SARS-CoV-2. PLoS Pathog. 18, e1010359 (2022).
Krishna, B. A. et al. Spontaneous, persistent, T cell-dependent IFN-γ release in patients who progress to long COVID. Sci. Adv. 10, eadi9379 (2024).
Riemondy, K. A. et al. Single cell RNA sequencing identifies TGFβ as a key regenerative cue following LPS-induced lung injury. JCI Insight 5, e123637 (2019).
Nabhan, A. N., Brownfield, D. G., Harbury, P. B., Krasnow, M. A. & Desai, T. J. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123 (2018).
Salama, C. et al. Tocilizumab in patients hospitalized with COVID-19 pneumonia. N. Engl. J. Med. 384, 20–30 (2020).
Stancil, I. T. et al. Interleukin-6-dependent epithelial fluidization initiates fibrotic lung remodeling. Sci. Transl. Med. 14, eabo5254 (2022).
Choi, J. et al. Release of Notch activity coordinated by IL-1β signalling confers differentiation plasticity of airway progenitors via Fosl2 during alveolar regeneration. Nat. Cell Biol. 23, 953–966 (2021).
Chen, B., Julg, B., Mohandas, S., Bradfute, S. B. & Force RMPT. Viral persistence, reactivation, and mechanisms of long COVID. eLife 12, e86015 (2023).
Proal, A. D. et al. SARS-CoV-2 reservoir in post-acute sequelae of COVID-19 (PASC). Nat. Immunol. 24, 1616–1627 (2023).
An EUA for baricitinib (olumiant) for COVID-19. Med. Lett. Drugs Ther. 62, 202–203 (2020).
An EUA for anakinra (kineret) for COVID-19. Med. Lett. Drugs Ther. 64, e203 (2022).
Goplen, N. et al. Combined sensitization of mice to extracts of dust mite, ragweed, and Aspergillus species breaks through tolerance and establishes chronic features of asthma. J. Allerg. Clin. Immunol. 123, 925–932 (2009).
Major, J. et al. Endothelial AHR activity prevents lung barrier disruption in viral infection. Nature 621, 813–820 (2023).
Weiner, A. I. et al. ΔNp63 drives dysplastic alveolar remodeling and restricts epithelial plasticity upon severe lung injury. Cell Rep. 41, 111805 (2022).
Huang, S. et al. PPAR-γ in macrophages limits pulmonary inflammation and promotes host recovery following respiratory viral infection. J. Virol. 93, e00030-19 (2019).
Hübner, R. H. et al. Standardized quantification of pulmonary fibrosis in histological samples. Biotechniques. 44, 507–511 (2008).
Wong, L.-Y. R. et al. Eicosanoid signalling blockade protects middle-aged mice from severe COVID-19. Nature 605, 146–151 (2022).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Acknowledgements
We thank the Lung Institute BioBank study staff at Cedars-Sinai Medical Center for providing human lung specimens and patient clinical data; and A. Vaughan and R. Zemans for protocols to isolate and culture AT2 cells. Schematics were created using BioRender. Data for this manuscript were generated using the Flow Cytometry and Research Histology Core Facilities at the University of Virginia. 10x Visium spatial transcriptomics on the human and mouse tissues were performed at the Applied Genomics, Computation and Translational Core at Cedars-Sinai Medical Center and the Advanced Genomics Core at the University of Michigan, respectively. The study was in part supported by the US National Institutes of Health grants AI147394, AG069264, AI112844, HL170961, AI176171 and AI154598 to J. Sun; R35GM133712 to C.Z.; R01HL132287, R01HL167202 and R01HL132177 to Y. M. Shim; R56 AI178669 to J.W.; R35HL150803 to J.E.K.; Emory Center of Excellence for Influenza Research and Response grant 75N93019R0028 to J.E.K.; F31HL164049-01A1 to C.L.R.M.; and F31HL170746 and T32AI007496 to H.N.
Author information
Authors and Affiliations
Contributions
H.N. and J. Sun conceived the overall project. H.N., I.S.C., W.Q., S.S.H., T.P., C.Z., P.C. and J. Sun designed the experimental strategy and analysed data. All of the authors performed experiments, analysed data, or contributed critical reagents to the study. H.N. and J. Sun wrote the original draft. All of the authors read, edited and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
J. Sun receives support for research unrelated to this project from Icosavax. J.W. receives support for research unrelated to this project from Regeneron.
Peer review
Peer review information
Nature thanks Stanley Perlman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Spatial transcriptomics of human PASC-PF lungs reveal persistent defects in alveolar regeneration and chronic inflammation.
(a) Representative RNA-ISH images staining for SARS-CoV-2 Spike RNA in acute COVID-19 (n = 5) and PASC-PF lung (n = 18) sections. (b) Representative H&E images of human control and PASC-PF lungs. (Scale bar = 2 mm) (c) UMAP visualization of spatial transcriptomics data from human control (n = 2) and PASC-PF (n = 3) lungs. (d) Gene expression distribution across all identified clusters of spots. (e) GSEA analysis of signalling pathways differentially regulated between control (n = 2) and PASC-PF (n = 3) lungs. (f) Spatial gene expression maps of epithelial and immune cell markers in control (n = 2) and PASC-PF (n = 3) lungs. (g) Spatial gene expression maps of CD4 in control (n = 2) and PASC-PF (n = 3) lungs. (h) Heatmap of the physical distribution of alveolar epithelium, dysplastic progenitors, and CD8+ T cells. (i) Representative immunofluorescence images staining alveolar epithelial cell markers (AT1 – AGER; AT2 – proSP-C) in control and PASC-PF lung sections, and (j) staining CD8+ T cells (CD8α+) and epithelial progenitors (KRT5+, KRT17+, and KRThigh) in control and (k) PASC-PF lung sections. (l) Higher magnification inset of PASC-PF lungs showing independent channels (red arrows – KRT5+, yellow arrows – KRT17+KRT5−, grey arrows – KRT8high). (m) Quantification of KRT5+ area in control and PASC-PF lung sections (n = 12 control, 8 PASC-PF) (***p = 0.0007). (n) Representative immunofluorescence images staining for myofibroblasts (αSMA) and aberrant epithelial progenitors (KRT5+, KRT17+KRT5−, KRT8high) in human control, PASC-PF, and IPF lung sections. (o) Representative immunofluorescence images staining CD8+ T cells and epithelial progenitors in idiopathic pulmonary fibrosis (IPF) lung sections. (p) Quantification of CD8+ T cell number in control, PASC-PF, and IPF lung sections (n = 11 control, 13 IPF, 19 PASC-PF) (*p = 0.0343; ***p = 0.0001). (q) Representative Masson’s Trichome (MT) image (Scale bar = 2 mm) and (r) immunofluorescence images staining alveolar epithelial cell markers (AT1 – AGER; AT2 – proSP-C) in influenza-infected human lung sections. (s) Quantification of AT1 (AGER+) (*p = 0.0261) and AT2 (proSP-C+) (*p = 0.0272) cells, and (t) representative immunofluorescence images staining CD8+ T cells (CD8α+) and KRT8high transitional cells in influenza-infected human lungs (n = 3 control, 4 influenza). Statistical analyses were conducted using a two-tailed unpaired t-test (m,s) and one-way ANOVA (p). *p < 0.05; ***p < 0.001. Data are expressed as mean ± SEM; ns – not significant.
Extended Data Fig. 2 Characterization of immune-epithelial progenitor interactions in human PASC-PF, influenza, and IPF lungs.
(a) Representative immunofluorescence images staining CD8+ T cells and (b) quantification of CD8+ T cell distribution within KRT8−/low and KRT8high areas in PASC-PF lungs (5-7 random fields/lung; n = 10) (****p < 0.0001). (c) Representative immunofluorescence images staining CD8+ T cells and (d) quantification of CD8+ T cell distribution within KRT17−KRT5− or KRT17+KRT5− areas in PASC-PF lungs (5-7 random fields/lung; n = 10) (****p < 0.0001). (e) Representative immunofluorescence images staining CD8+ T cells and (f) quantification of CD8+ T cell distribution within in KRT5− and KRT5+ areas in PASC lungs (5-7 random fields/lung; n = 10 lungs) (****p < 0.0001). (g) Unbiased analysis of CD8+ T cell distribution in an influenza-infected human lung section using QuPath. (h) Representative Masson’s Trichrome images of lungs from donors with prior influenza infection. (Scale bar – black = 500 μm; grey = 2 mm). (i) Simple linear regression of CD8+ T cell number and KRT8high area fraction in lungs with prior influenza infection (15 random fields/lung; n = 2 lungs). (j) Representative immunofluorescence images staining CD8+ T cells and (k) quantification of CD8+ T cells in KRT8−/low and KRT8high areas within IPF lungs. (n = 8) (l-m) Simple linear regression of CD8+ T cell number and (l) KRT8high area fraction in control lungs (n = 6-8 random fields; N = 7 lungs), (m) KRT5+ area fraction in PASC-PF (6-8 random fields/lung; n = 7) and (n) KRT+ area fraction in IPF (6-8 random fields/lung; n = 7) lung sections. Statistical analyses were conducted using a two-tailed unpaired t-test. ****p < 0.0001. Data are expressed as mean ± SEM; ns – not significant.
Extended Data Fig. 3 IAV but not SARS-CoV-2 infection induces chronic pulmonary pathology in aged mice.
(a) Percent change in bodyweight and survival (*p = 0.0291) of young and aged C57BL/6 following SARS-CoV-2 MA-10 infection (n = 10 per group). (b) Representative H&E images of young and aged C57BL/6 mice from acute (10 d.p.i.), and (c) H&E and MT at chronic (35 d.p.i.) phase post SARS-CoV-2 MA-10 infection. (d) Representative immunofluorescence images staining CD8+ T cells (CD8α) and epithelial progenitors (KRT5+ and KRT8high), and (e) quantification of CD8+ T cells in naïve and SARS-CoV-2 MA-10 infected (35 d.p.i.) aged C57BL/6 lungs. (n = 5 naïve, 4 MA-10) (**p = 0.0058). (f) Percent change in bodyweight and survival of aged BALB/c following SARS-CoV-2 MA-10 infection (n = 10 per group). (g) Representative H&E images of aged BALB/c mice from the acute (7 d.p.i.), and (h) H&E and MT chronic (35 d.p.i.) phase post SARS-CoV-2 MA-10 infection. (i) Representative immunofluorescence images staining CD8+ T cells (CD8α) and epithelial progenitors (KRT5+ and KRT8high), and (j) quantification of CD8+ T cells in naïve and SARS-CoV-2 MA-10 infected (35 d.p.i.) aged BALB/c lungs. (n = 4 naïve, 3 MA-10) (k) Percent change in bodyweight and survival data (*p = 0.0325) of aged K18-hACE2 mice following SARS-CoV-2 WA-1 virus infection (n = 3 PBS, n = 16 WA-1). (l) Representative H&E and MT images of aged K18-hACE2 lungs post SARS-CoV-2 WA-1 infection (35 d.p.i.) or PBS administration. (m) Representative immunofluorescence image staining CD8+ T cells (CD8α) and epithelial progenitors (KRT5+ and KRT8high), and (n) quantification of CD8+ T cells in naïve and SARS-CoV-2 WA-1 infected (35 d.p.i.) aged K18-hACE2 mice (n = 4 naïve, 4 WA-1). (o) Percent change in bodyweight and survival (*p = 0.0295) data of young and aged C57BL/6 following IAV virus infection (n = 10 per group). (p) Representative H&E and MT images of young and aged C57BL/6 lungs post IAV virus infection. (q) Representative image of young and aged IAV-infected mouse (35 d.p.i.) lung staining for CD8+ T cells and dysplastic progenitors (KRT8+ and KRT5+). Scale bar – black = 500 μm; grey = 2 mm. Data are representative of two (a-n) or three (o-p) independent experiments. Statistical analyses were conducted using a two-tailed unpaired t-test (e,j,n), two-way ANOVA (BW data – a,f,k,o), and log-rank test (survival data - a,f,k,o) *p < 0.05; **p < 0.01. Data are expressed as mean ± SEM; ns – not significant.
Extended Data Fig. 4 Chronic pathology and tissue sequelae after IAV infection mimics features of human PASC-PF.
(a) Representative immunofluorescence images staining CD8+ T cells (CD8α+) and epithelial progenitors (KRT5+ and KRT8high), and (b) quantification of KRT5+ area in young and aged lungs post IAV infection (n = 4-11 per time point) (*p = 0.0257). (c) Representative immunofluorescence images and (d) quantification of KRT8high transitional cells expressing PDLIM7 in young and aged C57BL/6 mouse lungs post-IAV infection (42 d.p.i.; n = 4) (*p = 0.0332). (e) Scatter plot comparing gene expression profiles of KRT8high transitional cells in young and aged mice following 14 days post-bleomycin injury from a previously published dataset (GSE157995). (f) Representative H&E and MT images (Scale bar – black = 500 μm; grey = 2 mm) and (g) evaluation of fibrotic disease (****p < 0.0001), and (h) evaluation of pathological lesions (**p = 0.0023, 0.0027; ****p < 0.0001) in aged naïve and IAV-infected C57BL/6 mice (250 d.p.i.) (n = 3 naïve, 8 infected). (i) Quantification of CD8+ T cells (*p = 0.0458), (j) KRT8high (*p = 0.0458), (k) KRT5+ areas (*p = 0.0327), (l) PDPN+ AT1 cells (*p = 0.0305) and (m) proSP-C+ AT2 cells in naïve and IAV-infected (250 d.p.i.) aged C57BL/6 mouse lungs (n = 3 naïve, 8 infected). Data are representative of two (d,g-m) or three (b) independent experiments. Statistical analyses were conducted using a two-tailed unpaired t-test (d,g,i-m), and two-way ANOVA (b,h). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Data are expressed as mean ± SEM; ns – not significant.
Extended Data Fig. 5 IAV infection of aged mice induces a sustained loss of alveolar epithelial cells unlike SARS-CoV-2.
(a) Representative immunofluorescence images of aged C57BL/6 and BALB/c mouse lungs infected with SARS-CoV-2 MA-10, staining alveolar epithelial cell markers (AT1- PDPN; AT2 – proSP-C). (b) Quantification of proSP-C+ AT2 (*p = 0.0141, 0.0168) and (c) PDPN+ AT1 area in aged C57BL/6 mice following SARS-CoV-2 MA-10 infection (n = 3-4 per time point). (d) Quantification of proSP-C+ AT2 (***p = 0.0005) and (e) PDPN+ AT1 area in aged BALB/c mice following SARS-CoV-2 MA-10 infection (n = 3-4 per time point). (f) Representative immunofluorescence images, and (g-i) quantification of proSP-C+ AT2 (Young - *p = 0.0333; **p = 0.0070. Aged - *p = 0.0391; **p = 0.0037; ***p = 0.0002) and PDPN+ AT1 (Young - **p = 0.0020, 0.0012; ***p = 0.0002. Aged - *p = 0.0441; **p = 0.0068) area in young and aged C57BL/6 mice following IAV infection (n = 3-4 per time point). (j) Representative immunofluorescence images of aged C57BL/6 mouse lungs infected with IAV or SARS-CoV-2 virus, staining for AT1 cells (PDPN) and viral nucleoprotein (NP). (k) Schematic of the FlexiVent system to evaluate pulmonary function in mice. The diagram was created using BioRender. (l) Evaluation of compliance of the respiratory system in young and aged mice post PBS administration or IAV infection (60 d.p.i.) (n = 3 per time point) (*p = 0.0135; **p = 0.0034). Data are representative of two (b-e,l) or three (g-h) independent experiments. Statistical analyses were conducted using a one-way ANOVA (b-e,h,i,l) and two-way ANOVA (g). *p < 0.05; **p < 0.01; ***p < 0.001. Data are expressed as mean ± SEM; ns – not significant.
Extended Data Fig. 6 Depletion of lung-resident CD8+ T cells improves fibrotic disease in aged but not young mice.
(a) Simple linear regression of CD8+ T cell number and KRT8high or (b) KRT5+ area fraction in aged IAV-infected mouse lungs at 30 d.p.i. (5-7 random fields/lung; n = 5) and (c) 60 d.p.i. (5-7 random fields/lung; n = 5). (d) Survival data of aged IAV-infected C57BL/6 mice following CD8+ T cell depletion prior to infection (n = 4 control IgG, 6 anti-CD8) (**p = 0.0046). (e) Representative H&E and MT images and (f) evaluation of fibrotic disease (n = 3 control IgG, 3 anti-CD8) (*p = 0.0207), and (g) pathological lesions in aged IAV-infected C57BL/6 mice (60 d.p.i.) following treatment with anti-CD8 or control IgG Ab. (n = 4 control IgG, 4 anti-CD8) (*p = 0.0125; **p = 0.0094; ***p = 0.0006). (h) Representative immunofluorescence images staining CD8+ T cells (CD8α+) and epithelial progenitors (KRT5+ and KRT8high), and (i) AT1 (PDPN+) and AT2 (proSP-C+) in aged C57BL/6 mouse lungs post IAV infection (60 d.p.i.), treated with anti-CD8 or control IgG Ab. (j) Representative H&E images (k) and immunofluorescence images of young C57BL/6 lungs post IAV infection (60 d.p.i.) treated with anti-CD8 or control IgG Ab. (l) Quantification of KRT8high, KRT5+ (*p = 0.0255), (m) AT1, and AT2 cells in young IAV-infected mouse lungs, treated with anti-CD8 Ab or control IgG Ab. (n = 5-6 IgG, 5-6 anti-CD8). (n) Representative immunofluorescence images of aged and young C57BL/6 lungs post-IAV infection (42 d.p.i.) following receipt of lung CD8+ T cells from young or aged infection-matched donors at 21 d.p.i. (o) Quantification of KRT8high area, (p) AT1 and (q) AT2 cells (**p = 0.0035) in aged or young C57BL/6 lungs post-IAV infection (42 d.p.i.) following receipt of lung CD8+ T cells from young or aged infection-matched donors at 21 d.p.i. (n = 3-6 per group). (r) Quantification of KRT8high (**p = 0.0021) and KRT5+ area (*p = 0.0454) in aged IAV-infected mouse lungs (60 d.p.i.), treated with control IgG Ab, low dose anti-CD8 or high dose anti-CD8 (n = 6-11 per group). Scale bar – black = 500 μm; grey = 2 mm. Data are representative of two (d,f,g,o) or three (l,m,r) independent experiments. Statistical analyses were conducted using a two-tailed unpaired t-test (f,l,m), two-way ANOVA (g), and an ordinary one-way ANOVA (o-r). *p < 0.05; **p < 0.01; ***p < 0.001. Data are expressed as mean ± SEM; ns – not significant.
Extended Data Fig. 7 CD8+ T cell depletion induces widespread changes in immune and epithelial cells gene expression when evaluated by spatial transcriptomics.
(a) Representative H&E images of aged IAV-infected mice (60 d.p.i.) treated with IgG Ab (n = 2) or anti-CD8 (n = 2) that were mounted on the 10X Visium slide. (b) UMAP visualization of spatial transcriptomics data from aged IAV-infected mice (60 d.p.i.), treated with IgG Ab or anti-CD8. (c) Heatmap of gene expression across all identified clusters of spots. (d) Evaluation of top DEGs enriched in human PASC-PF lungs within dysplastic (Krt) or healthy (AE) areas of aged IAV-infected mice. (e) Evaluation of top DEGs enriched in pathological areas of human PASC-PF lungs within dysplastic (Krt) or healthy (AE) areas of aged IAV-infected mice. (f) Evaluation of top signalling enriched in human PASC-PF lungs within dysplastic (Krt) or healthy (AE) areas of aged IAV-infected mice. (g) Evaluation of top DEGs enriched in human PASC-PF lungs within aged IAV-infected mice treated with IgG Ab or anti-CD8. (h) Evaluation of top DEGs enriched in pathological areas of human PASC-PF lungs (n = 3) within aged IAV-infected mice treated with IgG Ab or anti-CD8. (i) Evaluation of top signalling pathways enriched in human PASC-PF lungs (n = 3) within aged IAV-infected mice treated with IgG Ab or anti-CD8 (n = 2 each). (j) GSEA analysis of signalling pathways enriched in diseased (Krt5+ and Krt8hi-rich) areas compared to healthy alveolar epithelium. (k) Boxplots comparing gene expression signature of ADI/DATP/PATS and aberrant basaloid cells in diseased areas and healthy alveolar epithelium using spatial transcriptomics (n = 2 IgG). (l) Spatial map of the expression of dysplastic epithelial progenitors (Krt8, Krt5, Trp63), (m) alveolar epithelial (Sftpc1, Ager, Sftpa1), (n) CD8+ T cell (CD8a, CD8b1, Itgae), (o) monocyte-derived macrophages (Cx3cr1, Cd14, S100a6), and (p) alveolar macrophage (Flt1, Car4, Fabp5) marker genes in aged IAV-infected mice (60 d.p.i.), treated with IgG or anti-CD8. (q) Spatial map of the expression of genes associated with lung fibrosis in aged IAV-infected mice (60 d.p.i.), treated with IgG Ab or anti-CD8. Statistical analyses were conducted using a one-sided Wilcoxon rank sum test without any adjustment (d-I,k). Centerline, bounds of box, top line, and bottom line of the boxplots represent the median, 25th to 75th percentile range, 25th percentile − 1.5 × interquartile range (IQR), and 75th percentile + 1.5 × IQR, respectively.
Extended Data Fig. 8 Monocyte-derived macrophages but not alveolar macrophages are enriched in areas of dysplastic repair within human and mouse post-viral lungs.
(a) Representative immunofluorescence image of aged IAV-infected C57BL/6 mouse lungs staining for CXC3CR1+ macrophages and dysplastic progenitors (KRT5 and KRT8). (b) Representative immunofluorescence images of aged IAV-infected (60 d.p.i.) lungs treated with control IgG Ab or anti-CD8, staining monocyte-derived macrophages (CX3CR1) and dysplastic epithelial progenitors (KRT5 and KRT8). (c) Evaluation of CCL2 levels in bronchoalveolar lavage fluid from naïve or aged IAV-infected mice treated with control IgG Ab or anti-CD8 (n = 4 per group) (***p = 0.0002). (d) Quantification of Ccl2 (**p = 0.0020), Ccl3 (**p = 0.0052), and Cx3cl1 gene expression in lungs of aged IAV-infected mice treated with control IgG Ab or anti-CD8 (n = 3 IgG, 3 αCD8). (e) Quantification of the proportion of spots expressing the gene signature of monocyte-derived macrophages in human control and PASC-PF lungs. (f) Heatmap of the physical distribution of monocyte-derived macrophages (MDM), alveolar macrophages (AM), healthy alveolar epithelium (AE), and dysplastic areas (Krt) within human PASC-PF lungs. (g) Quantification of the proportion of spots expressing gene signatures characterizing monocyte-derived macrophages and alveolar macrophages in AGERhi and KRT17hi areas of human PASC-PF (n = 3) lungs. (h) Spatial gene expression maps of monocyte-derived macrophages, Krt-rich dysplastic areas, alveolar macrophages and healthy alveolar epithelium in human control (n = 2) and PASC-PF (n = 3) lungs. (i) Representative immunofluorescence image of human PASC-PF lungs staining for CD68+ macrophages and dysplastic progenitors (KRT5 and KRT8). (k) Quantification of CD68+ macrophages in human control and PASC-PF lungs (n = 12 control; 7 PASC-PF) (****p < 0.0001). Data are representative of two (c,d) independent experiments. Statistical analyses were conducted using a two-tailed unpaired t-test (d,k), and an ordinary one-way ANOVA (c). **p < 0.01; ***p < 0.001; ****p < 0.0001. Data are expressed as mean ± SEM; ns – not significant.
Extended Data Fig. 9 Areas of dysplastic repair are enriched with IL-1R signalling and inflammasome signatures in human PASC-PF and aged IAV-infected mouse lungs.
(a) Proportions of spots expressing gene signatures of IL-1R signalling and inflammasome activity in AGERhi and KRT17hi areas of human PASC-PF lungs. (b) Spatial gene expression maps of IL-1R signalling and inflammasome activity in aged IAV-infected mouse lungs (60 d.p.i.). (c) Gating strategy to identify proIL-1β+ cells IAV-infected mouse lungs by flow cytometry. (d) Histograms of caspase-1 levels in lungs of aged IAV-infected mice treated with IgG or Anti-CD8. (e) Gating strategy to identify FLICA+ cells in IAV-infected mice. (f) Spatial gene expression maps of inflammasome components in aged IAV-infected mice (60 d.p.i.) treated with IgG Ab or anti-CD8. (g) Representative immunofluorescence images staining for CD8+ T cells (CD8α) and TNF, and (h) IFN-γ in human control and PASC-PF lungs. (i) Quantification of TNF+ (*p = 0.0266) and IFN-γ+ (*p = 0.0378) lung-resident CD8+ T cells in naïve and IAV-infected (60 d.p.i.) aged mouse lungs (n = 5 naïve, 6 infected). (j) Proportions of spots expressing gene signatures of IFN-γ + TNF signalling in healthy (AGERhi) and diseased areas (KRT8hi or KRT17+) within human PASC-PF (n = 3) lungs. (k) Quantification of IL-1β gene expression (n = 3-4 wells/condition pooled from 4 mice) (*p = 0.0174; ***p = 0.0001), and (l) evaluation of IL-1β release following coculture of macrophages and CD8+ T cells from naïve aged C57BL/6 mice (n = 3 wells/condition pooled from 4 mice) (**p = 0.0037, 0.0089). (m) Gene expression of AT1 cell markers 3 days post AT2 cell culture and exposure to conditioned media (CM) (n = 2-3 wells/condition pooled from 5 mice) (*p = 0.0119, 0.0395; ****p < 0.0001). Data are representative of two (i,k,l,m) independent experiments. Statistical analyses were conducted using a two-tailed unpaired t-test (i), an ordinary one-way ANOVA (k,l), and a two-way ANOVA (m). *p < 0.05; **p < 0.01; *** p < 0.001; ****p < 0.0001. Data are expressed as mean ± SEM; ns – not significant.
Extended Data Fig. 10 Neutralization of IFN-γ and TNF or IL-1β activity in the post-acute phase of infection improves outcomes in aged IAV-infected mice.
(a) Representative H&E and MT images, (b) evaluation of fibrotic (n = 4 per group) (***p = 0.0003), (c) representative immunofluorescence images staining AT1 (PDPN+), AT2 (proSP-C+) and epithelial progenitors (KRT5+ and KRT8high), and (d) quantification of tissue damping (G) (*p = 0.0210) and elastance of the respiratory system (Ers) (*p = 0.0391) in aged IAV infected C57BL/6 mice (42 d.p.i.) treated with IgG Ab (n = 8) or anti-IFN-γ + anti-TNF neutralizing Ab (n = 9). (e) Experimental design for in vivo IL-1β blockade post IAV infection. The diagram was created using BioRender. (f) Representative H&E and MT images, (g) evaluation of fibrotic disease (n = 4 per group) (*p = 0.0400), (h) representative immunofluorescence images staining AT1 (PDPN+) and AT2 (proSP-C+) cells, and (i) quantification of tissue damping (G) (***p = 0.0008) and elastance of the respiratory system (Ers) (***p = 0.0001) in aged IAV infected C57BL/6 mice (42 d.p.i.) treated with control IgG Ab (n = 8) or anti-IL-1β neutralizing Ab (n = 9). (j) Evaluation of IL-1β levels in the BAL fluid of young and aged IAV-infected mice (42 d.p.i.; n = 6 young, 4 aged) (**p = 0.0024). (k) Representative H&E and MT images of young C57BL/6 lungs post IAV infection (42 d.p.i.) treated with anti-IL-1β neutralizing Ab or control IgG Ab. (l) Experimental design for in vivo IL-6 neutralization post IAV infection. The diagram was created using BioRender. (m) Representative H&E and MT images, and (n) evaluation of fibrotic disease in aged IAV-infected (42 d.p.i.) mice treated with anti-IL-6 neutralizing Ab or control IgG Ab (n = 3 per group), and (o) representative immunofluorescence images staining AT1 (PDPN+), AT2 (proSP-C+) and epithelial progenitors (KRT5+ and KRT8high), (p) quantification of KRT8high and KRT5+ area and (q) AT1 (PDPN+) and AT2 (proSP-C+) cells (n = 6 per group), and (r) evaluation of static compliance (Cst), resistance of the respiratory system (Rrs), and tissue elastance (H) in aged IAV-infected mice (42 d.p.i.) treated with IgG Ab (n = 5) or anti-IL-6 neutralizing Ab (n = 6). (s) An aberrant immune-epithelial progenitor nice drives post-viral lung sequelae. The diagram was created using BioRender. Scale bar – black = 500 μm; grey = 2 mm. Data are representative of two (b,g,j,n,p,r) or three (d,i) independent experiments. Statistical analyses were conducted using a two-tailed unpaired t-test. *p < 0.05; **p < 0.01; ***p < 0.001. Data are expressed as mean ± SEM; ns – not significant.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Narasimhan, H., Cheon, I.S., Qian, W. et al. An aberrant immune–epithelial progenitor niche drives viral lung sequelae. Nature (2024). https://doi.org/10.1038/s41586-024-07926-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41586-024-07926-8
- Springer Nature Limited