Abstract
Lung cancer is the leading cause of cancer deaths worldwide1. Mutations in the tumour suppressor gene TP53 occur in 50% of lung adenocarcinomas (LUADs) and are linked to poor prognosis1,2,3,4, but how p53 suppresses LUAD development remains enigmatic. We show here that p53 suppresses LUAD by governing cell state, specifically by promoting alveolar type 1 (AT1) differentiation. Using mice that express oncogenic Kras and null, wild-type or hypermorphic Trp53 alleles in alveolar type 2 (AT2) cells, we observed graded effects of p53 on LUAD initiation and progression. RNA sequencing and ATAC sequencing of LUAD cells uncovered a p53-induced AT1 differentiation programme during tumour suppression in vivo through direct DNA binding, chromatin remodelling and induction of genes characteristic of AT1 cells. Single-cell transcriptomics analyses revealed that during LUAD evolution, p53 promotes AT1 differentiation through action in a transitional cell state analogous to a transient intermediary seen during AT2-to-AT1 cell differentiation in alveolar injury repair. Notably, p53 inactivation results in the inappropriate persistence of these transitional cancer cells accompanied by upregulated growth signalling and divergence from lung lineage identity, characteristics associated with LUAD progression. Analysis of Trp53 wild-type and Trp53-null mice showed that p53 also directs alveolar regeneration after injury by regulating AT2 cell self-renewal and promoting transitional cell differentiation into AT1 cells. Collectively, these findings illuminate mechanisms of p53-mediated LUAD suppression, in which p53 governs alveolar differentiation, and suggest that tumour suppression reflects a fundamental role of p53 in orchestrating tissue repair after injury.
Similar content being viewed by others
Main
Multicellular organisms rely on intricate regenerative programmes to restore tissue homeostasis in response to injury5. These injury response mechanisms have been characterized in the epithelium of lung alveoli, the delicate sacs that mediate gas exchange in the mammalian lung5,6,7,8,9,10. The alveolar epithelium comprises AT1 cells—thin, quiescent cells that facilitate gas exchange—and AT2 cells—cuboidal cells that secrete pulmonary surfactants6,11. Following damage to the alveolar epithelium, AT2 cells serve as stem cells that both self-renew and differentiate to generate AT1 cells6,7,8,9,10. Dysregulation of homeostasis can lead to chronic inflammatory lung diseases, acute respiratory distress from viruses such as SARS-CoV-2, and LUAD6,12,13,14,15. During LUAD evolution, mutations in the tumour suppressor gene TP53 substantially derail homeostasis, which results in malignant progression and poor patient prognosis1,2,3,4. How p53 restrains LUAD development and whether these tumour suppression programmes relate to a more general role in governing lung homeostasis, however, remains unknown. Here we elucidate a new p53-directed mechanism of LUAD suppression through AT1 differentiation, which mirrors a physiological role of p53 in lung regeneration after injury.
p5353,54 is a super LUAD suppressor
To understand how p53 suppresses LUAD, we used mice carrying hyperactive, wild-type or loss-of-function Trp53 alleles (Fig. 1a, Extended Data Fig. 1a,b and Supplementary Fig. 1). Analyses of Trp5353,54 mice we generated that express p53(F53Q,F54S) (hereafter referred to p5353,54), a hyperactive tumour suppressor in pancreatic cancer16, provided an effective approach to delineate pathways that are fundamental to tumour suppression. We used genetically engineered mouse models harbouring Cre-inducible oncogenic KrasG12D and fluorescent reporter alleles (Kraslox-stop-lox (LSL)-G12D/+;Rosa26LSL-tdTomato/LSL-tdTomato (KT)) and homozygous for each Trp53 allele (KT;Trp53+/+ (hereafter referred to as KT), KT;Trp5353,54/53,54 (KFT) and KT;Trp53fl/fl (KPT); Fig. 1a and Extended Data Fig. 1c). These mouse models develop adenomas irrespective of Trp53 status but readily undergo malignant progression to adenocarcinomas following Trp53 loss17,18. Lung tumours were induced through Lenti-PGK-Cre instillation and examined 24 weeks later, a time point when adenocarcinomas predominate in KPT mice. We observed that wild-type p53 suppressed tumour burden, with reduced tumour sizes, as previously reported17,19, and decreased tumour numbers (Extended Data Fig. 1c–e). Tumour numbers and sizes were further reduced in KFT mice compared with KT mice, which demonstrates that p5353,54 is a super LUAD suppressor (Extended Data Fig. 1d,e). Histopathological analyses confirmed that p53 suppressed tumour progression, as increasing p53 activity correlated with a decreasing frequency of histological patterns associated with poor prognosis in humans (for example, solid) and reduced malignant features20 (Extended Data Fig. 1f–h). Notably, the reduced tumour numbers in p53-expressing mice suggested an unappreciated role for p53 in suppressing LUAD initiation. Next, we used a LUAD model in which KrasG12D expression was induced in AT2 cells, the predominant cell of origin for LUAD6,18,21,22. Using adenoviral-SPC-Cre (Ad-SPC-Cre) to initiate tumours, we observed that p5353,54 induced a greater reduction in tumour number, size and the overall tumour burden than wild-type p53 (Fig. 1b,c and Extended Data Fig. 2a,b). Proliferation was significantly reduced in KT tumours relative to KPT tumours and even more so in KFT tumours across cohorts (Extended Data Figs. 1i,j and 2c–e). Notably, a model in which p5353,54 was expressed only in cancer cells and not stromal cells showed that p5353,54 tumour suppressor activity was primarily cell-autonomous (Extended Data Fig. 2f–i). These results underscore the importance of p53 in suppressing LUAD initiation and progression and establish the enhanced potency of p5353,54 relative to wild-type p53. These mice also provide an important tool for deconstructing p53 pathways crucial for tumour suppression.
a, Allelic series of p53 mutants. p5353,54 is a hypermorphic transactivation domain 2 (TAD2) mutant allele. DBD, DNA-binding domain; fl, flox; TET, tetramerization domain; WT, wild type. b, Schematic of the lung cancer study. Histology cohort: KFT (n = 7), KT (n = 8) and KPT (n = 5) mice. RNA-seq cohort: KFT (n = 4), KT (n = 3) and KPT (n = 4) mice. ATAC-seq cohorts: (1) KT (n = 4) and KPT (n = 4) mice; and (2) KFT (n = 2) and KT (n = 2) mice. c, Lung tumour burden, number and size (n = 56, 311 and 617 tumours for KFT, KT and KPT cohorts, respectively). Data are the mean ± s.d. d, Principal component (PC) analysis of KFT, KT and KPT RNA-seq samples. e, Hyperactivated genes (n = 676) in KFT cells relative to KT cells (>1.2 fold change (FC);Methods). f, Top enriched terms in p5353,54-hyperactivated genes (Metascape51). g, Gene set enrichment analysis (GSEA) of gene ontology (GO) terms comparing KT and KPT cells. ES, enrichment score; FDR, false discovery rate; NES, normalized ES. h, Representative immunohistochemistry of NKX2-1 staining in 10-week old KFT (n = 3), KT (n = 3) and KPT (n = 3) mice. Scale bar, 20 μm. i, Quantification of NKX2-1 staining from h in n = 1,500 cells from KFT (n = 3), KT (n = 3) and KPT (n = 3) mice. Red line indicates the median. j, Left, differential accessibility versus log2(mean reads per region) in KT and KPT cells. Right, top TF motifs enriched in differentially accessible regions (log2(FC) > 2). k, Left, Differential accessibility versus log2(mean reads per region) in KT and KFT cells. (Right) TF motifs enriched in differentially accessible regions. l, Top, Venn diagram defining p5353,54 hyperaccessible regions (Methods). Bottom, Top terms enriched in genes associated with p5353,54 hyperaccessible regions. P values were calculated using one-way analysis of variance (ANOVA) with the Kruskal–Wallis test with multiple comparisons (two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli) (c), GSEA52 (g), or ordinary one-way ANOVA with multiple comparisons (Tukey’s multiple comparisons test) (i).
p53 enforces lung epithelial identity
We performed RNA sequencing (RNA-seq) and assay for transposase-accessible chromatin with sequencing (ATAC–seq) on sorted cancer cells from KFT, KT and KPT mice 10 weeks after Ad-SPC-Cre instillation, an early time point in tumour development, to reveal primary mechanisms of p53 action (Fig. 1b, Extended Data Fig. 3a and Supplementary Fig. 2). Cancer cells of each genotype displayed distinct transcriptional profiles, with >5,000 p53-dependent differentially expressed genes (Fig. 1d, Extended Data Fig. 3b,c and Supplementary Table 1). To pinpoint pathways pivotal for tumour suppression, we identified p53-dependent genes that were more highly activated in KFT cells than in KT cells (Fig. 1e). Functional annotation of p5353,54-hyperactivated genes identified high enrichment of development and differentiation pathways, including respiratory system development (Fig. 1f, Extended Data Fig. 3d and Supplementary Table 2). Although p5353,54 initially highlighted lung development and differentiation signatures, these signatures were also upregulated in KT cells relative to KPT cells (Fig. 1g, Extended Data Fig. 3e,f and Supplementary Tables 3 and 4). Immunostaining for NKX2-1, a pan-alveolar epithelial factor23,24,25, revealed increased expression of this marker in KT and KFT tumours relative to KPT tumours (Fig. 1h,i). These analyses suggest that p53 enforces a lung lineage programme during tumour suppression. ATAC-seq analysis of KT and KPT cancer cells revealed a global, p53-dependent reorganization of chromatin accessibility, and supported the role of p53 in promoting lung lineage fidelity (Fig. 1j, Extended Data Fig. 3g–i and Supplementary Table 5). Transcription factor (TF)-binding motifs associated with lineage specification TFs (for example, bHLH proteins)26 and oncogenic TFs (for example, RUNX proteins)27 were enriched in open chromatin regions of KT cells and KPT cells, respectively (Fig. 1j). Almost 3,000 regions with increased accessibility in KT cells relative to KPT cells were hyper-accessible in KFT cells and were associated with genes related to the lung and respiratory system, which mirrored our observations with p5353,54-hyperactivated genes (Fig. 1k,l, Extended Data Fig. 3g,j,k and Supplementary Table 5). Together, these data demonstrate that p53 enforces a chromatin landscape and transcriptional programmes reminiscent of normal lung cell identity.
p53 promotes AT1-like differentiation
To delineate mechanisms by which p53 enforces lung identity in alveolar adenomatous cells, we examined our RNA-seq dataset for signatures of AT1 and AT2 cells to determine whether p53 promotes a specific differentiation programme. Notably, despite the AT2 cell of origin for LUAD, gene expression programmes induced in KT cells relative to KPT cells were significantly enriched for AT1 cell signatures (Fig. 2a, Extended Data Fig. 4a,b and Supplementary Table 6). Moreover, upregulation of AT1-associated gene expression programmes was greater in KFT cells than in KT cells. This result provides support for the importance of this AT1-like programme in tumour suppression (Fig. 2a and Extended Data Fig. 4a,b). This upregulation did not result from wild-type p53 or p5353,54 affecting homeostatic alveolar cell proportions or proliferation (Extended Data Fig. 4c,d). By contrast, Trp53 status did not significantly affect the expression of most AT2-related signatures (Fig. 2a and Extended Data Fig. 4a). KPT cell expression profiles more closely resembled cells divergent from the lung, which provides support for the notion that Trp53 loss confers lineage plasticity in LUAD27,28,29 (Extended Data Fig. 4e). Reinforcing the idea that p53 drives AT1 identity in tumours, our epigenomic analyses revealed that AT1 gene cis-regulatory elements were increasingly accessible with increasing p53 activity (Extended Data Fig. 4f). Investigation of how p53 affects AT1 cell identity at the single-cell level by combined single-cell RNA-seq (scRNA-seq) and single-cell ATAC–seq (scATAC–seq; multiomics) analysis showed that KFT tumours had the highest proportion of AT1 cancer cells, followed by KT tumours and then KPT tumours (Extended Data Fig. 5 and Supplementary Fig. 2). Meanwhile, KPT tumours contained more cells with low lung lineage fidelity than p53-proficient tumours (Extended Data Fig. 5f,h,i). Accordingly, most tumours from KFT mice and KT mice displayed high AT1 cell marker protein expression, including AGER, HOPX and PDPN, whereas only rare tumours stained for AT1 cell markers in KPT mice (Fig. 2b,c and Extended Data Figs. 2j,k and 6a–f, h). By contrast, the AT2 cell marker SPC was expressed at similar levels among genotypes (Extended Data Fig. 6g,i,j). Of note, most AGER-expressing AT1-like cancer cells in KPT LUADs also expressed SPC, whereas few p53-proficient AT1-like cells stained for SPC. This result suggests that Trp53-null AT1-like cells exist in a more bipotent state (Extended Data Fig. 6k,l).
a, GSEA shows enrichment of alveolar cell signatures (Supplementary Table 6). NES > 0 indicates that the signature is enriched in KFT or KT cells relative to KPT cells. Asterisk indicates that the gene set is significantly enriched (P ≤ 0.05). Mouse and human alveolar signatures were taken from previously published studies12,24,39,53,54. b, Representative immunofluorescence (IF) images of AT1 cell markers and a cancer cell marker (tdTomato (tdT)) in 10-week-old KFT (n = 4), KT (n = 3) and KPT (n = 3) mice. c, AT1 cell marker quantification in n = 100 (KFT), 100 (KT) and 200 (KPT) tumours in 10-week-old KFT (n = 4), KT (n = 3) and KPT (n = 3) mice. Tumours were binned on AT1 cell marker positivity of tdT+ cells (Methods). Low, 0–5%; medium, 5–20%; high, 20–100%. d, Schematic of mouse LUAD cell line experiments. e, AGER quantification as a percentage of p53+ (n = 600) cells from p5353,54 (n = 3) and p53WT (n = 3) LUAD cell lines or of DAPI+ (n = 600) cells in p53null (n = 3) LUAD cell lines. f, Representative IF images of AGER and p53 from p5353,54 (n = 3), p53WT (n = 3) and p53null (n = 3) LUAD cells. g, Top, ChIP–seq31 tracks of p53 at AT1-associated genes. Bottom, ChIP–qPCR of p53 at AT1 cell genes in p53WT (n = 2) and p53null (n = 2) LUAD cell lines. h, AT1 cell signature expression in TP53WT (n = 249) and mutant TP53 (TP53MUT; n = 280) human LUADs (The Cancer Genome Atlas (TCGA)-LUAD cohort). Centre line, median; boxes, interquartile range (IQR); whiskers, 25% quantile – 1.5× IQR and 75% quantile + 1.5× IQR. Mouse and human alveolar signatures were taken from previously published studies12,39,54,55. i, Percentage of cytokeratin+ cancer cells that are AGER+ (n = 1,300) in TP53WT (n = 8) and TP53MUT (n = 5) human LUAD samples. j, Representative IF images of AGER and cytokeratin (CK) in TP53WT (n = 8) and TP53MUT (n = 5) human LUAD samples. Scale bar, 5 μm. Bar graphs are mean ± s.d. P values were calculated using GSEA52 (a), ordinary one-way ANOVA with Tukey’s multiple comparisons test (e), two-way ANOVA test (h; Methods) or two-tailed Student’s t-test (i). Scale bars, 5 μm (j) or 20 μm (b,f).
We next used multiple models of p53 reactivation to determine whether p53 directly induces AT1 cell identity. In LUAD cell lines with inducible Trp53 alleles, acute expression of wild-type p53 or p5353,54 was sufficient to induce AT1 cell marker expression and promote a thin, elongated morphology reminiscent of AT1 cells (Fig. 2d–f and Extended Data Fig. 7a,b). Furthermore, RNA-seq analysis of LUAD cells after wild-type p53 expression showed strong induction of AT1 cell, and not AT2 cell, signatures (Extended Data Fig. 7c,d and Supplementary Fig. 2). Reactivation of either wild-type p53 or p5353,54 in mouse LUADs in vivo drove the expression of the AT1 cell marker PDPN (Extended Data Fig. 7e–h). Finally, analyses of previously published gene expression data from mouse LUADs after p53 reactivation also revealed strong AT1 cell signature induction30 (Extended Data Fig. 7i). Thus, p53 is sufficient to drive AT1 cell phenotypes, which we proposed was due to direct activation of AT1 cell genes by p53. Indeed, p53 binds a significant fraction of AT1 cell genes, and not AT2 cell genes, in chromatin immunoprecipitation (ChIP) assays with sequencing (ChIP–seq) datasets31,32. Moreover, these p53-bound genes displayed greater chromatin accessibility and transcriptional induction in p53-proficient cancer cells than in p53-deficient cells (Fig. 2g and Extended Data Figs. 4g and 7j,k). ChIP with quantitative qPCR (ChIP–qPCR) confirmed p53 binding at numerous AT1 cell genes in our LUAD cell lines, which suggested that p53 directly induces AT1 cell identity (Fig. 2g).
p53 similarly promoted AT1 cell identity in human LUAD. Analyses of expression data from multiple lung cancer cohorts, after adjustment for tumour stage and proliferation, revealed that tumours with intact TP53 exhibited significantly higher AT1 cell marker expression than TP53 mutant tumours (Fig. 2h and Extended Data Fig. 8a). Moreover, immunostaining of EGFR-driven and KRAS-driven human LUADs of matched tumour grade revealed higher AT1 cell, and not AT2 cell, marker expression in wild-type TP53 cancer cells than in mutant TP53 cancer cells. This result demonstrates that intact TP53 status is associated with AT1-like cell identity in human LUADs (Fig. 2i,j, Extended Data Fig. 8b–g and Supplementary Table 7). Finally, expression of p53 in TP53-null human A549 LUAD cells was sufficient to induce the AT1 cell marker AGER (Extended Data Fig. 8j,k). Together, these findings suggest that p53 drives an AT1-like cell differentiation programme in mouse and human LUADs through direct transcriptional activation of AT1 cell genes.
p53 is active in multiple populations
To delve further into how p53 regulates cell identity during LUAD development, we used previously published scRNA-seq data from KT mice and KPT mice 2, 12, 20 and 30 weeks after Ad-SPC-Cre instillation29. This time course captured tumour evolution from normal AT2 cells to LUAD, with Trp53 expression largely restricting tumour development to late adenomas and Trp53 inactivation driving progression to advanced adenocarcinomas29. Although Trp53 loss was associated with increased cellular heterogeneity, how p53 influences cancer cell transcriptional programmes and AT1 cell differentiation were not examined.
Previous analyses of all samples defined 12 transcriptionally distinct cell clusters29, including those resembling normal alveolar cells—expressing AT2 cell markers (clusters 1, 2 and 4) or AT1 cell markers (cluster 3)—and others that emerged in advanced tumours and diverged from lung identity29 (clusters 6–12; Extended Data Fig. 9a–d). Clusters 1–5 predominated in KT tumours, whereas all cell states developed in KPT tumours (Fig. 3a and Extended Data Fig. 9b,c). The emergence of clusters 6–12, which resemble primordial gut derivatives29, in p53-deficient tumours recalls the gene signatures in our bulk RNA-seq data following Trp53 inactivation (Extended Data Fig. 4e).
a, t-distributed stochastic neighbour embedding (tSNE) plots of scRNA-seq data29. KT (n = 1,071) and KPT (n = 2,614) cells from all time points (2, 12, 20 and 30 weeks) are shown with previously defined29 clusters. b, Expression of Trp53 and p53 target genes in all KT cells. c, Proportions of early (<30 week) and late (30 week) KT and KPT cells in subclusters 5A, 5B and 5C. d, Hierarchical clustering of all cluster 5 cells identifies 3 subclusters. Top, genotype and time point composition of each cluster. Left, expression programme term chosen through functional annotation of programme genes. Right, top significant genes in each programme. e, For each programme identified in d, an aggregate score measuring the expression of all genes in the programme was calculated for each cluster 5 cell. Violin plots show the score for each cell along the previously described29 LUAD time course. DEG, differentially expressed gene. f, Violin plots of Cdkn2a and Hnf4a expression in cells from clusters 5A, 5B and 5C from KT and KPT mice. g, Representative IF images of CLDN4 and markers of clusters 5B/5C (p19(ARF), HNF4A and pERK) in tumours from 10-week-old KFT (n = 4), KT (n = 3) and KPT (n = 3) mice. Scale bar, 20 μm. h, PC analysis plot of the cancer cell subpopulations (clusters 3, 5 and 11) from KT and KPT mice across all time points. i, PC analysis plot from h coloured by CytoTRACE score. The CytoTRACE score shows the putative differentiation potential of cells. Least differentiated indicates that cells have the highest plasticity. j, Cell–cell transitions estimated using CellRank, based on CytoTRACE scores, reveals distinct trajectories from clusters 5A to 3 and from clusters 5B to 5C to 11. For the violin plots, the thicker dashed line indicates the median, the thinner dashed lines indicate the upper and lower quartiles.
To understand how p53 restrains LUAD development, we first identified KT cell populations with active p53 signalling. Early in tumorigenesis, at 2 weeks, we observed some expression of Trp53 along with classical p53 target genes, such as Cdkn1a, in AT2-like cells, which indicated an initial role for p53 in this population (Extended Data Fig. 9e). However, we observed strong p53 pathway induction across the time course of tumour development in AT1-like cells (cluster 3), as evidenced by the expression of Trp53 together with both canonical and AT1-associated p53 target genes, including Pdgfa and Fam174b (Fig. 3b). In support of our discovery that p53 activates an AT1-like cell differentiation programme, we observed an expansion of cluster 3 AT1-like cells and increased AT1 cell marker expression in KT tumours relative to KPT tumours (Extended Data Fig. 9f–g). Notably, cluster 5 displayed prominent expression of Trp53 and many classical p53 target genes (Fig. 3b). Cluster 5 was previously characterized as a high-plasticity, Cldn4+, transition-point state that gives rise to divergent cell states over time and is associated with tumour progression29. Why p53 would be activated in this high-plasticity state was puzzling; therefore we explored this question further.
p53 acts in transitional cells
To understand the significance of p53 pathway activation in cluster 5 transitional cells, we examined how p53 quantitatively and qualitatively regulates these cells. Although cluster 5 cells were present in both KT tumours and KPT tumours, which indicated that Trp53 expression is dispensable for their formation, they were expanded with Trp53 loss (Fig. 3a and Extended Data Fig. 9f–g). CLDN4 immunostaining in mouse and human LUADs validated that transitional cells were increased in p53-deficient tumours relative to p53-proficient tumours (Extended Data Figs. 8h,i and 9h,i). Hierarchical clustering of cluster 5 cells uncovered 3 distinct subpopulations: 5A, 5B and 5C (Fig. 3c,d and Extended Data Fig. 9j,k). Analysing the cell genotypes and time points within each subcluster revealed distinct paths of cluster 5 evolution depending on Trp53 status (Fig. 3c,d and Extended Data Fig. 9j,k). Cluster 5A almost entirely comprised KT cells and was characterized by a p53 pathway expression signature (Fig. 3c–e, Extended Data Fig. 9j,k and Supplementary Table 8). Cluster 5B comprised mostly KPT cells, with occasional late (30-week) KT cells, and was distinguished by growth and metabolic programmes correlated with tumour progression, including Kras signalling (Fig. 3c–e, Extended Data Fig. 9j,k and Supplementary Table 8). Cluster 5C contained only late KPT cells and was characterized by enhanced cluster 5B expression programmes, which suggested that cluster 5C cells arose from cluster 5B cells (Fig. 3c–e and Extended Data Fig. 9j,k). We observed induction of p19Arf in clusters 5B and 5C (5B/5C), which provides support for the idea that this state arises with Trp53 loss, as p19Arf is upregulated with Trp53 inactivation33 (Fig. 3f). CLDN4 and p19(ARF) immunostaining revealed that cluster 5B/5C cells predominate in KPT tumours, whereas cluster 5B/C cells were rare to non-detectable in KT and KFT tumours (Fig. 3g and Extended Data Fig. 10a,b). Another cluster 5B/5C marker, HNF4A, selectively marked most KPT CLDN4+ cells, but not KT or KFT CLDN4+ cells, and was expressed together with p19(ARF) (Fig. 3f,g and Extended Data Fig. 10c,d,g). Notably, Kras signalling was also induced in cluster 5B/5C cells, as evidenced by phosphorylated ERK staining. This result provides support for the that notion that growth pathways are increased in these subclusters (Fig. 3g and Extended Data Fig. 10e–g). Thus, p53 qualitatively regulates transitional cell identity, with Trp53-proficient cells primarily existing in the cluster 5A state and Trp53-deficient cells progressing to 5B and then 5C over time.
p53 activity in transitional cells led us to propose that p53 may promote their differentiation into AT1-like cells. To understand the evolution of transitional cells in KT and KPT LUADs, we inferred the trajectory of cluster 5 cells in relation to cluster 3 (AT1-like) cells and cluster 11 (epithelial–mesenchymal transition (EMT)-like) cells, an example of an advanced, late tumour state (Fig. 3h). We reconstructed cell trajectories using CytoTRACE34 and CellRank35, which revealed distinct trajectories from cluster 5A cells to cluster 3 AT1 cells and from cluster 5B cells to cluster 5C cells to EMT cells (Fig. 3i,j). Additional methods to study cell-fate transitions, such as pseudotime and tracking copy number alterations, further confirmed these trajectories (Extended Data Fig. 10h–k). Moreover, cluster 5A cells began to express cluster 3 AT1 cell markers, and gene expression programmes in clusters 5A and 3 displayed a strong positive correlation (Extended Data Fig. 10l,m). Thus, p53 activation in cluster 5A transitional cells limits their malignant progression and directs cells towards an AT1 cell fate, a notion supported by our observation that p53 reactivation in sorted LUAD transitional cells29 led to induction of the AT1 cell marker AGER (Extended Data Fig. 11a and Supplementary Fig. 2). Meanwhile, Trp53 deficiency facilitates progression to cluster 5B/5C, in which cells upregulate growth programmes and begin to assume divergent cellular identities, poising them for tumour progression.
p53 promotes lung injury repair
Our finding that p53 acts in transitional LUAD cells to direct AT1 cell differentiation recalls recent studies demonstrating that lung injury triggers AT2-to-AT1 cell differentiation through a Cldn4+Krt8+ transitional cell state characterized by activation of various signalling pathways (for example, NF-κB, HIFα and p53)12,13,14. Reinforcing this similarity, an injury-induced transitional cell expression programme was induced in LUAD transitional cells (Extended Data Fig. 11b–d) and became upregulated with Trp53 loss in tumours (Extended Data Fig. 9l). We aimed to understand whether p53-mediated AT1 cell differentiation during LUAD development may reflect a role for p53 in tissue repair. To that end, we used a model of alveolar regeneration after injury induced by butylated hydroxytoluene (BHT), an agent selected because of known parallels to KRAS-driven tumorigenesis. BHT damages AT1 cells, which in turn triggers activation of an EGFR–RAS axis in AT2 cells and subsequent AT2 cell proliferation and transdifferentiation into AT1 cells6,7 within days of treatment36,37.
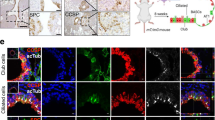
To deconstruct how p53 governs differentiation after injury, we used lineage tracing to analyse AT2 cell progeny 2–4 days after BHT treatment in the following mouse models: SftpccreERT2;Trp53+/+;Rosa26LSL-tdTomato/LSL-tdTomato (SpcT mice), SftpccreERT2;Trp5353,54/53,54;Rosa26LSL-tdTomato/LSL-tdTomato (SpcFT mice) and SftpccreERT2;Trp53fl/fl;Rosa26LSL-tdTomato/LSL-tdTomato (SpcPT mice) (Fig. 4a and Extended Data Fig. 11e,f). BHT induced AT2 cell proliferation in mice of all genotypes; however, SpcPT AT2 cells were more proliferative than SpcT AT2 cells, which in turn were more proliferative than SpcFT AT2 cells. This result suggests that p53 dampens AT2 cell proliferation after injury and mirrors the p53-mediated inhibition of AT2 cell proliferation in early-stage lung cancers (Fig. 4b and Extended Data Figs. 2c–e and 11h,i). CLDN4+ transitional cells emerged in all genotypes, but Trp53 loss triggered their significant accumulation, as in LUAD (Fig. 4c and Extended Data Fig. 11g). Finally, SpcPT mice displayed compromised AT1 cell differentiation relative to SpcT mice and SpcFT mice after injury, again reminiscent of the role of p53 in cancer (Fig. 4d and Extended Data Fig. 11g,i–k). Analyses of p53 ChIP–seq of transitional lung cells after injury14 revealed p53 binding to AT1 cell genes, which suggested that p53 directly induces AT1 cell identity in both regeneration and tumour suppression (Extended Data Fig. 11l). Notably, cellular changes observed in SpcPT mice were associated with substantial tissue damage (Fig. 4e,f and Supplementary Table 9). Thus, p53 is crucial for promoting tissue integrity after lung injury.
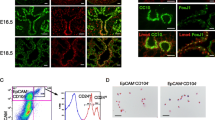
a, Schematic of the lung injury mouse models. Control cohort: SpcT (n = 3) and SpcPT (n = 3) mice. Treatment with BHT for both the 2 day and 4 day cohorts: SpcFT (n = 3), SpcT (n = 3) and SpcPT (n = 3) mice. scRNA-seq cohort: SpcT (n = 2) and SpcPT (n = 2) mice. b, SPC+ (n = 600) AT2 cells that are BrdU+ in SpcFT (n = 3), SpcT (n = 3) and SpcPT (n = 3) mice treated with BHT for 4 days. c, Left, representative IF images of CLDN4 and tdT staining of samples from SpcFT (n = 3), SpcT (n = 3) and SpcPT (n = 3) mice treated with BHT for 4 days. Right, tdT+ (n = 600) cells that are CLDN4+ in the same cohorts. d, Left, representative IF images of AGER and tdT in 4-day BHT-treated SpcFT (n = 3), SpcT (n = 3) and SpcPT (n = 3) mice. Right, tdT+ (n = 600) cells that are AGER+ in the same cohorts. Arrowheads indicate examples of AGER+tdT+ cells. e, Representative haematoxylin and eosin images of 4-day BHT-treated SpcT (n = 3) and SpcPT (n = 5) mice. Scale bar, 20 μm. f, Injury scoring of 4-day BHT-treated SpcT (n = 3) and SpcPT (n = 5) mice (Methods). g, Uniform manifold approximation and projection (UMAP) plot of tdT+ alveolar epithelial (n = 2,445) cells from scRNA-seq. h, p53 target gene expression across SpcT cells. i, Cluster frequency by genotype. j, Gene and cellular programme expression (Supplementary Table 6) in transitional SpcT (n = 332) and SpcPT (n = 189) cells. k, Representative IF images of CLDN4, VIM and tdT in 4-day BHT-treated SpcT (n = 3) and SpcPT (n = 3) mice. l, Model of p53 action. Bar graphs are mean ± s.d. For the box plots, centre line is the median; box limits, the upper and lower quartiles; whiskers, 2.5–97.5 percentile; points, outliers. P values were calculated using ordinary one-way ANOVA with Tukey’s multiple comparisons test (b–d), two-tailed Student’s t-test (f,j (bottom)) or two-tailed, non-parametric Wilcoxon rank sum test (j, top). Scale bars, Scale bar, 5 μm (k) or 20 μm (c,d).
To elucidate how p53 directs alveolar repair, we performed scRNA-seq on lungs from SpcT mice and SpcPT mice 4 days after BHT treatment (Fig. 4a and Supplementary Fig. 2). We identified five alveolar populations previously described in single-cell analyses of mouse lung injury: AT2; AT2 proliferating; AT2 primed or activated12,13; transitional12,13,14; and AT1 (Fig. 4g and Extended Data Fig. 12a,b). Pseudotime and CytoTRACE analyses suggested that AT2 cells generate AT1 cells through transitional cells (Extended Data Fig. 12c,d). We next identified alveolar epithelial populations with active p53 signalling in SpcT mice. We pinpointed strong expression of canonical and AT1-associated p53 target genes in transitional cells and AT1 cells (Fig. 4h and Extended Data Fig. 12e,f), which were reminiscent of p53 activation in these clusters in LUAD. Furthermore, as in cancer, p53 quantitively regulated alveolar epithelial cell populations. Although all alveolar populations were found irrespective of Trp53 status, transitional cells were substantially expanded in SpcPT mice (Fig. 4i). This observation was consistent with CLDN4 immunostaining, which showed an increased transitional cell population with Trp53 loss. This result reinforces the role of p53 in restricting this population (Fig. 4c). We observed a greater proportion of AT1 cells in SpcT mice than in SpcPT mice, although this expansion was smaller than expected (Fig. 4i). Notably, >40% of SpcPT AT1 cells retained expression of Trp53 RNA and a p53 transcriptional programme, which indicated that AT1 cell differentiation in SpcPT mice was enabled by escaping Cre-mediated Trp53 deletion, a phenomenon observed when Cre-mediated deletion of genes is detrimental to a particular population38 (Extended Data Fig. 12f,g). Collectively, these observations demonstrate that p53 modulates cell state transitions during alveolar repair by driving AT1 cell differentiation, as in LUAD.
Parallels in lung cancer and regeneration
We next assessed how Trp53 loss qualitatively affects transitional cells after injury. SpcPT transitional cells showed upregulation of genes for which expression is atypical for lung epithelium, such as Vim (which encodes vimentin), and of transcriptional signatures associated with divergent cell states (for example, EMT, liver)39,40 (Fig. 4j,k and Extended Data Fig. 12h–k). These results are indicative of lung lineage infidelity reminiscent of that seen with Trp53 inactivation in transitional cancer cells. To further explore parallels between injury and cancer, we asked whether transcriptional programmes associated with Trp53 inactivation in injury were seen with Trp53 loss in LUAD. Indeed, the top genes induced in SpcPT transitional cells were similarly upregulated in KPT LUAD cells (Extended Data Fig. 12l). Additionally, Vim was more highly expressed in transitional cells in KPT LUADs than in KT LUADs (Extended Data Fig. 12m–o). Conversely, transcriptional programmes associated with Trp53 loss in LUAD, including cluster 5B/5C signatures, were induced in SpcPT transitional cells (Extended Data Fig. 12h). Thus, p53 promotes lineage fidelity of transitional cells by directing their differentiation into AT1 cells in both cancer and injury. By contrast, Trp53 loss enables transitional cell expansion and lineage infidelity. Collectively, our analyses have uncovered parallels between p53 function in LUAD suppression and alveolar injury repair. These parallels begin with p53 dampening AT2 cell proliferation in LUAD and injury. Then, after AT2 cells progress to transitional cells, p53 becomes activated and promotes their differentiation into AT1 cells. These findings suggest that tumour suppression reflects the repurposing of a physiological role of p53 in governing lung regeneration (Fig. 4l). Indeed, studies of p53 in simpler eukaryotes, such as planaria and salamanders, have shown that p53 regulates stem cell self-renewal and differentiation, which suggests that this may be an ancestral p53 function41,42.
Discussion
Studies suggesting that canonical p53 functions are dispensable for tumour suppression19,43,44 have led to renewed investigation of p53 mechanisms that combat tumorigenesis. The role of p53 in cancer cell differentiation has begun to be appreciated, with recent studies suggesting that p53 restricts plasticity in pancreatic cancer and in liver cancer45,46,47. Here we discovered that p53 not only constrains plasticity in LUAD but also induces a highly specific AT1 cell differentiation programme unlike the cancer cell-of-origin. Moreover, acquisition of AT1 cell identity is associated with direct p53 induction of AT1-associated genes. Notably, the ability of p53 to govern cell fate relies on p53 acting in a transitional cancer cell state, where it induces AT1 cell differentiation. Unlike previous work14 suggesting that p53 acts by inducing transitional cell genes in a bleomycin lung injury model, we show here that p53 binds and activates AT1 cell genes. These findings, coupled with our observations that p53 reduces transitional cell accumulation in both LUAD and injury repair, clarifies that p53 is dispensable for entry into the transitional cell state but is essential for exit from this state. The importance of p53 in directing the fate of transitional cells is underscored by the deleterious patient outcomes associated with their accumulation. Accumulation of LUAD transitional cells, which occurs with Trp53 loss, is associated with aggressive tumour growth and poor patient survival, whereas in non-oncogenic contexts, inappropriate persistence of transitional cells is associated with idiopathic pulmonary fibrosis and lethal COVID-19 pneumonitis12,13,14,15,29.
Given the detrimental outcomes associated with TP53 mutation in LUAD, our findings suggest that there may be promise in differentiation therapy for patients with LUAD, a strategy being successfully exploited in other cancers such as acute promyelocytic leukaemia48. Moreover, methods to induce AT1 cell differentiation are under development49,50. Our work suggests that promoting AT1 cell differentiation may mimic pivotal functions of p53 function in tumour suppression and provide a promising new route of therapy in patients with LUAD.
Methods
Animal studies and ethics
All animal experiments were performed in accordance with the Stanford University Administrative Panel on Laboratory Animal Care (protocol number 10382) guidelines and regulations. Mice (Mus musculus) were maintained at Stanford University’s Comparative Medicine Pavilion and Research Animal Facility according to practices prescribed by the National Institutes of Health and the Institutional Animal Care and Use Committee (IACUC). The Association for Assessment and Accreditation of Laboratory Animal Care provides additional accreditation to Stanford University. We did not exceed limits specified in the Stanford IACUC guidelines for tumour monitoring, which state that the maximum measurable total tumour burden for multiple masses on any one animal should not exceed a combined mean diameter for the sum of all masses of 1.70 cm and 10% of the pre-induction body weight of the animal. Mice were housed at 22 °C ambient temperature with 40% humidity and a 12-h light–dark cycle (7:00–19:00). All experiments were performed on 8–10-week-old animals, and all mice used were of a 129/Sv-C57BL/6 mixed background. For mouse LUAD and injury studies, male and female mice were used in every experimental group with the exception of the single-cell multiomics experiment, which had two males for the KFT and KPT samples. Otherwise, mice of the same genotype were randomized into cohorts. For histological and IF analyses of mouse studies, investigators were blinded to group allocation.
Mouse LUAD studies
KrasLSL-G12D/+;Rosa26LSL-tdTomato/LSL-tdTomato (KT), KrasLSL-G12D/+;Trp5353,54/53,54;Rosa26LSL-tdTomato/LSL-tdTomato (KFT) and KrasLSL-G12D/+;Trp53flox/flox;Rosa26LSL-tdTomato/LSL-tdTomato (KPT) mice (mixed 129/Sv-C57BL/6) were used in the majority of LUAD experiments. KrasLSL-G12D/+;Trp53LSL-WT/+;Rosa26LSL-tdTomato/LSL-tdTomato (KT;Trp53LSL-WT/+) and KrasLSL-G12D/+;Trp53LSL-53,54/+;Rosa26LSL-tdTomato/LSL-tdTomato (KT;Trp53LSL-53,54/+) mice were used to assess the effects of the Trp5353,54 allele specifically in cancer cells, which would only be expressed in AT2 cells in the presence of Cre. In this study, non-transduced mice expressed one allele of wild-type Trp53 in every cell (Trp53−/+). After Ad5-SPC-Cre transduction, AT2 cells became Trp5353,54/+ (in KFT-LSL mice) or Trp53+/+ (in KT-LSL mice), whereas all other cells remained Trp53−/+. Throughout the text, Cre-treated Trp53fl/fl mice are still referred to as Trp53fl/fl for simplicity, although these mice were Trp53Δ/Δ. Lung tumours were induced as previously described61. In detail, 8–12-week-old male and female mice were anaesthetized by an intraperitoneal injection of avertin (2-2-2 tribromoethanol) and given a dose of either Lenti-PGK-Cre or Ad5-SPC-Cre intratracheally. Next, 50 µl of virus particles diluted in sterile PBS was delivered at a dose of 9 × 104 plaque-forming units. We first tested whether p5353,54 behaved as a super tumour suppressor 24 weeks after tumour initiation with Lenti-PGK-Cre, a time point when we knew wild-type p53 behaved as a potent tumour suppressor based on previous work in the field demonstrating the effect of p53 on tumour progression17. Next, because there was a shift in the LUAD mouse model field towards using AT2-specific Cre adenoviruses to initiate tumours, we switched from PGK-Cre lentiviruses to the AT2-specific Ad5-SPC-Cre virus, which allowed us to specifically initiate cancer in the primary cell of origin of LUAD to better mimic human cancer. Our most important experiments—the 10-week tumour study and corresponding genomics experiments—were therefore all done using Ad-SPC-Cre. For the Ad5-SPC-Cre studies, 1 × 109 virus particles (Ad5mSPC-Cre from the University of Iowa Viral Vector Core) were diluted in MEM, precipitated with CaCl2 and delivered into mice after a 20-min incubation. Lungs were collected 10–24 weeks after transduction. Lungs were either inflated and fixed with formalin for 24 h before paraffin embedding and processing or inflated with 2% low-melt agarose, fixed in ice-cold 4% paraformaldehyde (PFA) overnight, incubated with 30% sucrose in PBS overnight and then cryo-embedded in optimal cutting temperature compound (OCT).
Production, purification and titration of lentivirus
Lentivirus was produced by transfection of HEK293AH cells (a gift from M. Bassik at Stanford) with Lenti-PGK-Cre (a gift from M. Winslow at Stanford), VSVG and Δ8.2 vectors and Lipofectamine 2000 (ThermoFisher). Virus was collected from supernatant by ultracentrifugation at 25,000 r.p.m. at 4 °C for 2 h and resuspended in sterile PBS. Concentrated lentiviral particles were titred by infecting LSL-tdTomato mouse embryonic fibroblasts (Rosa26LSL-tdTomato mice), which determined the per cent of tdTomato+ cells by IF and comparing the infectious titre to a viral preparation of a known titre.
Immunostaining and microscopy
Haematoxylin and eosin (H&E) staining, immunohistochemistry and IF staining were performed on paraffin-embedded and OCT-embedded lungs using standard protocols. Immunostaining was performed using primary antibodies raised against the following antigens: Ki67 (mouse, BD Pharmingen AB_393778, clone B56, 1:200); phospho-histone H3 (pHH3, CST D7N8E, 1:200); TTF-1 (mouse, Leica NCL-L-TTF-1, clone SPT24, 1:100); AGER (goat, R&D Systems AF1145, 1:200; rat, R&D Systems MAB1179 clone 175410, 1:100); HT1-56 (mouse, Terrace Biotech TB29AHT1-56, 1:30); HOPX (rabbit, Proteintech 11419-AP-1, 1:100); SPC (proSP-C, rabbit, Sigma-Aldrich AB3786, 1:100); CLDN4 (rabbit, ThermoFisher AB_2533262, clone ZMD.306, 1:100; mouse, Santa Cruz Biotechnology, clone A-12, 1:100); PDPN (Syrian hamster, Developmental Studies Hybrdoma Bank, AB_531893, clone 8.1.1, 1:30); pan-cytokeratin (mouse, BioLegend, AB_2616960 clone AE-1/AE-3, 1:200; rabbit, Abcam, AB_273139, 1:200); vimentin (rabbit, Abcam, AB_45939, 1:100); phospho-ERK (rabbit, Cell Signaling Technologies, clone D13.14.4E, 1:100); HNF4A (mouse, Invitrogen, AB_2532197, clone H1415, 1:100); p19(ARF) (rat, Santa Cruz Biotechnology, clone 3-5C-1, 1:100); RFP (mouse, Thermo Scientific, AB_10999796, clone RF5R, 1:200); p53 (rabbit, Leica Biosystems, NCL-L-p53-CM5p, 1:100); and BrdU (mouse, BD Pharmingen AB_395993, clone 3D4, 1:50). The following secondary antibodies were used: anti-rat-488 (ThermoFisher, AB_2534074); anti-rabbit-FITC (Vector Laboratories, Fl-1000-1.5); anti-mouse-FITC (Vector Laboratories, Fl-2000-1.5); anti-mouse-546 (ThermoFisher, AB_2534089); anti-rabbit-546 (ThermoFisher, AB_2534093); biotinylated anti-mouse (Vector Laboratories, BA-9200-1.5); biotinylated anti-rabbit (Vector Laboratories, BA-1000-1.5); anti-Syrian hamster-488 (Abcam, AB_180063); anti-mouse 647 (ThermoFisher, AB_162542); and biotinylated anti-goat (Vector Laboratories, BA-9500-1.5). For immunohistochemistry experiments, paraffin sections were de-paraffinized, rehydrated, unmasked in 10 mM sodium citrate buffer with 0.05% Tween-20 in a pressure cooker for 10 min, quenched for 20 min in 3% H2O2, permeabilized for 10 min in Tris-buffered saline (TBS) with 0.3% Triton-X 100, and blocked for 30 min in a solution of TBS, 0.3% Triton-X 100, 10% serum and 10% BSA. Slides were incubated overnight at 4 °C with primary antibody diluted in blocking solution and subsequently incubated for 1 h at 37 °C with biotinylated secondary antibody compatible with the primary antibody (1:1,000, Vector Laboratories). Slides were then incubated using a Vectastain Elite ABC HRP kit (Vector Laboratories) according to the manufacturer’s instructions. A DAB peroxidase kit (Vector Laboratories) was used for staining, and Gill’s haematoxylin was used for counterstaining, after which slides were dehydrated and mounted with Permount. Slides were washed between steps with TBS. A NanoZoomer 2.0-RS slide scanner (Hamamatsu) was used for imaging. For IF experiments, slides sectioned on a Leica cryostat were air-dried for 10 min and then fixed for 15 min in 4% PFA. For sections containing the endogenous tdTomato reporter or being stained with the AGER antibody (rat, R&D systems), sections were not unmasked using the pressure cooker. For all other IF stains, sections were unmasked in the pressure cooker for 10 min. All slides were then permeabilized for 10 min in 3% Triton-X 100 in TBS followed by incubation in a solution of TBS, 3% Triton-X 100, 10% serum and 10% BSA and then incubated overnight at 4 °C with primary antibody diluted in blocking solution. Subsequently, slides were incubated for 1 h at 37 °C with Alexa Fluor-conjugated secondary antibody compatible with the primary antibody (1:200, Thermo Scientific). Slides were mounted in ProLong Gold antifade mountant with DAPI. Slides were washed with TBS between steps. Images were acquired with a Leica DM4B microscope (Leica Microsystems) or a DMi8 microscope (Leica Microsystems) and analysed using LAS X software (v.5.0.2, Leica Microsystems).
For quantifications throughout the article, tdTomato marks lineage-labelled cells in both the LUAD and injury models. For AT1 cell marker quantifications shown in Fig. 2c and Extended Data Fig. 6h, we binned tumours into three categories (high, medium or low) based on the percentage of lineage-marked tdTomato+ cells that stained for each given marker (AGER, HOPX and PDPN). Specifically, we graphed the per cent positivity for a given marker in 100 tumours in KFT and KT mice and 200 tumours in KPT mice across 3–4 mice in 2–3 fields per tumour and then defined natural breakpoints for high, medium and low staining (Extended Data Fig. 6d). Based on these graphs, high, medium and low staining were used to denote that low = 0–5%, medium = 5–20% and high = 20–100% of tdTomato+ cancer cells stained strongly for the given AT1 cell marker based on the distribution of marker positivity observed. Examples of such staining are shown in Extended Data Fig. 6a. For AT2 cell marker quantification in Extended Data Fig. 6i, high refers to staining in at least 70% of tdTomato+ cancer cells within a tumour and low refers to staining in less than 70% of cells, again based on a graph showing the staining trends in 100 tumours in KFT and KT mice and 200 tumours in KPT mice across 3–4 mice in 2–3 fields per tumour (Extended Data Fig. 6j). Examples of such staining are shown in Extended Data Fig. 6a. For marker quantification in human LUAD tissue in Fig. 2i,j and Extended Data Fig. 8c–i, we stained for cancer cells using pan-cytokeratin together with AT1 cell, transitional cell or AT2 cell markers. We then calculated the percentage of cytokeratin+ cells that stained for these alveolar cell markers. For marker quantification in mouse LUAD tissue shown in Extended Data Fig. 10b,d,f, tissue was stained for combinations of tdTomato, CLDN4 and markers of cluster 5B/C. The percentage of CLDN4+ cells staining for cluster 5B/C markers was quantified. For marker quantification in Extended Data Fig. 9i, we stained tumours for tdTomato and CLDN4 and quantified the percentage of tumours that were CLDN4+ or CLDN4–. CLDN4+ tumours had at least one tdTomato+CLDN4+ cell, whereas CLDN4– tumours had no such cells. For quantification of PDPN staining in p53 in vivo reactivation experiments in Extended Data Fig. 7g, cytokeratin+ tumours that were p53+ were assessed for PDPN+p53+cytokeratin+ cells. Tumours that displayed >10% of p53+cytokeratin+ cells also staining for PDPN were called as a positive tumour. Trp53null tumours were called positive if >10% of cytokeratin+ cells stained for PDPN.
Analysis of mouse LUAD studies
Paraffin embedded, H&E-stained slides were scanned using a NanoZoomer 2.0-RS slide scanner (Hamamatsu) and scored using NDP.view 2 (v.U12388-01, Hamamatsu). In brief, all lobes and lesions were outlined to quantify tumour number and size. Tumour burden was calculated as the percentage of lesion area compared with the entire lobe area using ImageJ (v.1.52, https://imagej.nih.gov/ij) and QuPath (v.0.3.2). For tumour proliferation quantifications, paraffin-embedded, Ki67-stained and pHH3-stained slides were scanned and processed using ImageJ. In brief, images were deconvoluted into DAB and haematoxylin channels and thresholds were set to gate haematoxylin-stained nuclei and DAB+ nuclei. The percentage of Ki67+ or pHH3+ was calculated using the ‘analyze particles’ feature of ImageJ. M.G.O. performed blinded histopathological analysis of tumour patterns based on H&E-stained slides. Each mouse was scored on the existence of at least one tumour of a given pattern in each mouse. For tumour pleomorphism analyses, tumours were assigned a score of 1–3, where 1 indicates that the tumour had little pleomorphism and 3 indicates substantial pleomorphism. Mice often displayed more than one tumour pattern and level of pleomorphism. Mitotic activity was determined by counting the number of mitotic figures per high power field in the most advanced nodule in each lung.
Human LUAD sample collection
Clinical samples were approved by the Stanford Institutional Review Board in accordance with the Declaration of Helsinki guidelines for the ethical conduct of research. All patients involved in the study provided written informed consent. Collection and use of human tissues were approved and in compliance with data protection regulations regarding patient confidentiality (Institutional Review Board protocol number 15166). Any self-selection bias was not anticipated to affect results. Following surgical resection of primary tumours from patients at Stanford Hospital, LUAD specimens were immediately embedded in OCT compound and stored at −80 °C. Samples used in subsequent analyses were chosen to best match tumour grade and differentiation status. No identifiable images of research participants were used in this study. For IF experiments of human samples, investigators were blinded to group allocation. LUAD samples were chosen based on their histological pattern (all LUADs). Furthermore, samples were chosen based on their mutational status. All samples chosen had mutations in either EGFR or KRAS and were from patients who underwent no previous treatment. Five of the samples also had a mutation in TP53. Finally, samples were matched, when possible, for tumour grade and differentiation status between TP53 wild-type and TP53 mutant samples.
FACS for bulk RNA-seq and ATAC–seq
KT, KFT and KPT mice were transduced with Ad5-SPC-Cre and collected for cell sorting at 10 weeks after transduction. Non-transduced lungs were used as a control. In brief, lungs were perfused with 10 ml of ice-cold PBS and placed in a sterile dish on ice. Lungs were finely minced with sterile scissors and razor blades and resuspended in 6 ml digestion medium (RPMI with 2.5 mg of Liberase TL (Sigma-Aldrich) and 25 µg ml–1 DNase (Sigma Aldrich)). Lung samples were rotated for 45 min at 37 °C. Tubes were briefly cooled on ice and then samples were passed through a 40 µm filter. Next, 5 ml of FACS buffer (10% FBS, 2 mM EDTA and 25 µg ml–1 DNase in DPBS) was added and tubes were spun for 5 min at 300g. Cells were resuspended in ACK lysing buffer (Gibco), incubated for 1 min on ice and quenched with 8 ml of FACS buffer. Cells were washed 2 times with FACS buffer, resuspended with biotinylated primary antibodies (CD45, BioLegend, 103104 30-F11; CD31, BioLegend, 102404 390; F4/80, BioLegend, 123106 BM8; Ter119, BioLegend, 116204 TER-119; 1:800 in FACS buffer) and incubated for 20 min on ice. After washing 2 times, cells were resuspended in streptavidin–APC secondary antibody (BioLegend 405207, 1:800) for 20 min on ice. Cells were washed 2 times and resuspended in FACS buffer with 1 µg ml–1 DAPI, filtered through a 40 µm filter and sorted using a Sony SH800S cell sorter. Data were analysed using the default Sony SH800S software (v.2.1.5) and FCS Express (v.7, De Novo Software).
RNA-seq
For RNA-seq, 1.5 × 105 FACS-isolated tumour cells were collected, pelleted and flash frozen in liquid nitrogen directly after sorting. Once all samples were collected, RNA was isolated using a RNeasy Micro kit (Qiagen). Quality and concentration of RNA were determined using a Bioanalyzer. cDNA libraries were constructed using a Trio RNA-seq library preparation kit (NuGEN) from samples with high-quality RNA according to the manufacturer’s instructions. Samples were sequenced on a HiSeq4000 (Illumina). High-performance computation was performed on the Stanford SCG Informatics Cluster, and subsequent bioinformatics analyses were performed in R (v.4.0.3) using various R packages unless otherwise noted. RNA-seq reads were aligned to the mouse genome (mm10) using HISAT2 (v.2.0.5, https://github.com/DaehwanKimLab/hisat2), sorted based on genomic location and indexed using Samtools (v.1.3.1, https://github.com/samtools/samtools), and counted and mapped to each gene using HTSeq-Count (v.0.6.1, https://github.com/simon-anders/htseq). DEGs were identified using DESeq2 with a cutoff of a P-adjusted value of <0.05 (v.1.24.0, https://github.com/mikelove/DESeq2). PC analysis was performed using DESeq2. Unsupervised hierarchical clustering and heatmap visualization of DEGs were performed using the ‘pheatmap’ package in R (v.1.0.12, https://github.com/raivokolde/pheatmap). p5353,54-hyperactivated genes were identified by identifying DEGs activated in KT cells and KFT cells compared with KPT cells to obtain p53-dependent genes and identifying those activated at least 1.3-times more (1.2-fold) in KFT cells relative to KT cells. Enrichr58,59,60 (v.3.2, https://maayanlab.cloud/Enrichr) was used to perform GO analyses and ChIP enrichment analysis (ChEA)57. PANTHER56 (v.16.0), through the GO Consortium (https://geneontology.org), was used for GO term enrichment, and Metascape51 (v.3.5, https://metascape.org) was used for functional enrichment analysis. In this article, ChEA analysis indicates the percentage of genes in a given gene set that are p53 bound in the dataset31.
GSEA
GSEA (v.4.0.3, https://github.com/GSEA-MSigDB/gsea-desktop) was performed using normalized RNA-seq counts in this study against the gene signatures from the MSigDB database40,52, the signatures derived herein and published expression signatures. Default parameters were used with the following exceptions: permutation type = gene_set; min size = 5. Plots were made using the ‘Rtoolbox’ package in R (https://github.com/PeeperLab/Rtoolbox). Custom gene sets were derived from various studies defining AT1 cell signatures12,14,24,39,53,54,55,62,63 and are described in Supplementary Table 6.
ATAC–seq
ATAC–seq was performed as previously described64. Two independent ATAC–seq experiments were performed: one with KT mice and KPT mice and the second with KT cells and KFT cells. In brief, 5 × 105 cells were sorted and washed 2 times in FACS buffer without DNase added. Nuclei were then isolated, lysed and incubated with Tn5 transposase exactly as described. Transposed DNA was then isolated using a MinElute Reaction Cleanup kit (Qiagen). Libraries were prepared by amplifying DNA for 5 cycles with NEBNext 2× MasterMix (NEB), after which qPCR was run on samples to determine the number of additional cycles needed. After any additional amplifications were performed, libraries were purified using a MinElute Reaction Cleanup kit (Qiagen). Samples were sequenced on a HiSeq4000 (Illumina) (KT versus KPT experiment) or a NextSeq500 (Illumina) (KT versus KFT experiment). High-performance computation was performed on the Stanford SCG Informatics Cluster, and subsequent bioinformatics analyses were performed in R (v.4.0.3) using various R packages unless otherwise noted. ATAC–seq reads were trimmed of adapter sequences using Skewer (v.0.2.2, https://github.com/relipmoc/skewer), aligned to the mouse genome (mm10) using Bowtie2 (v.2.4.0, https://github.com/BenLangmead/bowtie2) and sorted based on genomic location and indexed using Samtools (v.1.3.1, https://github.com/samtools/samtools). Peak calling and differential peak analysis were performed using the ‘ChrAccR’ package in R (v.0.9.17, https://github.com/GreenleafLab/ChrAccR) with the sorted BAM file as input using the default parameters. The setConfigElement ‘doPeakCalling’ was used with ‘annotationPeakGroupAgreePerc’ at 1 to generate the conserved peaks list. PC analysis was performed using ChrAccR. The genomic regions enrichment of annotations tool65 (GREAT, v.4.0.4, https://great.stanford.edu) was used to identify the most highly enriched GO terms of genes associated with differentially accessible peaks, the distance of peaks from transcriptional start sites and the distance of overlapping p53 ChIP and ATAC peaks from transcriptional start sites using standard parameters (https://great.stanford.edu). PAVIS66 was performed for peak annotation using standard parameters (version from 02-5-2018, https://manticore.niehs.nih.gov/pavis2/). Enrichr58,59,60 (https://maayanlab.cloud/Enrichr) was used to perform ChEA57 of p53 in various ChIP datasets31,32. p5353,54 hyperaccessible regions were defined as the overlap of regions that are more accessible in KT cells compared with KPT cells with the regions that are more accessible in KFT cells compared with KT cells.
HOMER analyses
TF motif analyses were performed individually on differentially accessible peaks from each experiment using the HOMER (v.4.11, https://homer.ucsd.edu/homer/) de novo motif discovery tool67 with the findMotifsGenome command and the following parameters: size = given; masked genome, background regions = all shared, non-differential peaks between genotypes.
Human cancer data analysis
Clinical lung cancer datasets used in this study were from TCGA-LUAD1, NCBI Gene Expression Omnibus (GEO) database identifier GSE13213 (ref. 68) and ArrayExpress database accession E-MTAB-8615 (ref. 69). The TCGA-LUAD dataset (280 TP53 mutated and 249 wild type) was downloaded from https://gdc.cancer.gov using the package TCGAbiolinks (https://github.com/BioinformaticsFMRP/TCGAbiolinks, v.2.18) and log2 transformed. GSE13213, which is composed of 38 TP53 mutated samples and 78 wild-type samples, was downloaded from GEO using the GEOquery Bioconductor package (https://github.com/seandavi/GEOquery, v.2.58). E-MTAB-8615 was downloaded from ArrayExpress using the function getAE from the ArrayExpress Bioconductor package (https://github.com/arrayexpress, v.1.32). The E-MTAB-8615 dataset is composed of 73 TP53 mutated and 34 wild-type samples. Single sample signatures were calculated using gene set variation analysis (GSVA) and the package GSVA from Bioconductor (https://github.com/rcastelo/GSVA, v.1.48) using Gaussian kernel (kcdf=“Gaussian”,mx.diff=T). We calculated the significant differences between the expression of the genes across TP53 mutated and wild-type tumours using a two-way ANOVA test in a linear model. To adjust for the possible contribution of TP53 to proliferation or stage, we included MKI67 expression in the linear model, and when available stage (E-MTAB-8615 did not have stage information available). ggpubr (https://cloud.r-project.org/web/packages/ggpubr/index.html, v.0.6.0) was used for plotting.
Single-cell sorting of lung tumour cells for multiomics experiments
KFT, KT and KPT mice were treated with Ad5-SPC-Cre as described above. After 10 weeks, lungs were collected and were perfused with 10 ml of ice-cold PBS and placed in a sterile dish on ice. Lungs were minced with sterile scissors and resuspended in 6 ml digestion medium (RPMI with 0.083 U ml–1 of collagenase type IV (Sigma-Aldrich), 0.6 U ml–1 dispase II (Sigma-Aldrich) and 25 µg ml–1 DNase (Sigma Aldrich)). Lung samples were rotated for 45 min at 37 °C. Tubes were briefly cooled on ice, and then samples were passed through a 70 µm filter followed by a 40 µm filter. Next, 5 ml of FACS buffer (10% FBS, 2 mM EDTA and 25 µg ml–1 DNase in DPBS) was added and tubes were spun for 5 min at 300g. Cells were resuspended in ACK lysing buffer (Gibco), incubated for 1 min on ice and quenched with 8 ml of FACS buffer. Cells were washed 2 times with FACS buffer, resuspended with biotinylated primary antibodies (CD45, BioLegend 103104 30-F11; CD31, BioLegend 102404 390; F4/80, BioLegend 123106 BM8; Ter119, BioLegend 116204 TER-119; 1:800 in FACS buffer) and incubated for 20 min on ice. After washing 2 times, cells were resuspended in streptavidin–APC secondary antibody (BioLegend 405207, 1:800) for 20 min on ice. Cells were washed 2 times and resuspended in FACS buffer with 1 µg ml–1 DAPI, filtered through a 40 µm filter and sorted using a Sony SH800S cell sorter. Data were analysed using the default Sony SH800S software (v.2.1.5) and FCS Express (v.7, De Novo Software).
Droplet-based scATAC and gene expression sequencing
Single-cell suspensions were prepared from mice with lung tumours as described above. Nuclei were isolated and libraries were prepared using a 10x Genomics Chromium Single Cell Multiome ATAC + Gene Expression kit according to the manufacturer’s protocols. An input of at least 2,000 nuclei were added to each 10x channel. Libraries were sequenced on an Illumina NovaSeq6000.
scATAC and gene expression analysis
Analyses were performed in R (v.4.0.3) using various R packages and Python (v.3.6) unless otherwise noted using the Stanford SCG Informatics Cluster. FASTQ files were processed using cellranger-arc count (10x Genomics). This pipeline aligns reads to the mm10 mouse reference genome and performs barcode error correction, PCR duplicate marking, peak calling and ATAC and gene expression molecule counting to produce expression and fragment count matrices. The gene expression matrices and ATAC fragments were loaded into R and processed using Seurat (v.4.0.1, https://github.com/satijalab/seurat) and Signac (v.1.7.0, https://github.com/timoast/signac) for downstream analyses. Cells were filtered to exclude cells that had the following parameters: (1) a nucleosome signal greater than 2; (2) transcription start site enrichment lower than 1; (3) ATAC counts of greater than 100,000; (4) RNA counts of greater than 25,000; (5) ATAC counts of fewer than 500; (6) RNA counts of fewer than 100.
Gene expression matrices were processed by normalizing the data, scaling the normalized expression matrix and performing FindVariableFeatures to identify the top 2,000 genes with the highest variance and expression using SCTransform in Seurat. DNA accessibility data were processed by finding the most frequently observed features, computing the term-frequency inverse-document frequency and running singular value decomposition. We then integrated our datasets using Harmony (v.1.0, https://portals.broadinstitute.org/harmony). Harmony was performed on both the gene expression and ATAC datasets. The variable used in the argument group.by.vars was the original sample identity, and the parameters used were block.size=0.05, max.iter.harmony=10, max.iter.cluster=20, epsilon.cluster=1e-05, epsilon.harmony=1e-04. The ‘pca’ reduction was used for the gene expression integration and the ‘lsi’ reduction was used for the ATAC integration. Joint UMAP visualization of gene expression and DNA accessibility data was done by using the weighted nearest neighbour methods (FindMultiModalNeighbors) in Seurat v.4. In brief, the weighted nearest neighbour graph was constructed by integrating the Harmony-integrated pca and Harmony-integrated lsi reductions of the gene expression and chromatin accessibility data. The following parameters were used: number of PCs = 50 (RNA), 39 (DNA accessibility, excludes first dimension), resolution = 1.0, k.nn = 20. Cancer cells were identified through analyses of genes and peak annotation of markers of each cluster using FindAllMarkers in addition to analysing the expression and accessibility patterns of lung epithelial markers. Cancer cells were subsetted, and the subsetted gene expression and DNA accessibility matrices were processed again as described above. A final joint UMAP was created using the weighted nearest neighbours method as described above. The following parameters were used: number of PCs = 50 (RNA), 39 (DNA accessibility, excludes first dimension), resolution = 0.4, k.nn = 20. Cancer cell clusters were identified using the FindAllMarkers function in Seurat to identify genes and ATAC peaks that were significantly enriched in each cluster in an unbiased manner. Additionally, the expression, accessibility and gene activity (computed from chromatin accessibility) of classical AT2, AT1 and transitional cell signatures and markers were assessed across the clusters to determine the identities of the clusters. Plots were generated using Seurat (v.4.0.1), Signac (v.1.7.0) and ggplot2 (v.3.4.2).
scRNA-seq analysis of a previously published dataset
A previously published scRNA-sequencing dataset of KT, KPT and T mice (GEO identifier GSE152607) was used29. Analyses were performed in R (v.4.0.3) using various R packages and Python (v.3.6) unless otherwise noted using the Stanford SCG Informatics Cluster. The gene expression matrices and clustering metadata were loaded into the R package Seurat (v.4.0.1, https://github.com/satijalab/seurat) for downstream analyses. t-SNE plots were reproduced using the previously defined dimensionality reduction and clustering metadata. Gene expression levels were displayed using the Seurat data visualization techniques FeaturePlot(), DotPlot() and DoHeatmap(). For heatmaps, the log-transformed transcripts per million (TPM) were rescaled and centred across all cells. For the visualization of direct overlap of the expression of two genes, the ‘blend’ feature of FeaturePlot was used with a blend threshold of 0.1. The top enriched Hallmark gene signatures from MSigDB in cluster 5 markers was performed using the full list of genes in supplementary table 1 from ref. 29 and Enrichr (https://maayanlab.cloud/Enrichr). To identify DEGs in cluster 5 between KT and KPT samples, the FindMarkers() function was run with default parameters on cluster 5 split by Trp53 status using log2-transformed TPM values. Cluster 5 subpopulations were identified by hierarchical clustering using Ward’s method, based on the relative expression of DEGs in cluster 5 (log2-transformed TPMs, rescaled across all cells). Only DEGs with differences greater than 2-fold at 10% FDR were considered (absolute log2(FC > 1 between KT and KPT cells). The top three subclusters were designated as 5A, 5B and 5C based on the relative proportion of cells from tumours at earlier (KT < 30 weeks) or later stages (KPT, 30 weeks) of LUAD progression. The main expression programmes, including DEGs involved in p53, metabolism, glycolysis and cellular growth, were characterized by functional annotation with WebGestaltR (https://webgestalt.org, v.0.4.4). The top ten enriched gene sets were identified for each DEG cluster by over-representation analysis using the GO Biological Process non-redundant collection or custom gene sets with Hallmark signatures from MSigDB52 (v.7.4), in which human genes with a one-to-one orthologue in mouse were mapped to the corresponding mouse homologue. To compare expression trends at consecutive stages of LUAD progression, we computed an aggregate score measuring the average expression of all genes in each DEG programme (log2-transformed TPMs, without rescaling) relative to a set of randomly sampled genes at comparable expression levels70,71. The same approach was used to evaluate the aggregate expression of cluster 3 markers in each cluster 5 subpopulation, using the full list of genes in supplementary table 1 from ref. 29. P values were determined using the Mann–Whitney U-test. The expression trend of genes in all DEG programmes were further compared between cluster 3 and cluster 5 subpopulations by evaluating the correlation between their mean expression levels in each cluster (log2-transformed TPMs, rescaled across all cells and averaged over cells in a given cluster). The Pearson correlation coefficient and P values were computed using SciPy (https://github.com/scipy/scipy, v.1.6.3), the regression line and 95% confidence interval were estimated using seaborn (https://github.com/mwaskom/seaborn, v.0.11.1) and statsmodels (https://github.com/statsmodels/statsmodels, v.1.12.2).
Cell-fate transitions were analysed in selected tumour subpopulations from the previously published dataset29, including all cells from transitional subclusters (5A, 5B and 5C), AT1-like cells (cluster 3) and cells undergoing EMT (cluster 11) at every stage of LUAD progression. Dimensionality reduction was performed by PC analysis, after re-scaling normalized counts across cells from the selected clusters and considering the top 5,000 highly variable genes. Cell–cell transition probabilities were estimated using CellRank (v.1.5.0) with two complementary approaches: one based on differentiation potential, using the CytoTRACE kernel, and one based on diffusion pseudotime, using the pseudotime kernel. For pseudotime calculations, one cell in cluster 5A was arbitrarily selected as root. In both cases, directed transition probabilities were computed in a k-nearest neighbour (kNN) graph comprising, for each cell, the top 20 neighbours in the first 30 PCs, using a hard threshold scheme with default parameters for generating the transition matrix. scVelo (v.0.2.4) was used for visualizing cell-fate trajectories along the first two PCs, after computing the transition matrix projections in PC space. Connectivities among cell clusters, computed by partition-based graph abstraction considering the top 20 nearest neighbours, were similarly visualized with scVelo, using a connectivity threshold of 0.05. Similar to AT1 cell markers from a previous study29, the aggregate expression of transitional cell markers from another study12 was computed across the selected clusters, using the full list of Krt8+ ADI marker genes in supplementary data 3 from ref. 12. Furthermore, for all clusters, the percentage of marker genes showing a significant upregulation relative to the original AT2 cluster (cluster 1) was determined by Wilcoxon rank-sum test. Only markers showing an upregulation of at least two-fold, at 5% FDR, were considered. Copy number alterations were identified by running inferCNV (v.1.2.1) on the raw counts matrix for the selected tumour subpopulations (subclusters analysis with default parameters). To compare the accumulation of copy number alterations over time, and across subpopulations, the percentage of cells harbouring one or more deletions or duplications, as predicted using the inferCNV 6-state HMM model, was computed for each cell cluster or at each time point for both KT and KPT samples. All analyses were carried out with Python using Scanpy (v.1.7.1), CellRank (v.1.5.0), scVelo (v.0.2.4), pandas (v.1.2.4), NumPy (v.1.20.3), SciPy (v.1.6.3) and fastcluster (v.1.2.4). Plots were generated using matplotlib (v.3.4.2) or seaborn (v.0.11.1). R packages were imported into Python using rpy2 (v.3.4.4). Plots were generated using matplotlib (https://github.com/matplotlib/matplotlib, v.3.4.2) or seaborn (https://github.com/mwaskom/seaborn, v.0.11.1).
BHT lineage tracing model
SPCcreERT2;Trp5353,54/53,54;Rosa26LSL-tdTomato/LSL-tdTomato, SPCcreERT2;Trp53+/+;Rosa26LSL-tdTomato/LSL-tdTomato and SPCcreERT2;Trp53flox/flox;Rosa26LSL-tdTomato/LSL-tdTomato mice were used in lineage tracing experiments in a model of BHT injury36,37,72. Tamoxifen was dissolved in corn oil with 2% ethanol at a concentration of 50 mg ml–1 by heating and sonicating until the solution was clear. BHT was dissolved in corn oil at a concentration of 9 mg ml–1 by gentle heating and vortexing. Male and female mice (8–10 weeks old) were treated on three consecutive days with 5 mg of tamoxifen by oral gavage. One week after the final dose, BHT (450 mg kg–1) or corn oil (vehicle) was intraperitoneally injected into the mice. At either 47 or 95 h after injection, BrdU dissolved at 10 mg ml–1 in PBS was intraperitoneally injected into mice at a concentration of 1 mg per 10 g of body weight. After 1 h, lungs were inflated with 2% low-melt agarose, fixed in ice-cold 4% PFA overnight, incubated with 30% sucrose in PBS overnight and then cryo-embedded in OCT.
Lung injury scoring
Lung injury scoring was done as previously described73,74. In brief, SpcT and SpcPT mice were treated with BHT for 4 days as described above. After 4 days, lungs were perfused with ice-cold PBS, inflated with 2% low-melt agarose and fixed in formalin (3.7% formaldehyde in PBS) for 24 h and removed for paraffin embedding. Slices were stained with H&E quantified by a pathologist (M.G.O.) blinded to the genotypes of the mice. The final injury score was calculated from 20 total fields per mouse using the equation: (20 × (A) + 14 × (B) + 7 × (C) + 7 × (D) + 2(E))/(number of fields × 100). Scoring parameters (A–E) can be found in Supplementary Table 9, which were adapted from previous studies73,74.
Single-cell sorting of BHT lineage tracing model
SpcT and SpcPT mice were treated as described above. Owing to mouse colony limitations, one SpcT mouse was SftpccreERT2;Trp53+/+;Rosa26Confetti (Brainbow 2.1)75. However, RFP is only expressed after Cre treatment, as in SpcT mice, and only red RFP+ cells were sorted from this mouse in an identical manner as the replicate SpcT mouse. At 96 h after BHT treatment, lungs were collected and perfused with 10 ml of ice-cold PBS and placed in a sterile dish on ice. Lungs were minced with sterile scissors and resuspended in 6 ml digestion medium (RPMI with 0.083 U ml–1 of collagenase type IV (Sigma-Aldrich), 0.6 U ml–1 dispase II (Sigma-Aldrich) and 25 µg ml–1 DNase (Sigma Aldrich)). Lung samples were rotated for 30 min at 37 °C. Tubes were briefly cooled on ice and then samples were passed through a 70 µm filter followed by a 40 µM filter. Next, 5 ml of FACS buffer (10% FBS, 2 mM EDTA and 25 µg ml–1 DNase in DPBS) was added and tubes were spun for 5 min at 300g. Cells were resuspended in ACK lysing buffer (Gibco), incubated for 1 min on ice and quenched with 8 ml of FACS buffer. Cells were washed 2 times with FACS buffer, resuspended with biotinylated primary antibodies (CD45, BioLegend 103104 30-F11; CD31, BioLegend 102404 390; F4/80, BioLegend 123106 BM8; Ter119, BioLegend 116204 TER-119; 1:800 in FACS buffer) and incubated for 20 min on ice. After washing 2 time, cells were resuspended in streptavidin–APC secondary antibody (BioLegend 405207, 1:800) for 20 min on ice. Cells were washed 2 times and resuspended in FACS buffer with 1 µg ml–1 DAPI, filtered through a 40 µm filter and sorted using a Sony SH800S cell sorter. Data were analysed using the default Sony SH800S software and FCS Express (v.7, De Novo Software).
Droplet-based scRNA-seq
Single-cell suspensions were prepared from mice treated with BHT as described above. Libraries were prepared using a 10x Genomics Single Cell 3′ Library v.3.1 kit according to the manufacturer’s protocols. An input of 10,000 cells were added to each 10x channel. Libraries were sequenced on an Illumina NovaSeq6000.
scRNA-seq analysis
Analyses were performed in R (v.4.0.3) using various R packages and Python (v.3.6) unless otherwise noted using the Stanford SCG Informatics Cluster. FASTQ files were processed using cellranger count (10x genomics). This pipeline aligns reads to the mm10 mouse reference genome and performs barcode error correction, PCR duplicate marking, barcode counting and unique molecular identifier counting to produce expression count matrices. The gene expression matrices were loaded into R and processed using Seurat (v.4.0.1, https://github.com/satijalab/seurat) for downstream analyses. Cells were filtered to exclude cells that had the following parameters: (1) fewer than 200 genes expressed; (2) over 7,000 genes expressed; and (3) over 25% mitochondrial reads. Genes were further filtered to eliminate those expressed in fewer than two cells. Gene expression matrices were processed by normalizing the data, performing FindVariableFeatures to identify the top 2,000 genes with the highest variance and expression, and centring and scaling the normalized expression matrix. PC analysis was run to identify major axes of variation. Clustering was performed by constructing a nearest neighbour graph and identifying clusters of cells by a shared nearest neighbour modularity optimization-based clustering algorithm (Seurat v.4). Clustering was visualized by UMAP dimensionality reduction. The following parameters were used: number of PCs = 30, k.param = 5, resolution = 0.75. Alveolar cells were identified through analysis of gene markers of each cluster using FindAllMarkers. Lineage-traced alveolar cells were subsetted, and the subsetted gene expression matrices were processed again as described above. Cells were filtered to have tdTomato expression of greater than or equal to 0.5. The following clustering parameters were used: number of PCs = 20, k.param = 30, resolution = 0.40. Differential expression analysis between genotypes was performed using a Wilcoxon rank sum test. Plots were generated using Seurat (v.4.0.1) and ggplot2 (v.3.4.2). Monocle 3 (v.1.0.0) was used to perform pseudotime analysis, and CytoTRACE34 (beta version, https://cytotrace.stanford.edu) was used to perform differentiation potential analysis.
LUAD cell generation
KrasLA2/+;Trp53LSL-WT/LSL-WT mice or KrasLA2/+;Trp53LSL-mut/LSL-mut mice76 were used to generate the LUAD cell lines. In brief, mice aged 11 weeks were killed and tumours were microdissected from the lungs. Tumours were dissociated using collagenase–dispase and DNase for 2 h at 37 °C. Cells were grown in N5 medium for at least 3 days. Once cells began to proliferate, cells were sorted based on EPCAM positivity to select for epithelial cancer cells (EPCAM-PE (rat, BioLegend G8.9, 1.0 μg per 1 million cells)) using a FACSymphony S6 cell sorter (BD Biosciences) and analysed using FlowJo (v.9) and FACSDiva (v.6.0). Cells were subsequently cultured in DMEM with 10% FBS. In experiments, cell lines were infected in culture with either Ad-Empty, as a p53 null control (denoted Trp53null), or Ad-Cre to reactivate either wild-type p53 or p5353,54.
Cell culture, adenoviral treatment and transfection
LUAD cell lines (generated in-house), A549 cells (from American Tissue Culture Collection) and HEK293AH cells (from M. Bassik at Stanford) were maintained in Dulbecco’s Modified Eagle medium (Gibco) supplemented with 10% FCS and incubated at 37 °C in a carbon dioxide incubator. No commonly misidentified cell lines were used in this study. The cell lines were not authenticated. These cell lines were routinely tested for mycoplasma contamination and were always negative. To reactivate p53, LUAD cell lines were transduced with Ad5-CMV-Cre (University of Iowa Viral Vector Core) or cell lines were transduced with Ad5-CMV-Empty (University of Iowa Viral Vector Core) to remain Trp53null. Cell lines were infected at a multiplicity of infection of 100. Cells were collected for IF, western blotting and qPCR with reverse transcription (RT–qPCR) between 24 and 96 h after infection. A549 cells were transfected with either an empty vector (pcDNA3.1) or pcDNA3.1-3XHA-TP53 that contains amino-terminal HA tagged full-length human p53 using Lipofectamine 3000 by following the manufacturer’s protocols. At 48 h after transfection, cells were fixed in 4% PFA for subsequent immunostaining. For experiments with TIGIT+ sorted LUAD cells, cells were sorted and transduced with Ad-Empty (to keep the cells Trp53null) or with Ad-Cre to reactivate p53. Samples were collected 1 and 3 days after adenoviral treatment.
LUAD cell line RNA-seq
For RNA-seq, LUAD cells were collected, pelleted and flash frozen in liquid nitrogen. Once all samples were collected, RNA was isolated for sequencing on a HiSeq4000 (Illumina). High-performance computation was performed on the Stanford SCG Informatics Cluster, and subsequent bioinformatics analyses were performed in R (v.4.0.3) using various R packages unless otherwise noted. The RNA-seq analysis pipeline is the same as described above.
LUAD cell line FACS
KrasLA2/+;Trp53LSL-WT/LSL-WT LUAD cells were collected, washed with PBS and then incubated with TIGIT-BV421 (mouse, BioLegend 1G9, 0.25 μg per 1 million cells) and EPCAM-PE (rat, BioLegend G8.9, 1.0 μg per 1 million cells) for 15 min on ice. Cells were washed 2 times with PBS for 5 min at 1,000 r.p.m. in a tabletop centrifuge. Cells were resuspended in FACS buffer (PBS and 2% FBS) and filtered through a 40 μm filter. TIGIT+ epithelial cells were sorted using a FACSymphony S6 cell sorter (BD Biosciences) and analysed using FlowJo (v.9) and FACSDiva (v.6.0).
ChIP–qPCR
KrasLA2/+;Trp53LSL-WT/LSL-WT LUAD cells were seeded at 2.5 × 106 cells (for Ad-Empty) or 5 × 106 cells (for Ad-Cre) per 10 cm dish and transduced with the indicated adenovirus. After 48 h, cells were crosslinked at room temperature by treatment with DMEM with 1% formaldehyde, and the reaction was quenched by the addition of glycine to a final concentration of 0.125 M. After washing with cold 1× PBS, cells were collected by scraping in lysis buffer (5 mM PIPES pH 8.0, 85 mM KCl and 0.5% NP-40) and pelleted. Cell pellets were processed by passage through a 21-gauge needle 20 times. Lysates were pelleted and resuspended in RIPA buffer. Sonication was performed in a Bioruptor sonicator (Diagenode) to shear chromatin to a size range of around 200–700 bp. Anti-p53 antibody (CM5, Leica Novocastra) was coupled to ChIP-grade protein A/G magnetic beads (Thermo Scientific) overnight. After saving 10% for an input sample, samples were immunoprecipitated for 1 h at room temperature and 1 h at 4 °C, and washes were performed 2 times with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl at pH 8.1 and 150 mM NaCl), 3 times with high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl at pH 8.1, and 500 mM NaCl) and 4 times with LiCl wash buffer (0.25 M LiCl, 1% IGEPAL CA630, 1% deoxycholic acid sodium salt, 1 mM EDTA and 10 mM Tris at pH 8.1). Input was reverse crosslinked by treatment with ProK and RNase A and incubation at 65 °C. All samples were purified using a PCR Purification kit (Qiagen). Chromatin-immunoprecipitated DNA was quantified by qPCR using SYBR Green (SA-Biosciences) and a 7900HT Fast Real-Time PCR machine (Applied Biosystems).
p53 reactivation experiments in mouse LUAD in vivo
KrasFSF-G12D/+;Trp53LSL-WT/frt;Rosa26creER mice and KrasFSF-G12D/+;Trp53LSL-53,54/frt;Rosa26creER mice were infected with 109 Ad5-CMV-Flpo virus intratracheally (University of Iowa Viral Vector core, VVC-U of Iowa-530-sa) to activate the oncogenic KrasG12D allele and delete Trp53 in cancer cells. The subsequent cancer cells formed are effectively Trp53null owing to the presence of the LSL cassette on the remaining Trp53 allele, whereas uninfected cells in the mouse remain heterozygous for Trp53. After 16 weeks, each mouse was treated with 100 µl of 5 mg ml–1 tamoxifen dissolved in corn oil each day for 3 days by oral gavage. The tamoxifen treatments were repeated after a week. Mice were killed and lungs were collected 2 weeks after the first dose of tamoxifen treatment.
Construction of A549 TP53 knockout cell lines
A549 TP53 knockout cells were generated using the pX330-based plasmid pX458 (gift from F. Zhang, Addgene plasmid 48138) expressing Cas9, GFP and sgRNA targeting human TP53. A549 cells were transfected with three pX458 TP53 plasmids expressing three distinct sgRNAs targeting TP53. Two days after transfection, the GFP+ population was sorted by FACS and clonally expanded. Individual cell clones were screened for p53 loss by immunoblotting.
Western blotting
Western blots were performed according to standard protocols. In brief, cells were lysed in NP-40 lysis buffer (ThermoFisher), extracts were run on SDS–PAGE gels and gels were transferred to PVDF membranes (Immobilon, Millipore). Membranes were blocked with 5% milk and probed with antibodies directed against p53 (CM5, 1:1000 Leica Novocastra) or GAPDH (1:20,000, Fitzgerald, clone 6C5), followed by anti-mouse or anti-rabbit HRP-conjugated secondary antibodies (Vector Laboratories). Blots were developed with ECL Prime (Amersham) and imaged using a ChemiDoc XRS+ (Bio-Rad). Gels were analysed using Image Lab (Bio-Rad, v.3.0).
RT–qPCR
TRIzol reagent (Invitrogen) was used for RNA preparation, and reverse transcription was performed with MMLV reverse transcriptase (Invitrogen). qPCR was performed in triplicate using gene-specific primers and SYBR Green (Life Technologies) in a 7900HT Fast Real-Time PCR machine (Applied Biosystems). Changes in transcript abundance were calculated using the standard curve method.
Statistical analysis
Data analysis and statistical tests were performed using GraphPad Prism software (v.9.0.1). Pearson correlation coefficient and related P values were computed using SciPy (https://github.com/scipy/scipy, v.1.6.3), the regression line and 95% confidence interval were estimated using seaborn (https://github.com/mwaskom/seaborn, v.0.11.1) and statsmodels (https://github.com/statsmodels/statsmodels, v.1.12.2). No statistical methods were used to predetermine sample sizes, and sample sizes were chosen on the basis of previously published studies in the field. All experiments were performed using at least three biological replicates, with the exception of the KT and KFT ATAC–seq experiment, the single-cell multiomics analysis and the ChIP–qPCR assays, all of which had two biological replicates. Additionally, experiments on TIGIT+ transitional LUAD cells were performed once as the results were replicated across both the day 1 and day 3 time points, and the phenotypes were stable and supported by the data from our in vivo models. All measurements were taken on discrete samples. All attempts at replication were successful. Student’s t-tests were two-tailed. All statistical tests are denoted in the figure legends, and data are presented as the mean ± s.d. unless otherwise noted.
Availability of materials
All unique biological materials are available from the corresponding author upon reasonable request.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data from in vivo LUAD RNA-seq, in vivo LUAD ATAC–seq, in vitro LUAD cell line RNA-seq, in vivo single-cell multiomics (RNA-seq and ATAC–seq) and in vivo lung injury scRNA-seq experiments are available from the GEO under the accession number GSE231681. The mouse LUAD microarray data have been previously published30 and are available at the GEO (GSE23875). The mouse p53 ChIP–seq datasets in mouse embryonic fibroblasts, mouse embryonic stem cells and mouse transitional cells after bleomycin injury have been previously published14,31,32 and are available at the GEO (GSE46240, GSE26361 and GSE141635, respectively). The mouse LUAD scRNA-seq data have been previously published29 and available at the GEO (GSE152607). Expression data for the human TGCA-LUAD cohort1 are available through the NCI GDC Data Portal (https://portal.gdc.cancer.gov/projects/TCGA-LUAD). Expression data for the human GSE13213 LUAD cohort68 is available at the GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse13213). Expression data for the human E-MTAB-8516 LUSC cohort69 is available at ArrayExpress (https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-8615?query=E-MTAB-8615). The MSigDB database (https://gsea-msigdb.org/gsea/msigdb/)40 was used to perform GSEA52 analyses. All other data supporting the findings of this study are in the article, supplementary information or are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
The Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Tammemagi, M. C., McLaughlin, J. R. & Bull, S. B. Meta-analyses of p53 tumor suppressor gene alterations and clinicopathological features in resected lung cancers. Cancer Epidemiol. Biomarkers Prev. 8, 625–634 (1999).
Jiao, X. D., Qin, B. D., You, P., Cai, J. & Zang, Y. S. The prognostic value of TP53 and its correlation with EGFR mutation in advanced non-small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer 123, 70–75 (2018).
Steels, E. et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur. Respir. J. 18, 705–719 (2001).
Vaughan, A. E. & Chapman, H. A. Regenerative activity of the lung after epithelial injury. Biochim. Biophys. Acta 1832, 922–930 (2013).
Desai, T. J., Brownfield, D. G. & Krasnow, M. A. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507, 190–194 (2014).
Nabhan, A. N., Brownfield, D. G., Harbury, P. B., Krasnow, M. A. & Desai, T. J. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123 (2018).
Barkauskas, C. E. et al. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036 (2013).
Adamson, I. Y. & Bowden, D. H. The type 2 cell as progenitor of alveolar epithelial regeneration. A cytodynamic study in mice after exposure to oxygen. Lab. Invest. 30, 35–42 (1974).
Evans, M. J., Cabral, L. J., Stephens, R. J. & Freeman, G. Renewal of alveolar epithelium in the rat following exposure to NO2. Am. J. Pathol. 70, 175–198 (1973).
Crapo, J. D., Barry, B. E., Gehr, P., Bachofen, M. & Weibel, E. R. Cell number and cell characteristics of the normal human lung. Am. Rev. Respir. Dis. 126, 332–337 (1982).
Strunz, M. et al. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun. 11, 3559 (2020).
Choi, J. et al. Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell 27, 366–382.e7 (2020).
Kobayashi, Y. et al. Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat. Cell Biol. 22, 934–946 (2020).
Melms, J. C. et al. A molecular single-cell lung atlas of lethal COVID-19. Nature 595, 114–119 (2021).
Mello, S. S. et al. A p53 super-tumor suppressor reveals a tumor suppressive p53-Ptpn14–Yap axis in pancreatic cancer. Cancer Cell 32, 460–473.e6 (2017).
Jackson, E. L. et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 65, 10280–10288 (2005).
Sutherland, K. D. et al. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc. Natl Acad. Sci. USA 111, 4952–4957 (2014).
Brady, C. A. et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145, 571–583 (2011).
Solis, L. M. et al. Histologic patterns and molecular characteristics of lung adenocarcinoma associated with clinical outcome. Cancer 118, 2889–2899 (2012).
Mainardi, S. et al. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc. Natl Acad. Sci. USA 111, 255–260 (2014).
Xu, X. et al. Evidence for type II cells as cells of origin of K-Ras-induced distal lung adenocarcinoma. Proc. Natl Acad. Sci. USA 109, 4910–4915 (2012).
Nakamura, N., Miyagi, E., Murata, S., Kawaoi, A. & Katoh, R. Expression of thyroid transcription factor-1 in normal and neoplastic lung tissues. Mod. Pathol. 15, 1058–1067 (2002).
Little, D. R. et al. Transcriptional control of lung alveolar type 1 cell development and maintenance by NK homeobox 2-1. Proc. Natl Acad. Sci. USA 116, 20545–20555 (2019).
Little, D. R. et al. Differential chromatin binding of the lung lineage transcription factor NKX2-1 resolves opposing murine alveolar cell fates in vivo. Nat. Commun. 12, 2509 (2021).
Jones, S. An overview of the basic helix-loop-helix proteins. Genome Biol. 5, 226 (2004).
LaFave, L. M. et al. Epigenomic state transitions characterize tumor progression in mouse lung adenocarcinoma. Cancer Cell 38, 212–228.e13 (2020).
Kastenhuber, E. R. & Lowe, S. W. Putting p53 in context. Cell 170, 1062–1078 (2017).
Marjanovic, N. D. et al. Emergence of a high-plasticity cell state during lung cancer evolution. Cancer Cell 38, 229–246.e13 (2020).
Feldser, D. M. et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature 468, 572–575 (2010).
Kenzelmann Broz, D. et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 27, 1016–1031 (2013).
Li, M. et al. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol. Cell 46, 30–42 (2012).
Zeng, Y., Kotake, Y., Pei, X. H., Smith, M. D. & Xiong, Y. p53 binds to and is required for the repression of Arf tumor suppressor by HDAC and polycomb. Cancer Res. 71, 2781–2792 (2011).
Gulati, G. S. et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 367, 405–411 (2020).
Lange, M. et al. CellRank for directed single-cell fate mapping. Nat. Methods 19, 159–170 (2022).
Adamson, I. Y., Bowden, D. H., Cote, M. G. & Witschi, H. Lung injury induced by butylated hydroxytoluene: cytodynamic and biochemical studies in mice. Lab. Invest. 36, 26–32 (1977).
Hirai, K. I., Witschi, H. & Cote, M. G. Electron microscopy of butylated hydroxytoluene-induced lung damage in mice. Exp. Mol. Pathol. 27, 295–308 (1977).
Valentin-Vega, Y. A., Okano, H. & Lozano, G. The intestinal epithelium compensates for p53-mediated cell death and guarantees organismal survival. Cell Death Differ. 15, 1772–1781 (2008).
Han, X. et al. Mapping the mouse cell atlas by Microwell-Seq. Cell 172, 1091–1107.e17 (2018).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Pearson, B. J. & Sanchez Alvarado, A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development 137, 213–221 (2010).
Yun, M. H., Gates, P. B. & Brockes, J. P. Regulation of p53 is critical for vertebrate limb regeneration. Proc. Natl Acad. Sci. USA 110, 17392–17397 (2013).
Valente, L. J. et al. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep. 3, 1339–1345 (2013).
Li, T. et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149, 1269–1283 (2012).
Morris, J. P. et al. α-Ketoglutarate links p53 to cell fate during tumour suppression. Nature 573, 595–599 (2019).
Tschaharganeh, D. F. et al. p53-dependent Nestin regulation links tumor suppression to cellular plasticity in liver cancer. Cell 158, 579–592 (2014).
Rivlin, N., Koifman, G. & Rotter, V. p53 orchestrates between normal differentiation and cancer. Semin Cancer Biol. 32, 10–17 (2015).
Gocek, E. & Marcinkowska, E. Differentiation therapy of acute myeloid leukemia. Cancers (Basel) 3, 2402–2420 (2011).
Zhao, L., Yee, M. & O’Reilly, M. A. Transdifferentiation of alveolar epithelial type II to type I cells is controlled by opposing TGF-β and BMP signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 305, L409–L418 (2013).
Sugimoto, K., Takayasu, H., Nakazawa, N., Montedonico, S. & Puri, P. Prenatal treatment with retinoic acid accelerates type 1 alveolar cell proliferation of the hypoplastic lung in the nitrofen model of congenital diaphragmatic hernia. J. Pediatr. Surg. 43, 367–372 (2008).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Angelidis, I. et al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 10, 963 (2019).
Travaglini, K. J. et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625 (2020).
Marconett, C. N., Zhou, B., Siegmund, K. D., Borok, Z. & Laird-Offringa, I. A. Transcriptomic profiling of primary alveolar epithelial cell differentiation in human and rat. Genom. Data 2, 105–109 (2014).
Thomas, P. D. et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003).
Lachmann, A. et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26, 2438–2444 (2010).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Xie, Z. et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 1, e90 (2021).
DuPage, M., Dooley, A. L. & Jacks, T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 4, 1064–1072 (2009).
Treutlein, B. et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014).
Wang, Y. et al. Pulmonary alveolar type I cell population consists of two distinct subtypes that differ in cell fate. Proc. Natl Acad. Sci. USA 115, 2407–2412 (2018).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Huang, W., Loganantharaj, R., Schroeder, B., Fargo, D. & Li, L. PAVIS: a tool for Peak Annotation and Visualization. Bioinformatics 29, 3097–3099 (2013).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Tomida, S. et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J. Clin. Oncol. 27, 2793–2799 (2009).
Haakensen, V. D. et al. Molecular characterisation of TP53 mutated squamous cell carcinomas of the lung to identify putative targets for therapy. Int. J. Cancer 147, 2957–2966 (2020).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Kehrer, J. P. & DiGiovanni, J. Comparison of lung injury induced in 4 strains of mice by butylated hydroxytoluene. Toxicol. Lett. 52, 55–61 (1990).
Matute-Bello, G. et al. An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am. J. Respir. Cell Mol. Biol. 44, 725–738 (2011).
Patel, B. V., Wilson, M. R. & Takata, M. Resolution of acute lung injury and inflammation: a translational mouse model. Eur. Respir. J. 39, 1162–1170 (2012).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Johnson, L. et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410, 1111–1116 (2001).
Acknowledgements
We thank P. Chu for processing and embedding all paraffin-embedded mouse tissue samples; staff at the Stanford Functional Genomics Facility for help with the library preparation and sequencing of LUAD cell lines; K. Nuno and A. Spencley for technical advice regarding the ATAC–seq experiments; and T. Desai, M. Winslow, J. Sage, S. Ferreria, L. Cong, D. Bhattacharya and A. Boutelle for advice and critical reading of the manuscript. Research in this article was supported by NCI PHS grant CA009302, NCI R35 grant CA197591 (to L.D.A.) and the NSF (GRFP, to A.M.K.). A.M.K was also supported by the Stanford Graduate Fellowship.
Author information
Authors and Affiliations
Contributions
A.M.K. designed and performed experiments, interpreted and analysed data and wrote the manuscript. A.M.K. performed all mouse tumour studies, lung injury studies, in vivo sequencing experiments and the subsequent analyses for these experiments. A.M.K., A.G. and K.J.H. analysed and interpreted the previously published29 scRNA-seq data analyses. A.G. performed hierarchical clustering to identify clusters 5A, 5B and 5C, defined gene expression programmes within cluster 5, performed CytoTRACE, pseudotime and CellRank analyses, performed copy number variation analyses, and identified transitional cell markers across cancer cell clusters. K.J.H. and M.W. sorted TIGIT+ LUAD cells. R.L.Z. performed IF, ChIP–qPCR and LUAD cell line reactivation experiments, and assisted with BHT lung injury experiments. N.R. performed ChIP–qPCR experiments. J.A.S. performed all human tumour genomics analyses. K.T.B.-R. generated LUAD cell lines and performed RNA-seq on them. M.W. generated the A549 TP53 knockout cell lines, performed p53 reactivation experiments in vivo and performed IF experiments. I.L., W.L.T., D.Z.L., J.B.S. and S.K.P. collected and curated the human LUAD samples. A.M.N. assisted with trajectory analyses, and C.V.R. provided support for scRNA-seq analyses. L.D.A. designed experiments, interpreted data and wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 p5353,54 suppresses late-stage lung adenocarcinoma better than wild-type p53.
(a) Representative Western blot for p53 and GAPDH in p53LSL-wt/p53LSL-wt (n = 3) and p53LSL-53,54/LSL-53,54 (n = 3) LUAD cell lines infected with adenoviral (Ad)-Empty or Ad-Cre for 48 h. Samples were run on the same gel. For gel source data, see Supplementary Figure 1. LSL, lox-stop-lox. (b) RT-qPCR of a panel of p53 target genes in p53LSL-wt/p53LSL-wt (n = 3) and p53LSL-53,54/LSL-53,54 (n = 3) cell lines infected with Ad-Empty or Ad-Cre for 48 h. (c) Schematic for the 24-week lung cancer study. Histology cohort: KFT (n = 6), KT (n = 5), and KPT (n = 5) mice. (d) Representative histological H&E images of 24-week lungs from KFT (n = 6) and KT (n = 5) mice. Scale bar, 100 μm. (e) Quantification of 24-week lung tumor burden, number, and size (n = 112, 268 and 1,429 tumors for KFT, KT, and KPT cohorts, respectively). (f) Histopathological analysis of tumor patterns in 24-week KFT (n = 8), KT (n = 8), and KPT (n = 5) mice. Chart displays the percentage of mice that displayed tumors of a given pattern. NE, neuroendocrine; AAH, atypical adenomatous hyperplasia. (g) Histopathological analysis of tumor pleomorphism (see Methods) in 24-week KFT (n = 8), KT (n = 8), and KPT (n = 5) mice. The chart displays the percentage of mice that displayed tumors of a given score. (h) Histopathological analysis of mitotic activity in the most advanced lesion in 24-week KFT (n = 8), KT (n = 8), and KPT (n = 5) mice. HPF, high powered field. (i) Representative Ki67 IHC in tumors from 24-week KFT (n = 4) and KT (n = 4) mice. Scale bar, 50 μm. (j) Percentage Ki67-positive nuclei in (n = 20) tumors in 24-week KFT (n = 4) and KT (n = 4) mice. All data are mean ± s.d. P values were calculated by two-tailed Student’s t-test (b, j) and Kruskal-Wallis test with multiple comparisons (two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli) (e, h).
Extended Data Fig. 2 p5353,54-mediated tumor suppression is dependent upon its expression in AT2 cells.
(a) Schematic for the 10-week AT2 cell-of-origin tumor study. (b) Representative histological H&E images of lungs from 10-week KFT (n = 8), KT (n = 8), and KPT (n = 5) mice. Images below are a higher magnification view of the image above. Scale bars: top, 5 mm; middle, 500 μm; bottom, 50 μm. (c) Representative Ki67 and phosphohistone H3 (pHH3) IHC in tumors from 10-week KFT (n = 4), KT (n = 4), and KPT (n = 4) mice. Scale bar, 50 μm. (d) Percentage Ki67-positive nuclei in (n = 20) tumors in 10-week KFT (n = 4), KT (n = 4), and KPT (n = 4) mice. (e) Percentage pHH3-positive nuclei in (n = 20) tumors in 10-week KFT (n = 4), KT (n = 4), and KPT (n = 4) mice. (f) Schematic for the 10-week cancer cell autonomy study. Histology cohort: KT;p53LSL-53,54/+ (KFT-LSL) (n = 8) and KT;p53LSL-wt/+ (KT-LSL) (n = 8) mice. The results of this study demonstrate that p5353,54 expression in AT2 cells results in enhanced tumor suppression. (g) Quantification of lung tumor burden, number, and size (n = 83 and 213 tumors for KFT-LSL and KT-LSL cohorts, respectively). (h) Representative Ki67 IHC in tumors from KFT-LSL (n = 4) and KT-LSL (n = 4) mice. Scale bar, 50 μm. (i) Percentage Ki67-positive nuclei in (n = 20) tumors from KFT-LSL (n = 4) and KT-LSL (n = 4) mice. (j) Representative IF of HOPX or PDPN and tdTomato (tdT) in KFT-LSL (n = 3) mice and KT-LSL (n = 3) mice. Scale bar, 20 μm. (k) HOPX and PDPN quantification in (n = 50 [KFT-LSL] and 100 [KT-LSL]) tumors in KFT-LSL (n = 3) mice and KT-LSL (n = 3) mice (see Methods). Tumors were binned based on the percentage AT1 marker positivity of tdTomato+ cells. Low = 0–5%, medium = 5–20%, high = 20–100%. All data are mean ± s.d. P values were calculated by ordinary one-way ANOVA with Tukey’s multiple comparisons test (d-e), two-tailed Mann Whitney U test (g), and two-tailed Student’s t-test (i).
Extended Data Fig. 3 p5353,54 and wild-type p53 induce transcriptional and epigenetic profiles distinct from p53-deficient programs.
(a) FACS-sorting and library preparation schematic (see Methods). RIN, RNA integrity number. (b) Unsupervised hierarchical clustering of RNA-sequencing samples from KFT (n = 4), KT (n = 3), and KPT (n = 4) mice. (c) Heatmap of the 5,065 differentially expressed genes (DEGs, P-adjusted ≤ 0.05) between p53-proficient (KFT and KT) and p53-deficient (KPT) cells. DEGs identified in the analysis of KFT vs. KPT cells and KT vs. KPT cells are shown. (d) Significantly enriched gene ontology (GO) biological process terms in p5353,54 hyperactivated genes, identified by PANTHER56. (e) GO term analysis by PANTHER56 of genes upregulated in KT or KPT cells. (f) GSEA shows enrichment of the indicated GO biological process term in KT cells relative to KPT cells. NES, normalized enrichment score. (g) Percentage of all called peaks that are differentially accessible in the ATAC-sequencing experiments. (h) Principal component analysis of the KT (n = 4) vs. KPT (n = 4) ATAC-sequencing samples. (i) Peak annotation of differentially accessible regions in KT vs. KPT ATAC-sequencing experiment. UTR, untranslated region. (j) Principal component analysis of the KFT (n = 2) vs. KT (n = 2) ATAC-sequencing samples. (k) Peak annotation of differentially accessible regions in KFT vs. KT ATAC-sequencing experiment. P values were calculated by GSEA52 (f).
Extended Data Fig. 4 p5353,54 and wild-type p53 activate AT1 cell transcriptional and epigenetic programs.
(a) Expanded version of GSEA shown in Fig. 2a of alveolar epithelial cell signatures (Supplementary Table 6). Normalized enrichment score (NES) > 0 indicates the signature is enriched in KFT or KT cells relative to KPT cells. *Indicates gene sets significantly enriched (P ≤ 0.05). (b) GSEA shows enrichment of the indicated AT1 signatures in specific genotypes. (c) AT1 cells (HOPX+) as a ratio of all alveolar epithelial cells (NKX2-1+) (n = 600) at homeostasis in p5353,54/53,54 (n = 3), p53+/+ (n = 3), and p53−/− (n = 3) mice (no oncogenic Kras). (d) Proliferating (BrdU+) cells as a ratio of all alveolar epithelial cells (NKX2-1+) (n = 600) at homeostasis in p5353,54/53,54 (n = 3), p53+/+ (n = 3), and p53−/− (n = 3) mice (no oncogenic Kras). (e) GSEA shows enrichment of non-alveolar cell signatures (Supplementary Table 6) in KPT cells. NES < 0 indicates the signature is enriched in KPT cells. *Indicates gene sets significantly enriched (P ≤ 0.05). (f) (Top) Plot from Fig. 1j showing differentially accessible regions (P-adjusted ≤ 0.05) at AT1-associated gene promoters (± 1kb from the transcription start site, Angelidis_Mouse_AT1 signature, denoted by red/blue dots). (Bottom) Plot from Fig. 1k showing differentially accessible regions (P-adjusted ≤ 0.05) at AT1-associated gene promoters (Angelidis_Mouse_AT1 signature). (g) Counts per million (CPM) indicating AT1 gene expression in KT (n = 3) and KPT (n = 4) RNA-sequencing samples. All data are mean ± s.d. P values were calculated by GSEA52 (a, b, e), ordinary one-way ANOVA with Tukey’s multiple comparisons test (c, d) and using the DESeq2 R package (two-tailed Wald test adjusted for multiple testing using the procedure of Benjamini and Hochberg) (g).
Extended Data Fig. 5 Single-cell analyses show that p53 promotes AT1 cancer cell formation.
(a) Uniform Manifold Approximation and Projection (UMAP) plot of single-cell multiome (scRNA and scATAC) profiles of cancer cells (n = 6,929) collected from 10-week KFT (n = 2), KT (n = 2), and KPT (n = 2) mice. (b) Dot plot showing the RNA expression of alveolar epithelial cell signatures (Supplementary Table 6, AT1 and AT2 signatures are derived from the Mouse Cell Atlas) and markers in all cells across clusters. Sig = signature, Trans. sig. = transitional cell signature. (c) Dot plot showing the computed gene activity from ATAC data of alveolar epithelial cell signatures (Supplementary Table 6, AT1 and AT2 signatures are derived from the Mouse Cell Atlas) and markers in all cells across clusters. (d) Heatmap showing expression of AT2 and AT1 markers (log-transformed transcripts per million rescaled across all cells) in all cells. (e) Feature plots show the RNA expression/computed gene activity of (top) AT2 cell signatures and markers and (bottom) AT1 cell signatures and markers in all cells (Supplementary Table 6, AT1 and AT2 signatures are derived from Mouse Cell Atlas). (f) Feature plot shows the RNA expression of GO lung differentiation term genes (Supplementary Table 6) in all cells. (g) ATAC-sequencing tracks showing chromatin accessibility at Sftpc (AT2 gene), Ager (AT1 gene), and Cldn4 (transitional cell gene) in all cells split by clusters. (h) UMAP plot from (a) split by KFT (n = 1,200), KT (n = 4,318), and KPT (n = 1,411) cells. (i) Relative proportions of all clusters split by genotype. (j) Relative proportions of AT1-like cancer cells in each genotype. KFT was set to 1.0.
Extended Data Fig. 6 p53 promotes an AT1 differentiation program in LUAD.
(a) Representative IF of tdTomato (tdT) and AGERhigh/medium/low, HOPXhigh/medium/low, and SPChigh/low tumors. Scale bar, 20 μm. (b) Uncropped images from Fig. 2b of AGER and tdTomato in 10-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. Scale bar, 200 μm. (c) Expanded view of images from Fig. 2b of AGER and tdTomato in 10-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. Scale bars, 20 μm. (d) Percentage tdTomato+ cancer cells positive for AGER in individual (n = 100 [KFT], 100 [KT], and 200 [KPT]) tumors in KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. Red lines; break points for high, medium, and low staining. (e) Expanded view of images from Fig. 2b of HOPX and tdTomato in 10-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. Scale bars, 20 μm. (f) Representative IF of PDPN and tdTomato in 10-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. Scale bars, 20 μm. (g) Representative IF of SPC and tdTomato in 10-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. Scale bar, 20 μm. (h) PDPN quantification in (n = 100 [KFT], 100 [KT], and 200 [KPT]) tumors in KFT (n = 4), KT (n = 3), and KPT (n = 3) mice (see Methods). The graph shows the percentage of tumors that are classified as AT1 marker high, medium, and low. Low = 0–5%, medium = 5–20%, high = 20–100%. (i) SPC quantification in (n = 100 [KFT], 100 [KT], and 200 [KPT]) tumors in 10-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice (see Methods). The graph shows the percentage of tumors that are classified as AT2 marker high and low. Low = 0–70%, high = 70–100%. (j) Percentage tdTomato+ cancer cells positive for SPC in individual (n = 100 [KFT], 100 [KT], and 200 [KPT]) tumors in KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. Red lines; break points for high and low staining. (k) Representative IF of AGER, SPC, and tdTomato in 10-week KFT (n = 3), KT (n = 3), and KPT (n = 3) mice. Arrowheads indicate AGER+ cells in KPT mice that co-stain with SPC. Scale bar, 10 μm. (l) Quantification of SPC-positivity of AGER+ cancer cells (n = 150) in 10-week KFT (n = 3), KT (n = 3), and KPT (n = 3) mice. The percentage of AGER+tdTomato+ cells that co-stained with SPC were quantified. All data are mean ± s.d. P values were calculated by ordinary one-way ANOVA with Tukey’s multiple comparisons test (d, j).
Extended Data Fig. 7 p53 directly promotes AT1 identity in vitro and in vivo.
(a) PDPN quantification as a percentage of p53+ (n = 600) cells from p5353,54 (n = 3) and p53wt (n = 3) LUAD cell lines or as a percent of DAPI+ (n = 600) cells in p53null (n = 3) LUAD cell lines. (b) Representative IF of PDPN and p53 from p5353,54 (n = 3), p53wt (n = 3), and p53null (n = 3) LUAD lines. Scale bar, 20 μm. (c) GSEA shows enrichment of AT1 cell signatures (Supplementary Table 6) in p53wt LUAD cells. NES > 0 indicates the signature is enriched in p53wt cells. *Indicates gene set is significantly enriched (P ≤ 0.05). (d) GSEA shows enrichment of alveolar signatures in specific genotypes. (e) Schematic for in vivo p53 restoration experiments. (f) Representative IF of PDPN and p53 from p5353,54-restored (n = 3) and p53wt-restored (n = 3) mice. Scale bar, 20 μm. (g) PDPN quantification as a percentage of p53+CYTOKERATIN (CK)+ tumors (n = 150) from p5353,54-restored (n = 3) and p53wt-restored (n = 3) mice or as a percent of CK+ tumors (n = 150) from p53null (n = 3) mice. (h) Representative IF of PDPN, p53, and CK in tumors from p5353,54-restored (n = 3), p53wt-restored (n = 3), and p53null (n = 3) mice. Scale bar, 20 μm. (i) GSEA shows p53 restoration induces AT1-associated gene expression profiles (Supplementary Table 6). Gene expression data30 comprised p53null (n = 8) and p53restored (n = 10) lung tumor samples. NES > 0 indicates signature enrichment in p53restored cells. *Indicates gene sets significantly enriched (P ≤ 0.05). (j) Percentage of genes in each AT1 signature that fulfill the stated conditions. The P values indicate the enrichment of p53 bound genes31 in each signature. (k) ChIP-seq31 tracks show p53 binding at AT1-associated genes. All data are mean ± s.d. P values were calculated by ordinary one-way ANOVA with Tukey’s multiple comparisons test (a, g), by GSEA52 (c, d, i), and ChIP enrichment analysis57 (j).
Extended Data Fig. 8 p53 promotes AT1-like identity in human LUAD.
(a) (Left) AT1 marker expression in human (left) TP53WT (n = 249) and TP53MUT (n = 280) LUADs (TCGA), (middle) TP53WT (n = 78) and TP53MUT (n = 38) LUADs (GSE13213), and (right) TP53WT (n = 34) and TP53MUT (n = 73) lung squamous cell carcinomas (LUSC, E-MTAB-8615). Center line, median; boxes, interquartile range (IQR); whiskers, 25% quantile – 1.5*IQR & 75% quantile + 1.5*IQR; points, outliers. (b) Representative IF of AGER, HT1-56, and SPC in (n = 3) normal human lung samples. Scale bar, 50 μm. (c) Representative IF of HT1-56 and CYTOKERATIN (CK, cancer cells) in TP53WT (n = 8) and TP53MUT (n = 5) human LUADs. Scale bar, 5 μm. (d) Percentage of CK+ (n = 1,300) cells positive for HT1-56 in TP53WT (n = 8) and TP53MUT (n = 5) human LUADs. (e) Representative IF of SPC and CK in TP53WT (n = 5) and TP53MUT (n = 5) human LUADs. Scale bar, 5 μm. (f) Percentage of CK+ (n = 1,000) cells positive for SPC in TP53WT (n = 5) and TP53MUT (n = 5) human LUADs. (g) Representative IF of AGER, HT1-56, and CK in TP53WT (n = 8) and TP53MUT (n = 5) human LUADs. Scale bar, 20 μm. (h) Representative IF of CLDN4 and CK in TP53WT (n = 5) and TP53MUT (n = 5) human LUADs. Scale bar, 5 μm. (i) Percentage CK+ (n = 1,000) cells positive for CLDN4+ in TP53WT (n = 5) and TP53MUT (n = 5) human LUADs. (j) Representative IF of AGER and p53 in TP53knockout (KO) A549 cells transfected with an empty vector or a P53 expression vector (n = 3). Scale bar, 20 μm. (k) AGER quantification as a percentage of p53+ (n = 300) cells in (n = 3) A549 lines transfected with P53 or as a percentage of DAPI+ (n = 300) cells in (n = 3) A549 lines transfected with empty vector. All bar graphs are mean ± s.d. P values were calculated by two-way ANOVA test (a, see Methods) and two-tailed Student’s t-test (d, f, i, k).
Extended Data Fig. 9 p53 is activated in transitional and AT1-like cells.
(a) t-distributed stochastic neighbor embedding (tSNE) plot of (n = 3,891) cells29 across all clusters across all genotypes (KT [n = 1,071], KPT [n = 2,614], and normal AT2 [T, n = 206] cells) and all timepoints. (b) t-SNE plots of cancer cells split by both genotype and timepoint. (c) Proportion of cells in each cluster split by genotype and timepoint. (d) Heatmap showing expression of AT2, transitional (cluster 5), and AT1 cell markers (log-transformed TPMs rescaled across all cells) in all cells in (a). (e) (left) t-SNE plot of 2-week KT cells from the scRNA-sequencing dataset29 color-coded by cluster shows the predominance of clusters 1 and 2, which resemble AT2 cells. (Right) Feature plots showing the expression of AT2 markers (Sftpc and Sftpd), p53, and a panel of p53 target genes in 2-week KT cells. (f) Dot plot showing the expression of transitional (cluster 5) and AT1 cell markers in all cells in (a) split by genotype (T, KT, and KPT). (g) Relative proportions of cluster 3 and 5 cells in KT and KPT tumors. (h) Representative IF of CLDN4 and tdTomato in 10-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. Scale bars, 50 μm (top) & 20 μm (bottom). (i) Quantification of CLDN4+ (n = 100 [KFT], 100 [KT], and 200 [KPT]) tumors (see Methods) in 10-week and 24-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. Data are mean ± s.d. (j) t-SNE plots split by genotype shown in Fig. 3a with the cluster 5 sub-clusters color-coded. (k) Relative composition of subclusters 5A, 5B, and 5C split by genotype and timepoint. (l) GSEA shows enrichment of an injury-induced transitional cell signature (Supplementary Table 6) in KPT mice. P values were calculated by GSEA52 (l).
Extended Data Fig. 10 p53 restricts transitional cancer cell formation and promotes their differentiation into AT1-like cells.
(a) Expanded view from Fig. 3g of p19ARF, CLDN4 and tdTomato in tumors from 10-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. Scale bars, 20 μm. (b) Quantification of p19ARF staining in (n = 100 [KFT], 100 [KT], and 200 [KPT]) CLDN4+ cancer cells in 10-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. (c) Expanded view from Fig. 3g of HNF4A and CLDN4 in tumors from 10-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. Scale bars, 20 μm. (d) Quantification of HNF4A staining in (n = 100 [KFT], 100 [KT], and 200 [KPT]) CLDN4+ cancer cells in 10-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. (e) Expanded view from Fig. 3g of phosphorylated-ERK (pERK) and CLDN4 in tumors from 10-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. Scale bars, 20 μm. (f) Quantification of pERK staining in (n = 100 [KFT], 100 [KT], and 200 [KPT]) CLDN4+ cancer cells in 10-week KFT (n = 4), KT (n = 3), and KPT (n = 3) mice. (g) Representative IF of CLDN4, p19ARF, HNF4A, and pERK in tumors from 10-week KPT (n = 3) mice. Scale bar, 20 μm. (h) Copy number variations (CNVs) (red, amplifications; blue, deletions) inferred from the scRNA-sequencing data. (i) (Left) Frequency of CNVs across tumor progression or (right) across clusters. (j) Partition based graph abstraction of indicated clusters. (k) Cell-cell transitions estimated by CellRank based on pseudotime. (l) Scatter plots displaying the expression trend of programs identified in Fig. 3d. (m) Expression of cluster 3 markers29 in cluster 5 cells. Center line is the median with the upper and lower quartiles shown. Bar graphs are mean ± s.d. P values were calculated by two-tailed Pearson correlation test (df = 851) (l) and two-tailed Mann Whitney U test (m).
Extended Data Fig. 11 p53 promotes AT1 differentiation after lung injury.
(a) (Left) Representative IF of AGER and p53 in p53LSL-wt/LSL-wt transitional (TIGIT+)29 LUAD cells 1 and 3 days after Ad-Empty (p53null) or Ad-Cre (p53wt) treatment. Scale bar, 20 μm. (Right) AGER quantification as a percent of DAPI+ (n = 200) p53null cells or p53+ (n = 200) p53wt cells 1 (top) and 3 (bottom) days post-adenoviral treatment. (b) Plot from Fig. 3h depicting an expression score of transitional cell markers12. (c) Transitional cell marker12 induction across cancer clusters. (d) Enriched gene sets in cluster 5 markers29 identified by Enrichr58,59,60. (e) (Left) tdTomato+ (n = 600) cells that are SPC+ in control SpcT (n = 3) and SpcPT (n = 3) mice. (Right) SPC+ (n = 600) cells that are tdTomato+ in the same cohort. (f) Representative IF of SPC and tdTomato in control SpcT (n = 3) and SpcPT (n = 3) mice. Scale bar, 20 μm. (g) Expanded view of Fig. 4c, d of AGER/tdTomato or CLDN4/tdTomato in 4-day BHT-treated SpcFT (n = 3), SpcT (n = 3), and SpcPT (n = 3) mice. Scale bar, 20 μm. (h) Representative IF of BrdU and SPC in 4-day BHT-treated SpcFT (n = 3), SpcT (n = 3), and SpcPT (n = 3) mice. Scale bar, 20 μm. (i) tdTomato+ (n = 600) cells that are (left) AGER+, (left-middle) CLDN4+, or (right-middle) SPC+ in 2-day BHT-treated SpcT (n = 3) and SpcPT (n = 3) mice. (Right) Proliferating (BrdU+) SPC+ (n = 600) cells in the same cohort. (j) tdTomato+ (n = 600) cells that are SPC+ in 4-day BHT-treated SpcFT (n = 3), SpcT (n = 3), and SpcPT (n = 3) mice. (k) Representative IF of SPC and tdTomato in 4-day BHT-treated SpcFT (n = 3), SpcT (n = 3), and SpcPT (n = 3) mice. Scale bar, 20 μm. (l) ChIP-sequencing14 tracks of p53 binding at AT1 genes in transitional cells after injury. Data are mean ± s.d. P values were calculated by two-tailed Student’s t-test (e, i) and ordinary one-way ANOVA with Tukey’s multiple comparisons test (j).
Extended Data Fig. 12 p53 promotes alveolar lineage fidelity during lung injury repair.
(a) Expression of the top 100 markers (by P-adjusted value) for each cluster. (Right) Representative genes. (b) Expression of indicated marker/signatures (Supplementary Table 6) across clusters. (c) UMAP from Fig. 4g colored by pseudotime. (d) UMAP from Fig. 4g colored by CytoTRACE score. (e) p53 pathway signature expression (Supplementary Table 6) across clusters. (f) (Left) UMAP from Fig. 4g split by genotype. (Middle) p53 expression across clusters. (Right) p53 pathway signature expression (Supplementary Table 6). (g) Overlap of p53 and p53 target gene expression from (f). (h) (Top-left) Expression of Dclk1 or (right, bottom) liver and LUAD cluster (cluster 5B/C, 10, 11, or 12) programs (Supplementary Table 6) in SpcT (n = 332) and SpcPT (n = 189) transitional cells from BHT injury scRNA-sequencing data. (i) Top enriched Hallmark gene sets58,59,60 in genes upregulated in SpcPT compared to SpcT transitional cells. (j) CLDN4+tdTomato+ (n = 100) cells that are VIMENTIN-positive in 4-day BHT-treated SpcT (n = 3) and SpcPT (n = 3) mice. (k) Expanded view from Fig. 4k of CLDN4, VIMENTIN, and tdTomato in 4-day BHT-treated SpcT (n = 3) and SpcPT (n = 3) mice. Scale bar, 20 μm. (l) SpcPT transitional cell signature (Supplementary Table 6) expression in scRNA-sequencing data29 from KT (n = 1,071) and KPT (n = 2,614) LUAD cells. (m) Representative IF of CLDN4 and VIMENTIN in tumors from 10-week KT (n = 3) and KPT (n = 3) mice. Scale bar, 5 μm. (n) Expanded view of (m). Scale bar, 20 μm. (o) CLDN4-positive (n = 100) tumor cells that are VIMENTIN-positive in 10-week KT (n = 3) and KPT (n = 3) mice. Bar graphs are mean ± s.d. Box plots: center line, median; box limits, upper/lower quartiles; whiskers, 2.5–97.5 percentile; points, outliers. P values were calculated by two-tailed non-parametric Wilcoxon rank sum test (h, bar graph) and two-tailed Student’s t-test (h [box plots], j, l, o).
Supplementary information
Supplementary Figures
This file contains two supplementary figures. Supplementary Fig. 1 has three panels showing uncropped western blot gels and a legend. Supplementary Fig. 2 has six panels showing FACS plots and a legend.
Supplementary Table 1
Differentially expressed genes between KT cells and KPT cells (sheet 1) and between KFT cells and KPT cells (sheet 2).
Supplementary Table 2
Functional enrichment analysis showing the GO Biological Process terms enriched in genes hyperactivated in KFT cells.
Supplementary Table 3
Functional enrichment analysis showing the GO Biological Process terms enriched in KT upregulated genes.
Supplementary Table 4
Functional enrichment analysis showing the GO Biological Process terms enriched in KPT upregulated genes.
Supplementary Table 5
Differentially accessible regions in KT cells and KPT cells (sheet 1) and KT cells and KFT cells (sheet 2).
Supplementary Table 6
Custom gene signatures used in GSEA and other analyses.
Supplementary Table 7
Human LUAD sample characterizations.
Supplementary Table 8
Genes comprising each signalling programme in cluster 5 (sheets 1–3).
Supplementary Table 9
Lung injury scoring parameters (sheet 1) and histology scoring of SpcT and SpcPT mice (sheet 2).
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kaiser, A.M., Gatto, A., Hanson, K.J. et al. p53 governs an AT1 differentiation programme in lung cancer suppression. Nature 619, 851–859 (2023). https://doi.org/10.1038/s41586-023-06253-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-023-06253-8
- Springer Nature Limited
This article is cited by
-
A novel approach to the analysis of Overall Survival (OS) as response with Progression-Free Interval (PFI) as condition based on the RNA-seq expression data in The Cancer Genome Atlas (TCGA)
BMC Bioinformatics (2024)
-
Tumor initiation and early tumorigenesis: molecular mechanisms and interventional targets
Signal Transduction and Targeted Therapy (2024)
-
An atlas of epithelial cell states and plasticity in lung adenocarcinoma
Nature (2024)
-
AT1 cells appear centre stage
Nature Reviews Cancer (2023)