Abstract
The liver is bathed in bacterial products, including lipopolysaccharide transported from the intestinal portal vasculature, but maintains a state of tolerance that is exploited by persistent pathogens and tumours1,2,3,4. The cellular basis mediating this tolerance, yet allowing a switch to immunity or immunopathology, needs to be better understood for successful immunotherapy of liver diseases. Here we show that a variable proportion of CD8+ T cells compartmentalized in the human liver co-stain for CD14 and other prototypic myeloid membrane proteins and are enriched in close proximity to CD14high myeloid cells in hepatic zone 2. CD14+CD8+ T cells preferentially accumulate within the donor pool in liver allografts, among hepatic virus-specific and tumour-infiltrating responses, and in cirrhotic ascites. CD14+CD8+ T cells exhibit increased turnover, activation and constitutive immunomodulatory features with high homeostatic IL-10 and IL-2 production ex vivo, and enhanced antiviral/anti-tumour effector function after TCR engagement. This CD14+CD8+ T cell profile can be recapitulated by the acquisition of membrane proteins—including the lipopolysaccharide receptor complex—from mononuclear phagocytes, resulting in augmented tumour killing by TCR-redirected T cells in vitro. CD14+CD8+ T cells express integrins and chemokine receptors that favour interactions with the local stroma, which can promote their induction through CXCL12. Lipopolysaccharide can also increase the frequency of CD14+CD8+ T cells in vitro and in vivo, and skew their function towards the production of chemotactic and regenerative cytokines. Thus, bacterial products in the gut–liver axis and tissue stromal factors can tune liver immunity by driving myeloid instruction of CD8+ T cells with immunomodulatory ability.
Similar content being viewed by others
Main
The highly tolerogenic properties of the liver are exemplified by the successful transplantation of this organ across MHC barriers. Homeostatic downregulation of immunity in the liver at steady state is underscored by its predilection to harbour persistent pathogens and tumours, although this can be overridden during resolution of hepatic viral infection, transplant rejection or inflammatory liver diseases. The cellular mechanisms that mediate hepatic immunoregulation, while enabling the switch to immunity or immunopathology, are poorly understood1,2. As the liver receives the majority of its blood supply from the portal vein, it is heavily exposed to bacteria and bacterial products such as lipopolysaccharide (LPS) that are able to translocate the gut and permeate the liver through the extensive fenestrated sinusoidal vasculature2,3,4. The gut microbiome and LPS content can influence the progression of liver diseases, including hepatocellular carcinoma (HCC), but the cellular basis for this connection also remains poorly defined3,5. We recently described a population of tissue-resident memory CD8+ T (CD8+ TRM) cells in the human liver and noted that some express CD146—a component of the LPS receptor that is usually expressed on myeloid cells. Here we investigate the unique features of CD14+CD8+ T cells within the human liver, providing a mechanistic link between the stromal/myeloid network, bacterial LPS and hepatic immunity.
CD14+CD8+ T cells in the human liver
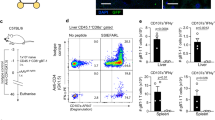
We observed that a variable fraction of CD8+ T cells compartmentalized within human liver tissue stained for CD14, TLR4 and myeloid differentiation factor 2 (MD-2), components of the LPS receptor that are typically found on myeloid cells (Fig. 1a, assessed using a REAfinity Fc-engineered antibody and an FcR blocking step (to exclude non-specific FcR staining) and the gating strategy with doublet exclusion shown in Extended Data Fig. 1a,b). We further excluded the possibility that CD14+CD8+ T cells represented T cell/monocyte doublets7 or atypical myeloid cells by visualizing single cells co-staining for αβT cell receptor (αβTCR), CD8, CD14 and TLR4 using imaging flow cytometry (Fig. 1b) and by confirming that CD14-expressing CD8+ T cells were equivalent in size, morphological appearance and expression of αβTCR, CD45 and CD3ζ chain to conventional intrahepatic CD8+ T cells without CD14 (Fig. 1c and Extended Data Fig. 1c,d). CD14+CD8+ T cells were similarly enriched for other surface receptors that are highly expressed by myeloid cells, including TLR2, CD36 and HLA-DR (Fig. 1d). CD14+CD8+ T cells accounted for a highly variable fraction of intrahepatic CD8+ T cells (median, 8.3%; maximum, 45.7%), whereas they were typically barely detectable in paired peripheral blood samples from the same donors (Fig. 1e,f). CD14 tended to be detectable on a higher proportion of CD8+ T cells than CD4+ T cells or natural killer cells in the same liver samples (Extended Data Fig. 1e). CD14+CD8+ T cells were also found at higher frequencies in healthy human skin and lymphoid organs (spleen and lymph nodes) compared with that in the blood (Fig. 1f).
a, Representative flow cytometry analysis of human peripheral and intrahepatic CD8+ T cells stained with a REAfinity anti-CD14 monoclonal antibody compared with the fluorescence minus one (FMO) control (using the gating strategy in Extended Data Fig. 1a), and assessment of intrahepatic CD14−CD8+ T cell (black outline) and CD14+CD8+ T cell (blue filled) expression (mean fluorescence intensity (MFI)) of TLR4 (n = 39) and MD-2 (n = 10). b, Single-cell images of intrahepatic CD8+ T cells that were co-stained for αβTCR, CD14 and TLR4 (ImageStreamX MKII; ×60; representative of three samples). Scale bars, 7 μm. c, Representative cytospin images of FACS-sorted intrahepatic CD14−CD8+ and CD14+CD8+ T cells (representative of two samples). Scale bars, 10 μm. d, Assessment of intrahepatic CD14−CD8+ T cell and CD14+CD8+ T cell TLR2 (n = 23), CD36 (n = 35) and HLA-DR (n = 22) expression (MFI). e–g, The frequency of CD14+CD8+ T cells in paired blood and non-diseased liver tissue from the same donor (n = 46) (e), and blood (n = 97), explanted/resected liver tissue (n = 106), skin biopsies (n = 6), splenic tissue (n = 7), freshly isolated lymph nodes (n = 9) (f) and explanted/resected liver tissue (n = 106) compared with transplant vascular perfusates (perf.; n = 25) (g). h, CD14+CD8+ T cell longevity in liver allografts explanted after 8 months to 11 years from HLA-haplotype-disparate recipients (n = 5). HLA staining distinguishing donor- or recipient-derived leukocytes in the blood and intrahepatic leukocytes. Plots and summary data of CD14 expression on recipient-derived (donor-HLA−) and donor-derived (donor-HLA+) CD8+ T cells are shown. i,j, Representative plots and summary data (MFI) of CXCR3 (n = 34) and CXCR4 (n = 14) (i) and the percentage of CD49a (integrin α1β1; n = 28) and CD49b (integrin α2 n = 17) on intrahepatic CD14−CD8+ T cells or CD14+CD8+ T cells (j). Each dot (or pair of dots) represents a study participant sample that was processed, stained and analysed independently. Data are mean. For f and g, data are median (centre line) ± interquartile range (IQR) (dotted lines). P values were determined using Kruskal–Wallis tests (analysis of variance (ANOVA)) with Dunn’s multiple-comparisons test (f), two-tailed Wilcoxon t-tests (a, d–e and i–j) and two-tailed Mann–Whitney U-tests (g).
The substantially reduced proportion of CD8+ T cells staining for CD14 in the circulation pointed to this being a feature of tissue residency; consistent with this, liver CD14+CD8+ T cells expressed high levels of the prototypic residency markers CD69 and CD1036 (Extended Data Fig. 1f). To further investigate the propensity for CD14 to preferentially accumulate on CD8+ TRM cells, we examined MHC-mismatched liver allografts, distinguishing donor and recipient populations by their disparate HLA haplotype staining. In this setting, we recently showed that a small pool of donor-derived CD8+ TRM cells can persist for more than a decade8, suggesting that this fraction has specialized adaptations rendering it resistant to removal by pretransplant liver perfusion and able to undergo homeostatic proliferation to maintain its progeny in the long term. Global CD8+ TRM cells (defined by CD69 and CD103 co-expression) are flushed out during liver transplant donor organ perfusion at similar frequencies to those isolated from digested liver tissue, consistent with a predominantly intravascular localization6,9. By contrast, CD14+CD8+ T cells were less abundant in pretransplantation perfusates than in tissue digests (Fig. 1g). Consistent with reduced removal by perfusion, CD14+CD8+ T cells were markedly enriched among the small pool of long-lived donor-derived lymphocytes in HLA-mismatched allografts, ranging from 86% at 8 months to 27–45% around 11 years after transplantation (Fig. 1h and Extended Data Fig. 1g,h). By contrast, recipient T cells, which can increase expression of CD69 but not CD103 or CXCR3 after infiltrating liver allografts8, were not able to acquire substantial CD14 expression, even after 11 years (Fig. 1h and Extended Data Fig. 1h). These data indicate that CD14 expression was preferentially induced on a well-tethered proliferative CXCR3high tissue-resident population but could not be imprinted on HLA-mismatched peripheral CD8+ T cells infiltrating the liver vasculature.
To probe a role for chemokine/stromal-dependent signals, we examined whether CD14-staining CD8+ T cells had differential expression of prototypic liver-homing chemokine receptors and integrins. CD14+CD8+ T cells freshly isolated from liver tissue were selectively enriched for expression of CXCR3 (promoting CXCL10 signals from liver sinusoidal endothelial cells and hepatocytes10) and CXCR4 (favouring binding to CXCL12 (also known as stromal-cell-derived factor 1) produced by endothelial and stromal cells11,12) (Fig. 1i). CD14+CD8+ T cells also expressed markedly more of the extracellular matrix (ECM)-binding integrins CD49a and CD49b (Fig. 1j). Taken together, the allograft and homing profile of CD14+CD8+ T cells suggested that they were selectively tethered within the liver stromal network, dictating the local cues that they receive.
CD14 marks myeloid-instructed CD8+ T cells
Next, we tested whether hepatic stromal cells (activated stellate cells/myofibroblasts) that produce ECM proteins and CXCL12 (relevant ligands for CD49a/b and CXCR4, respectively)11 could have a role in instructing CD8+ T cells to express CD14. Primary hepatic stellate cells (pHSCs) were isolated from resected liver tissue from three donors and differentiated into activated myofibroblasts, able to release CXCL12 (Fig. 2a). Transdifferentiated pHSCs promoted CD8+ T cells that co-stained for CD14 within peripheral blood mononuclear cell (PBMC) co-cultures in a time-dependent manner, accompanied by an increase in the expression of TLR4 and TLR2 (Fig. 2b and Extended Data Fig. 2a–c). Notably, only a proportion of PBMCs acquired CD14 in vitro, with the mean and range approximately mimicking that seen ex vivo in human liver (Fig. 2b and Extended Data Fig. 2b). Stellate cell induction of CD14 on CD8+ T cells was significantly reduced by blocking the CXCL12 receptor CXCR4 (Fig. 2c). The role of CXCL12 was confirmed by its ability to recapitulate the increase in CD14 expression on CD8+ T cells in the absence of stellate cells—an effect that was not seen after the addition of other candidate prototypic liver-derived cytokines/chemokines (Fig. 2d). Culture in three-dimensional decellularized liver scaffolds13 similarly induced CD14+CD8+ T cells from PBMCs (Extended Data Fig. 2d), implicating a role for the ECM, which is known to contain TLR4-binding proteins and retain chemokines such as CXCL1214.
a, pHSC production of CXCL12 (pg ml−1; n = 3) after culture for 4 days with increasing cell numbers. b,c, The percentage of CD14 staining on CD8+ T cells over time after co-culture of PBMCs (n = 4 donors) with or without pHSCs (derived from n = 3 donor livers) (b) and after co-culture for 4 days with or without anti-CXCR4 antibodies or isotype-matched control (n = 11) (c). For b, data are mean ± s.e.m. d, The fold change in the percentage of CD14+CD8+ T cells in PBMCs supplemented with recombinant IL-6, IL-8, IL-15, TGFβ, CXCL9, CXCL10, CXCL11, CXCL12 or CXCL16 (n = 7, 7, 8, 16, 12, 14, 12, 24 and 8, respectively) compared with medium alone. e, The percentage of CD14+CD8+ T cells after co-culture of PBMCs (n = 27) or magnetic-bead-isolated CD8+ T cells (n = 27) with pHSCs for 4 days. f–k, The percentage of CD14+CD8+ T cells after co-culture of purified CD8+ T cells with or without freshly isolated autologous MNPs (1:1 ratio; n = 27) (f), soluble CD14 (sCD14; n = 12) (g), MNPs with or without separation by a 0.4 µm Transwell insert (n = 8) (h), after MNP fixation with 4% paraformaldehyde (n = 8) (i), MNP with or without latrunculin B (n = 10) (j) and the MNP effect on the fold change in the percentage of CD14+CD8+ T cells after treatment with anti-ICAM-1 antibodies or isotype-matched control (n = 15) (k). l, Representative single-cell images of biotin-labelled autologous MNPs cultured for 4 days alone (bottom 2 rows) or with magnetic-bead-isolated peripheral CD8+ T cells (top 4 rows), all co-stained with monoclonal antibodies against αβTCR, CD3, CD8, CD14 and Streptavidin (Strep.; ImageStreamX MKII; ×60, representative of three samples). Scale bars, 10 μm. m, Expression (MFI) of MD-2, TLR4, HLA-DR and TLR2 (n = 6, 6, 4 and 8, respectively) on peripheral T cells with or without staining for CD14 and Streptavidin after exposure to biotin-labelled MNPs. n, Representative imaging mass cytometry analysis of macroscopically healthy liver sections (representative of three samples), co-stained for CD8 (red), CYP1A2 (purple), CD14 (green), HLA-DR (cyan) and DNA (blue). Scale bars, 200 µm (large image) and 50 µm (magnified images). o, Summary data of the distribution of CD14−CD8+ T cells and CD14+CD8+ T cells across liver zones (defined by expression of CYP1A2 intensity; Extended Data Fig. 2q). p, UMAP analysis of 72,000 single cells clustered by expression profile (left) and scaled by CD14 intensity, with magnification of the CD8+ T cell cluster (right). LSEC, liver sinusoidal endothelial cells. q, Summary data of the distance of CD14−CD8+ T cells and CD14+CD8+ T cells to the nearest Iba1+ cell. Each dot represents a PBMC or CD8+ T cell/MNP donor sample that was processed, stained and analysed independently. Data are mean. For a,c–j,o and q, data are median (centre line) ± IQR (dotted lines). P values were determined using Kruskal–Wallis tests (ANOVA) with Dunn’s multiple-comparisons test compared with the medium alone (d) or compared with the MNP + T cell condition (g–j), and two-tailed Wilcoxon t-tests (c,e–f,k,m,o and q).
In vitro induction of CD14+CD8+ T cells was abrogated when pHSCs were co-cultured with isolated CD8+ T cells rather than PBMCs (Fig. 2e). This raised the possibility that CD14 on CD8+ T cells in the liver could be transferred from other cell types such as mononuclear phagocytes (MNPs), which were recently postulated to deposit CD14+ ‘particles’ onto T cells after in vivo conjugate formation7. Consistent with this, the addition of purified monocytes to T cell/stellate cell co-cultures recapitulated, in a dose-dependent manner, the expansion of CD14+CD8+ T cells observed with whole PBMCs, whereas the addition of soluble CD14 (at physiological concentrations) did not (Fig. 2f,g and Extended Data Fig. 2e). In support of CD14+CD8+ T cells predominantly acquiring CD14 protein rather than transcribing it, examining sorted intrahepatic CD14+CD8+ T cells (Extended Data Fig. 2f (gating strategy)) revealed that CD14 mRNA was at the threshold of detection by quantitative PCR with reverse transcription (RT–qPCR) (Extended Data Fig. 2g), whereas single-cell RNA sequencing (scRNA-seq) identified classical CD8+ T cell transcripts (Extended Data Fig. 2h) but no detectable CD14 or overlap with the transcriptional profile of MNPs sorted from the same livers (Extended Data Fig. 2h,i).
We investigated the mechanism by which CD8+ T cells acquire prototypic myeloid markers using autologous purified T cells and MNPs co-cultured in contact with pHSCs. Separation of T cells and MNPs by Transwell membranes abrogated the ability of MNPs to transfer CD14 to CD8+ T cells (Fig. 2h), indicating a contact-dependent mechanism. Active transfer was supported by prior fixation of MNPs preventing CD14 acquisition by CD8+ T cells (Fig. 2i). Addition of latrunculin B to block the actin polymerization required for immune synapse formation reduced both the percentage and intensity of CD14 staining on CD8+ T cells, whereas blockade of microtubule polymerization (involved in lytic granule transport) using nocodazole did not (Fig. 2j and Extended Data Fig. 2j,k). LFA-1 is a key feature of liver TRM cells, enabling their patrolling function15 and the LFA-1–ICAM-1 axis is central to T cell synapse formation; ICAM-1 blockade also decreased CD14 acquisition compared with the isotype control (Fig. 2k).
To more closely mimic the myeloid landscape of the liver, we co-cultured CD8+ T cells with monocyte-derived macrophages (MDMs) as hepatic macrophages (Kupffer cells) have a central role in dictating the function of liver T cells16,17. MDMs, like circulating monocytes, were able to transfer CD14 to CD8+ T cells, on the basis of flow cytometry analysis, with the acquisition of punctate CD14 staining on individual CD8+ T cells (co-cultured with MDMs) confirmed by confocal imaging (Extended Data Fig. 2l,m). The previous Transwell data argued against a role for exosomes (as these are 30–100 nM diameter, small enough to pass through the 400 nM pore inserts; Fig. 2h). Exosome transfer was further excluded by specifically purifying CD14-containing exosomes from the supernatant of MDMs derived from three donors (Extended Data Fig. 2n (left)) and showing that these were unable to recapitulate the transfer of CD14 to CD8+ T cells achieved by direct contact with their MDMs (Extended Data Fig. 2n (right)). To further examine the process by which CD14 was acquired by CD8+ T cells, we incorporated a biotin label into the plasma membrane of the myeloid cells before co-culture. Imaging cytometry confirmed the capture of CD14 from the biotin-labelled membrane of the myeloid cells by CD8+ T cells, with fluorescently labelled Streptavidin co-staining CD8+ T cells that had acquired CD14 and not those lacking CD14 (Fig. 2l). Flow cytometry phenotyping of CD8+ T cells labelled with both CD14 and Streptavidin revealed that CD8+ T cells had acquired the entire CD14–TLR4–MD-2 complex, along with additional prototypic MNP markers HLA-DR and TLR2 from the biotin-labelled myeloid cell membrane (Fig. 2m and Extended Data Fig. 2o).
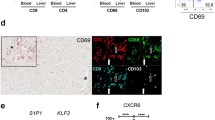
To examine the topological relationship between CD14+CD8+ T cells and MNPs in vivo, we visualized them in macroscopically healthy human liver sections using imaging mass cytometry. Images were segmented for single-cell analysis and cell clustering analysis was performed (two regions of interest per liver from three donors; Fig. 2n); quantitative high-dimensional spatial profiling of liver sections identified a proportion of intrahepatic CD8+ T cells co-expressing CD14 (mean, 20%; Extended Data Fig. 2p), a frequency consistent with the range that we had quantified using flow cytometry. Whereas CD14−CD8+ T cells tended to accumulate in the periportal region (zone 1), CD14+CD8+ T cells had a significantly different liver zonation index, being relatively enriched in the midlobular zone 2, the region in which hepatocytes maintain liver homeostasis18 (Fig. 2o and Extended Data Fig. 2q). Using uniform manifold approximation and projection (UMAP), we visualized a cluster with increased CD14 expression within CD8+ T cells as well as a cluster of CD14highHLA-DRhigh MNPs (Fig. 2p). Importantly, CD14+CD8+ T cells were observed in close proximity to CD14highHLA-DRhigh MNPs (Fig. 2n), with quantitative analysis confirming that they were significantly closer to the nearest MNPs compared with the distance between CD14−CD8+ T cells and MNPs (Fig. 2q).
Taken together, these data suggest that the population of CD14+CD8+ T cells identified ex vivo from human liver, with a distinct zonal distribution, represented the fraction that had acquired cellular material from CD14high myeloid cells in a contact-dependent manner that was facilitated by CXCL12/stromal interactions.
Functional profile of CD14+CD8+ T cells
We postulated that liver CD8+ TRM cells distinguished by the acquisition of CD14 would have distinct functional features instructed by myeloid cells. Hepatic myeloid cells can shape the tolerance and/or activation of neighbouring T cells through local cytokine release or transpresentation, exchange of metabolites, antigen presentation and co-stimulation16,19,20,21,22,23. A gain-of-function screen recently identified several myeloid-restricted genes as the most potent at remodelling T cells to promote their activation and effector function24. We therefore investigated whether the fraction of human liver CD8+ T cells staining for CD14 had a distinct activation status and functional potential compared with their CD14−CD8+ T cell counterparts directly ex vivo. A higher proportion of human liver CD14+CD8+ T cells expressed the chronic activation marker CD57 and the proliferation marker Ki-67 compared with the remaining intrahepatic CD8+ T cells (Fig. 3a), in contrast to prototypical CD8+ TRM cells, which have a low turnover6,25. Ex vivo human liver CD14+CD8+ T cells constituted a highly activated population, with significantly increased CD38 and a marked upregulation of CD25 but an equivalent percentage of PD-1 expression compared with CD14−CD8+ T cells (Fig. 3b). Congruent with the increased nutrient uptake required to maintain a highly activated state, CD14+CD8+ T cells had a consistent increase in system L-amino acid transporters marked by CD98, and in the transferrin receptor CD71 (Extended Data Fig. 3a). Other cardinal features of highly activated/immunomodulatory T cells that segregated exclusively with the CD14+ fraction of CD8+ T cells were FOXP3, CTLA-4, and co-expression of the ectonucleotidases CD39 and CD73, whereas expression of LAG-3, GITR and ICOS was also increased on CD14+CD8+ T cells (Extended Data Fig. 3b,c). Intrahepatic CD14+CD8+ T cells were potent constitutive producers of the immunoregulatory cytokine IL-10 directly ex vivo (without stimulation; mean, 23%; maximum, 67%; Fig. 3c (left)). Cell-autonomous IL-10 has been shown to protect CD8+ T cells from excessive stimulation26, thereby promoting their function against chronic hepatotropic viral infections27. Similarly, abundant CD8+ T cell production of IL-2, which can counteract PD-1-mediated tolerance in the liver6,28, was seen in the CD14-expressing fraction without the need for ex vivo stimulation, frequently co-expressed with IL-10 (Fig. 3c (right) and 3d). This combination of functional features characterized CD14+CD8+ T cells as a constitutively proliferative, activated population with an immunomodulatory cytokine profile associated with maintenance of effector function in the liver.
a,b, Representative flow cytometry plots and summary data showing the percentage or MFI of CD57 (n = 23) and Ki-67 (n = 34) (a) and CD25 (n = 13), CD38 (n = 34) and PD-1 (n = 36) (b) on intrahepatic CD14−CD8+ T cells (black outline) or CD14+CD8+ T cells (blue filled) ex vivo. c,d, Representative flow cytometry plots and summary data showing the percentage of intracellular staining for IL-10 (n = 19) and IL-2 (n = 21), by CD14−CD8+ T cells or CD14+CD8+ T cells ex vivo (without exogenous stimulation or brefeldin A) (c), and co-production of IL-10 by IL-2-positive or IL-2-negative CD14+CD8+ T cells (n = 16) (d). e, Assessment of CD14+CD8+ T cell intracellular cytokine production with or without (unstim.) stimulation for 4 h with anti-CD3/CD28 antibodies in the presence of brefeldin A. n = 19 (IFNγ), n = 15 (TNF), n = 12 (MIP1β) and n = 5 (CD107a). f, Intrahepatic HBV-specific CD8+ T cells were identified using ex vivo staining with a panel of HLA-A2+ dextramers loaded with immunodominant peptides, gated using a control HLA-A2+ dextramer loaded with an irrelevant peptide (ctrl/dex). Example plots and summary data show CD14 expression on the global (HBV dex-negative) or HBV-specific (n = 11) CD8+ T cells. g, The frequency of CD14+CD8+ T cells in HCC TILs in comparison to paired non-cancerous tissue (margin) from the same study participant (n = 16). h,i, The functionality of FACS-purified TCR-redirected HBVenv183-specific CD14±CD8+ T cells against HepG2-preS1 presenting endogenously processed peptide at an effector:target ratio of 1:2.5 for 16 h by intracellular cytokine staining analysis of IFNγ and TNF production (n = 7) and CD107a mobilization (n = 6) (h) and specific target cell lysis (n = 9) (i). Each dot (or pair of dots) represents a study participant sample that was processed, stained and analysed independently. Data are mean. For e and h–i, data are median (centre line) ± IQR (dotted lines). P values were determined using Kruskal–Wallis tests (ANOVA) with Dunn’s multiple-comparisons test (e) and two-tailed Wilcoxon t-tests (a–d and f–i).
After short-term (4 h) TCR engagement, CD14+CD8+ T cells maintained their high constitutive production of IL-10 and IL-2 (Extended Data Fig. 3d), while simultaneously further increasing their high steady-state production of the effectors IFNγ, TNF and MIP1β, and their capacity for cytotoxic degranulation (CD107a), all of which are important anti-pathogen and anti-tumour functions (Fig. 3e). This was not simply a generic feature of tissue residence, as the liver CD8+ TRM cell fraction expressing CD14 was more functional than the remainder of CD8+ TRM cells lacking CD14 (Extended Data Fig. 3e). Thus, CD14 staining demarcates a population of human liver CD8+ TRM cells that are enriched for high constitutive production of immunoregulatory cytokines and the ability to also produce proinflammatory effectors immediately after TCR engagement. The disease relevance of the heightened immunomodulatory potential of CD14+CD8+ T cells was underscored by the finding that hepatitis B virus (HBV)-specific responses (identified by HLA/peptide multimer staining of intrahepatic leukocytes (IHLs) extracted from HBV-infected livers) were more than twofold enriched for the expression of CD14 compared with the residual global CD8+ T cells (Fig. 3f). CD14+CD8+ T cells were also around threefold enriched among tumour-infiltrating leukocytes (TILs) that were freshly isolated from resected HCC compared with paired unaffected liver margins (Fig. 3g), consistent with this tumour type being rich in the stromal factors (ECM proteins and CXCL1211) that we had found promoted CD14 acquisition by CD8+ T cells.
We next examined whether CD8+ T cells interacting with MNPs in vitro, marked by the acquisition of CD14, could recapitulate the enhanced functionality that we observed in this fraction of liver CD8+ T cells when sampled directly ex vivo. CD8+ T cells that acquired CD14 after in vitro culture with pHSCs and myeloid cells had enhanced effector function compared with the fraction that did not co-stain for CD14, with increased IL-2, IFNγ, TNF and CD107a on the basis of flow cytometry (Extended Data Fig. 3f (top)) and increased IL-2, IFNγ, TNF, IL-10, granzyme B on the basis of Luminex profiling of sorted CD14+CD8+ T cells that were stimulated with anti-CD3/CD28 antibodies (Extended Data Fig. 3f (bottom)). To test whether MNP instruction, marked by CD14 acquisition, has therapeutic potential for antigen-specific T cells, we used TCR-gene-transduced T cells specific for an HBV tumour antigen that are being trialled for immunotherapy treatment of HCC29. T cells genetically engineered with a TCR specific for the HBVenv183/HLA-A*02 epitope were expanded in the presence of MNPs and stellate cells. The CD14-expressing fraction of TCR-transduced CD8+ T cells induced by this in vitro protocol (Extended Data Fig. 3g) had the hyperfunctionality after TCR engagement that we had previously observed ex vivo; significantly more of the CD14+ TCR-gene-transduced CD8+ T cells produced the anti-viral/anti-tumour effectors IFNγ and TNF and degranulated within 16 h of stimulation with an HBsAg-expressing hepatoma cell line compared with their CD14-negative counterparts (Fig. 3h). Moreover, specific lysis of the HBsAg-expressing hepatoma cell line by HBVenv183/HLA-A2 TCR-redirected T cells was enhanced by those that had acquired CD14 after addition of MNPs (Fig. 3i). These data suggest that CD14 expression marks a fraction of ‘super-responder’ CD8+ T cells that have undergone MNP reprogramming, both in situ in human liver and in vitro in TCR-gene-engineered populations.
Bacterial LPS shapes CD14+CD8+ T cells
We postulated that the substantial exposure to bacterial wall LPS and shed LPS in the liver may promote and/or differentially skew MNP interactions with locally resident T cells to increase CD14+CD8+ T cells and/or alter their functional profile. Previous reports have shown that binding of exogenous CD14 can confer LPS responsiveness on cells lacking endogenous expression30 as well as suggesting that human CD8+ T cells can upregulate sufficient functional TLR4 to respond directly to LPS after anti-CD3 activation in vitro31. First, we showed that fluorescently labelled LPS preferentially bound to the CD14-expressing fraction of ex vivo liver CD8+ T cells and to stromal-cell-induced CD14+CD8+ T cells, with a stepwise increase in binding over time (Fig. 4a and Extended Data Fig. 4a). Stimulation of fluorescence-activated cell sorting (FACS)-sorted CD14+CD8+ T cells with ultraviolet-killed Escherichia coli (as a source of LPS and other bacterial pathogen-associated molecular patterns sensed by CD1432,33) produced a distinct profile of soluble mediators compared with that seen at rest or after TCR engagement (Fig. 4b and Extended Data Fig. 4b). The CD14+CD8+ T cell response to E. coli was dominated by the production of innate-like hepatoprotective cytokines (IL-6, IL-33)34,35 as well as chemokines driving neutrophil recruitment, neutrophil recruitment of T cells36 and wound healing, including CXCL8 (IL-8) (Fig. 4b), which was previously noted to be produced by a fraction of HBV-specific CD8+ T cells37. These data raised the possibility that CD14+CD8+ T cells acquire some direct responsiveness to LPS after CD14 acquisition as described for other cell types previously30. In vivo, the dominant response of CD14+CD8+ T cells to LPS is probably also influenced by the activation of interacting MNPs. In the presence of MNPs, the addition of ultraviolet-killed E. coli again induced CD14+CD8+ T cell production of CXCL8, CXCL1 (Gro-α) and CCL2, which were already increased in CD14+ compared with CD14−CD8+ T cells (Fig. 4c). Thus CD14+CD8+ T cells, with or without addition of MNPs and using two different readouts, had a propensity to produce profibrogenic/wound-healing chemokines that are able to drive an influx of innate responders such as neutrophils, and this property was further enhanced by re-exposure to bacterial LPS.
a, The MFI of LPS–AlexaFluor488 bound to CD14−CD8+ T cells or CD14+CD8+ T cells (n = 5) after addition for 30 min, 60 min or 120 min compared with medium alone for 120 min. b, Heat map showing the expression profiles of soluble mediators produced by FACS-sorted pHSC-induced CD14+CD8+ T cells after no stimulation (purple) or overnight stimulation with anti-CD3/CD28 antibodies (orange) and 0.3 × 106 ultraviolet (UV)-killed E. coli (green) using unbiased hierarchical clustering. The sample similarity (left dendrogram) and mediator similarity based on average-linkage using Euclidian distance (top dendrogram) are shown. c, pHSC- and MNP-induced CD14−CD8+ T cell and CD14+CD8+ T cell functionality on the basis of intracellular cytokine staining analysis of CXCL8 (n = 12), CXCL1 (n = 15) or CCL2 (n = 15) with or without exposure for 16 h to ultraviolet-killed E. coli in the presence of brefeldin A. d, The fold increase in CD14 on intrahepatic CD8+ T cells after 24 h (n = 13), 48 h (n = 7), 72 h (n = 12) or 96 h (n = 6) in vitro culture with 0.3 × 106 ultraviolet-killed E. coli compared with medium alone. e, Representative plots and summary data showing the frequency of CD14+CD8+ T cells isolated from paired peripheral blood and ascitic fluid (n = 11). f, The frequency of CD14+CD8+ T cells in the ascitic fluid of a study participant with cirrhosis with spontaneous bacterial peritonitis (SBP) before antibiotics (baseline) and after antibiotics (after ABX). g, Schematic of the human forearm skin blister model for in vivo exposure to ultraviolet-killed E. coli. h, The frequency of CD14+CD8+ T cells in the blood or skin blister aspirate (grey filled) with (green dots) or without (black dots) intradermal injection of 1.5 × 107 ultraviolet-killed E. coli 24 h before (n = 8). i, Expression of HLA-DR on CD14−CD8+ T cells or CD14+CD8+ T cells from blister exudates with or without previous intradermal injection of ultraviolet-killed E. coli (n = 7). Each dot (or pair of dots) represents a study participant sample, which was processed, stained and analysed independently. Data are mean (e), mean ± s.e.m. (h and i) and median (centre line) ± IQR (dotted lines) (a,c and d). P values were determined using Kruskal–Wallis tests (ANOVA) with Dunn’s multiple-comparisons test (c,h and i) and Wilcoxon t-tests (e).
Finally, we examined whether exposure to bacterial LPS could activate MNPs and promote membrane transfer to T cells to partially account for the increased frequencies of CD14+CD8+ T cells in tissues like the liver. Exposure of IHLs to ultraviolet-killed E. coli augmented CD8+ T cell staining for CD14 over time (Fig. 4d and Extended Data Fig. 4c). In the presence of ultraviolet-killed E. coli, proliferating CD14+CD8+ T cells preferentially acquired new CD14 expression (distinguished by staining with a different CD14 antibody before and after CFSE division during culture) (Extended Data Fig. 4d,e). Moreover, LPS preferentially enhanced CD49b and CXCR4 expression on CD14+CD8+ T cells (Extended Data Fig. 4f), thereby rendering them more sensitive to stromal signals.
The peritoneal cavity is another body site that is rich in ECM and CXCL12-producing mesothelial cells38,39 and is heavily exposed to Gram-negative bacteria under the surveillance of liver capsular macrophages40. Consistent with these features, CD14+CD8+ T cells (co-expressing TLR4 and TLR2; Extended Data Fig. 4g) were markedly increased in the ascites of patients with cirrhosis or non-cirrhotic portal hypertension compared with that in their blood, accounting for up to 57% (mean, 25%) of all ascitic CD8+ T cells (Fig. 4e). Notably, the proportion of CD14+CD8+ T cells within the ascitic fluid of a patient with cirrhosis with the common complication of spontaneous bacterial peritonitis halved on repeat sampling after a course of antibiotics (Fig. 4f), suggesting that reduced LPS activation of MNPs was reflected in decreased CD8+ T cell acquisition of CD14.
To more definitively test the postulate that LPS activation could drive CD8+ T cell acquisition of CD14, we turned to a human challenge model. We examined the effect of E. coli in vivo using a human skin suction blister challenge model41, in which we found that CD14+CD8+ T cells were detectable at a similar frequency to skin (Extended Data Fig. 4h). Seven healthy volunteers had suction blisters raised on both forearms, after intradermal injection of ultraviolet-killed E. coli 24 h previously in one arm but not the contralateral arm (Fig. 4g). CD14+CD8+ T cells increased within the cellular exudate aspirated from the blister raised 24 h after injection with ultraviolet-killed E. coli compared with the control blister, and remained barely detectable in paired blood samples, indicating a localized in vivo response (Fig. 4h and Extended Data Fig. 4i). The bacterial-induced increase in CD8+ T cell CD14 was accompanied by an increase in HLA-DR, TLR4 and CD38 (Fig. 4i and Extended Data Fig. 4j,k). Thus in vivo tissue-localized introduction of E. coli was able to induce the acquisition of MNP-derived molecules by CD8+ T cells, thereby reshaping local T cell immunity at the site of bacterial exposure.
Discussion
Memory T cells undergo extensive reprogramming after acquiring organ residence, but the local tissue components that can drive these distinct features are poorly understood. Examining CD8+ T cells directly isolated from human liver tissue, we found that those cells distinguished by co-staining for CD14, and other prototypic myeloid markers, have increased turnover, stromal homing potential and immunomodulatory effector function. CD14+CD8+ T cells have high constitutive production of the immunoprotective cytokine IL-10, mount rapid and potent effector function after TCR engagement, and switch to chemotactic/hepatoprotective function after LPS exposure (Extended Data Fig. 4l). This profile can be recapitulated by CD8+ T cell ICAM-1/actin-dependent acquisition of membrane proteins, including the LPS receptor from MNPs, consistent with the closer proximity of CD14+CD8+ T cells to MNPs in situ in human liver. Our data therefore suggest that MNP instruction drives the increased turnover and hyperfunctionality of tissue CD8+ TRM cells marked by CD14; this concept is supported by a recent study showing highly efficient remodelling of T cell proliferation and effector function by overexpression of membrane receptors such as lymphotoxin β receptor (LTBR) that are usually restricted to myeloid cells24. MNPs can also reshape T cell functionality through cell–cell transfer of specific metabolites19. Furthermore, a subset of Kupffer cells (KC2) has recently been defined in mouse and human liver that can be activated by IL-2 to cross-present antigen to convert tolerized into functional CD8+ T cells; notably, the latter are distributed more widely within the liver than the periportal-localized CD8+ T cells primed by hepatocytes, reminiscent of the zonal differences that we observed for CD14+ versus CD14−CD8+ T cells16,17. Thus, further mechanistic studies are needed to investigate whether MNP-derived cytokines, receptors like LTBR, metabolites and/or antigen presentation contribute to the distinct topology and functional profile of hepatic CD8+ T cells marked by CD14. The accumulation of CD14+CD8+ T cells in zone 2 of the liver suggests that interactions with CD14high myeloid cells are favoured in this midlobular region. It remains to be clarified what downstream signalling changes occur in tissue CD8 T cells that have been reprogrammed by myeloid cells to account for their altered constitutive, TCR- and LPS-dependent functions.
Tissue stroma and chemokines in the ECM are increasingly recognized to have a fundamental role in dictating organ-specific immune responses42,43,44,45. Consistent with this, we find that CD14+CD8+ T cells express receptors for ECM and CXCL12—stromal-derived factors that we demonstrate can promote CD8+ T cell acquisition of CD14. Hepatic stromal (stellate) cells are known to have an essential role in imprinting Kupffer cell identity on MNPs46 and coordinating their response to LPS47. We now demonstrate that stellate cells also collaborate with myeloid cells to render liver CD8+ TRM cells responsive to LPS. LPS, and stromal factors such as CXCL12, are likely to predominantly expand CD14+CD8+ T cells through increased MNP activation promoting the transfer of the LPS receptor complex to T cells. We show that inactivated E. coli further increase the capacity of CD8+ T cells for the stromal interactions that we found support their CD14 acquisition, congruent with a previous report showing that LPS drives T cell adherence to fibronectin48. Our data reveal that E. coli exposure increases CD14+CD8+ T cell frequencies in vitro and in vivo in a skin blister model, probably contributing to their preferential accumulation at sites that are rich in bacterial LPS such as the liver and cirrhotic ascites. The LPS-induced myeloid collaboration with CD8+ T cells provides a means by which bacterial products can mould a highly immunomodulatory fraction of hepatic CD8+ TRM cells, including virus-specific responses. The recognition that LPS can also skew CD14+CD8+ T cells towards rapid production of chemotactic cytokines points to an unappreciated early role for tissue T cells in inflammation and hepatoprotective responses triggered by Gram-negative bacteria or LPS that breach the initial firewall formed by periportal MNPs to reach the midlobular zone responsible for hepatocyte homeostasis18,45.
Our data imply that antibiotics or other drugs that decrease the amount of gut bacterial LPS in the portal circulation will reduce CD14+CD8+ T cells, while TLR4-based adjuvants such as monophosphoryl-lipid A or stromal factors such as CXCL12 could be targeted to the liver to promote myeloid reprogramming. The recognition that MNPs can reprogramme T cell effector function can be exploited therapeutically; for example, we show that myeloid co-cultures can be used to generate TCR gene-edited T cells with superior immunotherapeutic potential for adoptive cell therapy. Further studies are needed to explore the immunomodulatory potential of CD14+CD8+ T cells preferentially accumulating among the long-lived donor T cells surviving in HLA-mismatched allografts and within HCC TILs and whether they become tolerant to LPS, as described for liver MNPs after repetitive high-dose exposure49. Notably, studies of human CD8+ TRM cells typically gate out CD14-expressing cells, precluding previous analysis of this population. The finding that CD14+CD8+ T cells also accumulate in cirrhotic ascites suggests that they may have a role in monitoring bacterial invasion of the peritoneal cavity and underscores the need to study them in other human barrier organs that are heavily exposed to LPS. Our study exemplifies the importance of sampling and monitoring human TRM cells to uncover the tissue-imposed adaptations that enable them to respond to local microenvironmental cues.
Methods
Study samples, ethics and inclusion
Each study participant provided written informed consent before inclusion. The storage of any samples/clinical information conformed to the requirements of the Declaration of Helsinki, the Data Protection Act 1998 and the Human Tissue Act 2004.
Liver samples were obtained from non-diseased distal margins of colorectal metastases or margins of HCC surgical resections (on the background of chronic HBV/HCV infection, non-alcoholic steatohepatitis or alcoholic liver disease), explants obtained after solid-organ transplantation or perfusion fluid (perfusates) obtained from the donor livers used for solid-organ transplantation. Spleen and lymph node samples were obtained from the NHSBT organ donation pathway. The majority of tissue and blood samples were obtained through the Tissue Access for Patient Benefit Initiative (TAPb) at The Royal Free Hospital (approved by the UCL-Royal Free Hospital BioBank Ethical Review Committee references: 11/WA/0077, 16/WA/0289 and 21/WA/0388); healthy donor blood samples were covered by REC 11//LO/0421 (approved by South East Cost Brighton and Sussex Ethics Committee). Further liver tissue samples were obtained in Freiburg for imaging mass cytometry (approved by the ethics committee of the Albert-Ludwigs-University; Freiburg reference: 21-1372) and from The Royal London Hospital (approved by the London Brent & The City Research Ethics Committee references: 16/LO/1699 or the London Bridge Research Ethics Committee reference 17/LO/0266). Skin experiments were taken under UCL Research Ethics Committee project numbers 87878 for biopsies and 1309/005 for skin blister/E. coli challenge studies. Ascitic fluid was obtained from study participants with cirrhosis or IVC obstruction requiring an ascitic tap at either the Royal Free Hospital (approved by the UCL Research Ethics Committee reference: 15/LO/0800), the Hospital Clínic de Barcelona (approved by Ethics Committee Hopsital Clínic de Barcelona, committee reference: 2019-0267) or the Royal London Hospital (approved by the London Bridge Research Ethics Committee reference 17/LO/0266), assessed for spontaneous bacterial peritonitis and, in one case, treated with norfloxacin before resampling. HIV seropositive donors were excluded from the study but there were no other exclusions; all available samples with sufficient viable cell yields for the experimental protocol were included. No statistical method was used to predetermine sample size. In addition, no sample randomization was used in this study; all participants were pseudo-anonymized and sampled ad hoc.
Human cell isolation
IHLs were isolated from resected/explanted liver material as previously described50. In brief, tissue was cut up and incubated at 37 °C for 30 min in HBSS+/+ (Life Technologies; Thermo Fisher Scientific) containing collagenase IV (Thermo Fisher Scientific) and DNase I (Roche; Sigma-Aldrich; Merck), before mechanical disruption using the GentleMACS (Miltenyi Biotech) dissociator, with debris removed by filtration through a 70 µm filter (Greiner). Parenchymal cells were removed by centrifugation on a 30% Percoll gradient (GE Healthcare; VWR) followed by further leukocyte isolation by density centrifugation using a Pancoll gradient (PAN Biotech). IHLs were isolated from perfusion liquid (perfusates) or small core-biopsy samples as previously described51. In brief, the perfusate fluid was first concentrated by centrifugation. Concentrated cells were then resuspended in RPMI-1640 (Life Technologies; Thermo Fisher Scientific) and isolated by density centrifugation on a Pancoll gradient.
For the isolation of human leukocytes from the skin, spleen and lymph nodes, tissue was first cut into small pieces and washed with an EDTA-containing buffer (2 mM EDTA (Sigma-Aldrich; Merck) + 0.5% heat inactivated fetal bovine serum (HI-FBS; Sigma-Aldrich; Merck) in 1× phosphate-buffered saline (PBS; Life Technologies; Thermo Fisher Scientific)). For the skin, an additional enzymatic digestion (using the Whole Skin Digestion kit; Miltenyi Biotech) was required overnight. Mechanical disruption was performed using the barrel of a 5 ml sterile syringe to pass the cells through a 70 µm filter. Concentrated cells were resuspended, and leukocytes were isolated by density centrifugation on a Pancoll gradient. Leukocytes obtained from the spleen and lymph nodes were further subjected to a red blood cell lysis step (BioLegend), on ice, before use. All intrahepatic, skin, splenic and lymphoid cell samples were used immediately.
For the isolation of human leukocytes from ascitic fluid, the fluid drained from the patient was concentrated by centrifugation and washed twice with RPMI-1640. Where necessary leukocytes were further subjected to a red blood cell lysis before use and frozen in 10% DMSO (Sigma-Aldrich; Merck)-HI FBS if not used immediately.
Where possible, peripheral blood was obtained from the same study participants for comparison. PBMCs were isolated from heparinized peripheral blood by density centrifugation using a Pancoll gradient.
Multiparametric flow cytometry
Multiparametric flow cytometry was used for phenotypic and functional analysis. For analysis of the CD14-expressing T cells, a strict gating criterion was used throughout. Cells were stained with a blue fixable live/dead dye (Invitrogen; Thermo Fisher Scientific) for the exclusion of dead cells. Doublets, CD45−, CD56+, CD19+ and CD3+CD4+ cells were also excluded from the analysis (the sequential gating strategy is shown in Extended Data Fig. 1a). In functional and sorting experiments, and for confirmatory phenotyping, additional gating excluded CD3+Vα7.2+ mucosal-associated invariant T cells and γδ T cells (the gating strategy is shown in Extended Data Fig. 2f). Surface markers were stained with saturating concentrations of monoclonal antibodies for 30 min at 4 °C in 50% diluted brilliant violet buffer (BD Bioscience). To avoid non-specific antibody binding to Fc receptors in ex vivo stains of tissue lymphocytes, an additional Fc receptor blocking step (Miltenyi Biotech, 130-059-901) was included according to the manufacturer’s instructions before surface antibody labelling. In the majority of the experiments, CD14 was detected using a Miltenyi Biotech REAfinity Recombinant Antibody (IgG1 Fc region mutated to abolish binding to Fcγ receptors). Cells were fixed using Cytofix/Cytoperm (BD Bioscience) or the Foxp3 buffer kit (BD Bioscience). Intracellular/intranuclear markers were subsequently stained in a 0.1% saponin-based buffer (Sigma-Aldrich; Merck) or 1× PBS for 30 min at 4 °C. Full details of the monoclonal antibodies, including dilutions and catalogue numbers, are provided in Supplementary Table 1. All of the samples were acquired in 1× PBS on the BD Bioscience LSR-FortessaX20-SORP system running DIVA (v.8.0.1) and analysed using FlowJo (v.9.9.4 or v.10.8.1; TreeStar; BD Bioscience).
Imaging flow cytometry
For confirmation of single-cell expression of αβTCR, CD3, CD8, CD14 and TLR4 using imaging cytometry, global CD3+ T cells were first isolated using the magnetic-bead-based human pan T cell negative selection kit (MACS-bead isolated; Miltenyi Biotech) from three liver samples, and stained as described in the ‘Multiparametric flow cytometry’ section with the addition of a DAPI nuclear dye (Sigma-Aldrich; Merck). For confirmation of in vitro membrane acquisition by CD8+ T cells, MACS-bead-isolated CD8+ T cells (isolated according to the manufacturer’s instruction using the human CD8+ T cell isolation kit (Miltenyi Biotech) were assessed for expression of αβTCR, CD3, CD8 and CD14, and further stained with a PE-labelled Streptavidin antibody to detect the biotin acquired directly from MNPs or MDMs as described in the ‘Isolation of MNPs and the in vitro generation of MDMs’ section. Samples were acquired in 1× PBS on the ImageStreamX MKII (Merck) system running INSPIRE (v.200.1.388.0) with a ×60 optic and analysed using Amnis Ideas. Doublets were discriminated by gating area versus aspect ratio by detecting high-aspect singlets, with a histogram gradient RMS (root mean square) to define in-focus single cells.
CD14+CD8+ T cell cytospin
Intrahepatic leukocytes were sorted by FACS for CD14±CD8+ T cells as follows: singlets, CD3+CD56−CD19−CD4−CD8+Vα7.2− and split in to CD14± based on an isotype control. Cells were isolated into chilled FACS-tubes coated with FBS and prefilled with 500 μl sterile 1× PBS for cytospinning. Giemsa staining (Sigma-Aldrich; Merck) was used to determine the morphology of sorted cells on cytospins. Slides were viewed using the Zeiss AxioImager microscope, images were taken of four fields from n = 2 samples using the ×100 objective, and viewed using Zen (v.2.3).
Detection of CD14-expressing CD8+ T cells in the setting of HLA-mismatch transplantation
An additional cohort of five study participants undergoing retransplantation (that is, receiving a second liver transplant) for disease recurrence (not chronic rejection; obtained through the TAPb Initiative, as above). Detection of donor and recipient leukocytes from isolated IHLs were determined on the basis of an HLA class-I mismatch by flow cytometry. HLA haplotyping was further confirmed by PCR, undertaken by A. Nolan or the staff at the MRC Weatherall Institute of Molecular Medicine Sequencing Facility. The samples were further profiled as described in the ‘Multiparametric flow cytometry’ section to determine CD14 expression.
Isolation of primary human hepatic stellate cells and co-cultures with PBMCs and CD8+ T cells
To generate CD14-expressing CD8+ T cells in vitro, freshly isolated PBMCs from healthy control donors were co-cultured in complete RPMI-1640 (cRPMI; containing 10% FBS, 100 IU ml−1 penicillin–streptomycin, 20 mM HEPES, 0.5 mM sodium pyruvate, MEM non-essential amino acids, MEM essential amino acids and 50 µM β−mercaptoethanol; all Life Technologies; Thermo Fisher Scientific) for 4 days in the presence of 40 IU ml−1 recombinant human IL-2 (PeproTech) at 37 °C with primary human hepatic stellate cells from three donors. Hepatic stellate cells were isolated as previously described51,52 from liver tissue samples using density centrifugation with an Optiprep (Sigma-Aldrich; Merck) gradient. Pre-isolated hepatic stellate cells were thawed and cultured in 25 or 75 cm2 tissue culture flasks in stellate cell medium (ScienCell Research Laboratories) to approximately 90% confluency before co-culture. For the co-culture experiments, hepatic stellate cells were detached using trypsin-EDTA (Life Technologies; Thermo Fisher Scientific) and reseeded at a density of 0.15 × 105 cells per well in cRPMI and left to adhere before the addition of 0.5 × 106 freshly isolated PBMCs. After co-culture for 4 days, PBMCs were collected and profiled as described in the ‘Multiparametric flow cytometry’ section for CD14 expression and phenotype. Where indicated, T cells were purified from freshly isolated PBMCs before hepatic stellate cell co-culture using the pan T cell isolation kit or the CD8+ T cell isolation kit for comparison with bulk PBMCs. In certain indicated T cell–hepatic stellate cell co-culture experiments, the cells were further supplemented with 5 µg ml−1 anti-CXCR4 (BioTechne) or IgG isotype control (BioTechne).
CXCL12 ELISA
Cell-free supernatants from primary human hepatic stellate cells from three donors cultured in 24-well flat-bottom plates for 4 days in stellate cell medium at various cell densities were collected and assessed for CXCL12 production using the human CXCL12/SDF-1α Quantikine ELISA kit (BioTechne) according to the manufacturer’s instructions.
In vitro derivation of CD14-expressing CD8+ T cells with cytokines
To generate CD14-expressing CD8+ T cells in vitro, freshly isolated PBMCs from healthy control donors were co-cultured in cRPMI for 4 days in the presence of 40 IU ml−1 recombinant human IL-2 (PeproTech) at 37 °C with the addition of 50 ng ml−1 recombinant human IL-6 (R&D Systems), IL-8 (R&D Systems), IL-15 (PeproTech), TGFβ (R&D Systems), CXCL9 (BioLegend), CXCL10 (R&D Systems), CXCL11 (BioLegend), CXCL12 (BioTechne) or CXCL16 (R&D Systems). After co-culture for 4 days, PBMCs were collected and profiled as described in the ‘Multiparametric flow cytometry’ section for CD14 expression and phenotype.
Isolation of MNPs and the in vitro generation of MDMs
For the isolation of autologous MNPs, PBMCs were freshly isolated from heparinized blood and isolated using the pan monocyte isolation kit (Miltenyi Biotech) according to the manufacturer’s instructions.
To generate MDMs, 5 × 106 PBMCs per ml were plated in 24-well plates for 2 h in RPMI-1640 supplemented with 10% HI-FBS to allow for MNP adherence before being washed thoroughly in 1× PBS to remove non-adherent cells. After the initial adherence step, MNPs were exposed to macrophage differentiation medium (RPMP-1640 supplemented with 10% human AB-serum and 20 ng ml−1 recombinant human MSCF (BioLegend)) for 6–10 days, with medium changes around every 3 days.
Biotin labelling of MNPs and MDMs
Where indicated, purified MNPs or in vitro-derived MDMs were labelled with EZ-Link Sulfo NHS-LC-LC-Biotin (Thermo Fisher Scientific) as previously described53. In brief, MNPs or MDMs were incubated with 1× PBS containing 10 µg ml−1 biotinylation reagent for 10 min at 37 °C. To quench, an equal volume of HI-FBS was added per well before the MNPs or MDMs were washed thoroughly and used in co-culture experiments with autologous CD8+ T cells. Autologous CD8+ T cells were co-cultured with labelled MNPs/MDMs in cRPMI for 4 days in the presence of 40 IU ml−1 recombinant human IL-2 at 37 °C at an approximate 1:1 ratio. After co-culture for 4 days, CD8+ T cells were collected and profiled as described in the ‘Multiparametric flow cytometry’ section for CD14 expression and phenotype.
CD14 acquisition by peripheral CD8+ T cells
To assess the ability of CD8+ T cells to acquire CD14 from MNPs, a series of in vitro trogocytosis assays were performed using freshly isolated peripheral CD8+ T cells. Autologous magnetic-bead-isolated CD8+ T cells and magnetic-bead isolated MNPs were co-cultured with primary human hepatic stellate cells in cRPMI supplemented with 100 IU ml−1 IL-2 in the presence or absence of the following inhibitors/antibodies/reagents: 25 µM latrunculin B (actin polymerization inhibitor), 100 nM nocodazole (microtubule polymerization inhibitor), 1 µg ml−1 anti-ICAM or the matched isotype control or 5 µg ml−1 soluble CD14 (BioLegend). In other experiments, where indicated, the MNPs and CD8+ T cells were physically separated with 0.4 µm Transwell inserts or with previous fixation of the MNP fraction for 10 min with 4% paraformaldehyde. After fixation, MNPs were washed thoroughly in 1× PBS to remove any residual fixative. After co-culture for 4 days, CD8+ T cells were collected and profiled as described in the ‘Multiparametric flow cytometry’ section for CD14 expression and phenotype.
Exosome purification and analysis from MDM supernatants and re-addition to CD8+ T cells
To exclude a role for T cell acquisition of CD14 by exosome-mediated transfer, exosomes were isolated and purified from MDMs from three donors (generated as described in the ‘Isolation of MNPs and the in vitro generation of MDMs’ section) using the CD63 exosome isolation kit (Miltenyi Biotech) according to the manufacturer’s instructions. In brief, cellular debris was removed from the cell-free MDM supernatants by centrifugation, before incubation for 1 h with the kit-provided exosome isolation microbeads. After incubation, exosomes were purified from the supernatant by magnetic bead isolation and quantified using the bicinchoninic acid assay (Thermo Fisher Scientific). The MACSPlex Exosome Detection kit (Miltenyi Biotech) was used according to the manufacturer’s instructions to phenotype and analyse the MDM-isolated exosomes by flow cytometry after high-speed centrifugation.
Once purified, 5 µg ml−1 primary human MDM-derived exosomes were co-cultured with isolated autologous CD8+ T cells in cRPMI supplemented with 40 IU ml−1 IL-2. After co-culture for 4 days, CD8+ T cells were collected and profiled as described in the ‘Multiparametric flow cytometry’ section for CD14 expression and phenotype.
Confocal imaging of in vitro-derived CD14+CD8+ T cells
Autologous MDM/CD8+ T cell co-culturing was performed in 24-well plates containing poly-lysine-coated coverslips (refer to the ‘Isolation of MNPs and the in vitro generation of MDMs’ section). After co-culture, the coverslips were removed and mounted onto slides for image acquisition. Before staining, the slides were processed for a 1 h blocking step using 10% goat serum/1% BSA/0.001 Triton X-100 in 1× PBS at room temperature. After blocking, the slides were incubated with unconjugated antibodies against rat anti-human CD8 and rabbit anti-human CD14 (Thermo Fisher Scientific) for a further 18 h and washed thoroughly in 1× PBS. Primary antibody detection required further staining with secondary fluorochrome-labelled antibodies: goat anti-rat AlexaFluor-488 and anti-human Alexa647 conjugates (Jackson Immuno Research) for 1 h. All of the slides were also labelled with Hoechst 33342 (Thermo Fisher Scientific).
Images were acquired using the Zeiss LSM880 Airyscan confocal microscope at ×63, ×4 zoom (PLAN-Apochromat ×63/1.4 NA) and ×20 (EC Plan-Neofluar 0.5 NA) magnification. For representative images, ×63/1.4 NA z-stacks were acquired and visualized as maximum-intensity projections. For quantification of CD14, confocal ×20 magnification z-slices were acquired in the medial plane of the CD8 signal in the CD8+ T cell alone and MDM-associated CD8+ T cell populations. Image analysis was performed using FIJI ImageJ, with 30 CD8+ T cells quantified per condition54. Nuclear segmentation occurred through segmentation of Hoechst 33342 fluorescence. T cell populations were identified, and their cell boundaries were segmented using the extent of CD8 expression. The mean fluorescence of CD14 was then quantified within the segmented CD8+ T cell populations.
Mass cytometry imaging of CD14+CD8+ T cells in human liver
Imaging mass cytometry was conducted as reported previously55. In brief, relevant antibodies were conjugated to lanthanide metals using the Maxpar X8 antibody labelling kit. Formaldehyde-fixed paraffin-embedded (FFPE) sections (thickness, 4 µm) were deparaffinized and incubated for 40 min in EnVision FLEX Target Retrieval Solution High pH (DAKO). Before staining, the liver sections were blocked by incubation with SuperBlock Blocking Buffer (Thermo Fisher Scientific). Once blocked, the slides were incubated with the relevant conjugated antibodies at room temperature overnight (Supplementary Table 2) in 0.5% BSA, 1% Triton-X-100 in Tris. A 30 min exposure to Iridium Cell-ID Intercalator (Fluidigm) was used to visualize DNA. Ablation/imaging was conducted using the Hyperion Imaging Mass Cytometry (Fluidigm) system.
Data analysis was performed using ImageJ, R Studio with imctools, QMIQ and cytomapper55. The liver zonation index was calculated by importing relevant tissue section images into ImageJ (v.1.52) using the imctools plugin, and custom code, which is available at GitHub (https://github.com/ljpallett/pallettetal_2022_imc). A Gaussian blurr filter was applied to the CYP1A2 signal with a sigma of 100, and the intensity of each pixel was measured. Background subtraction was performed by subtracting the minimum intensity per image from all values. Areas with a liver zonation index of ≥0.5 were classified as zone 3; <0.5 and ≥0.25 as zone 2; and <0.25 as zone 1. Enrichment scores were calculated by comparing the absolute cell counts in a certain zone to a hypothetical cell count if there was an equal distribution of the respective cells across all zones. The enrichment ratio was calculated by dividing the enrichment score of one cell population by the enrichment score of another cell population in a given zone. The distance to the closest myeloid structure was obtained by calculation of the distance to the closest Iba1+ myeloid structure in ImageJ.
3D decellularized liver scaffolds
For the preparation of decellularized scaffolds, liver tissue was obtained through the TAPb Initiative (study approval reference: 11/WA/0077). The full protocol for the development of 3D acellular biological scaffolds was previously described13. In brief, whole human livers were perfused with 1× PBS to remove blood and frozen at −80 °C before decellularization. The right lobe was sectioned and agitated with deionized water (Merck-Millipore) containing 3% sodium deoxycholate (Sigma-Aldrich; Merck), 0.5% sodium dodecyl sulfate (Sigma-Aldrich; Merck), 0.3% Triton X-100 (Thermo Fisher Scientific) and 4.5% sodium chloride (Sigma-Aldrich; Merck) and 1× PBS until the tissue was translucent with the dissolution of cells. The absence of cells in the ECM protein scaffolds was confirmed by the absence of DNA material quantified using the DNeasy Blood and Tissue Kit (Qiagen). All scaffolds were confirmed by proteomics and immunohistochemical analysis to express natural ECM including collagen-I, -III and -IV, fibronectin, laminin, lumican, mimecan and vitronectin56. PBMCs (0.5 × 106–1 × 106) were cultured in cRPMI in the presence 40 IU ml−1 recombinant human IL-2 and 5 ng ml−1 IL-15 (BioTechne) for 3 days in 96-well round-bottom plates with or without an ECM scaffold. After culture, 3D scaffolds (and cells cultured in the absence of a 3D scaffold) were digested for 30 min with collagenase to recover leukocytes for phenotypic analysis as described in the ‘Multiparametric flow cytometry’ section.
Quantification of gene expression by RT–qPCR
For the quantification of mRNA transcripts for CD14, TLR4 and LY96 (also known as MD2), total RNA was extracted from highly pure paired ex vivo human intrahepatic CD14−CD8+ T cells or CD14+CD8+ T cells using TRIzol and RNAeasy Plus Micro Kit (Qiagen). Subsequently RNA was reverse-transcribed to cDNA with the High-Capacity RNA-to-cDNA Kit (Applied Biosystems) or Quantitecht RT Kit (Qiagen). Gene expression was measured by RT–qPCR using the 7500 Real-Time PCR System (Applied Biosystems). KiCqStart PCR primers used were as follows: 18S, CD14, TLR4, MD-2 (Sigma-Aldrich; Merck). The relative expression level of specific transcripts was normalized to the internal standard 18S, and calculated using the ΔCt method57.
scRNA-seq samples
For the analysis of the single-cell transcriptome, two intrahepatic perfusate samples were isolated and frozen at −80 °C for storage before transportation to Newcastle, UK. Once thawed in prewarmed HBSS containing 0.001% DNase I, intrahepatic leukocytes were sorted by FACS to index classical MNPs, defined as singlets, CD3−CD56−CD19−CD14highHLA-DR+ or CD14±CD8+ T cells as follows: singlets, CD3+CD56−CD19−CD4−CD8+Vα7.2− and split in to CD14± based on an isotype control. Single cells were sorted into 96-well skirted LoBind plates (Eppendorf) containing 5 µl lysis buffer (TCL buffer (Qiagen), supplemented with 1% β-mercaptoethanol (Sigma-Aldrich; Merck)) using the BD Bioscience FACSFusion system. The sorted plates were subsequently immediately sealed, centrifuged and placed onto dry ice to flash-freeze the cell lysate and stored at −80 °C.
scRNA-seq Smart-Seq2 library preparation and sequencing
A modified SMART-seq2 protocol58 was performed on the flow-cytometry-sorted single cells as previously described59. After cDNA generation, libraries were prepared (384 cells per library) using the Illumina Nextera XT kit (Illumina). Each library was sequenced to achieve a minimum depth of 1–2 million raw reads per cell on the Illumina HiSeq 4000 system using v.4 SBS chemistry to generate 75 bp paired-end reads.
scRNA-seq alignment, quantification and quality control
SMART-seq2 sequencing data were aligned with STAR (v.2.5.1b) using the GRCh38 human reference genome. Quantification was performed using htseq-count (v.0.10.0). Cells with fewer than 200 detected genes, more than 2,750 genes or for which the total mitochondrial gene expression exceeded 20% were removed as previously described59. Genes expressed in fewer than three cells were removed. The simulated doublet histogram generated using Scrublet was unimodal, probably due to cellular homogeneity60. Harmony data integration61 was used to correct for any batch effects between samples.
scRNA-seq clustering and annotation
After quality control, gene expression was normalized by cell to correct for cell-to-cell variation in total reads and log-transformed using the NormalizeData tool in Seurat. The top 2,000 highly variable genes were identified using the vst method in Seurat. Linear dimensional reduction was performed by principal component analysis after expression levels were scaled to mean expression 0 and variable 1 across all cells. The dimensionality of the data was determined using the JackStraw procedure in Seurat and the top 30 principal components were used to generate the neighbourhood graph. Clustering was performed using the Leiden algorithm (0.6 resolution) and embedded using UMAP.
Dextramer staining for the identification of virus-specific CD8+ T cells
In study participants known to be HLA-A*02+ (confirmed by monoclonal HLA-A*02-FITC antibody (BioRad; Abcam) staining), HBV-specific APC-conjugated HLA-A*02/dextramers (Immudex) of the following specificities were used: HBVcore(18–27) (FLPSDFFPFV), HBVenvelope(183–191) HBVenvelope(335–342) (WLSLLVPFV), HBVenvelope(348–357) (GLSPTVWLSV), HBVpolymerase(455–463) (GLSRYVARL) and HBVpolymerase(502–510) (KLHLYSHPI). To identify antigen-specific CD8+ T cells, isolated intrahepatic leukocytes were stained with the corresponding dextramers at 37 °C in 1× PBS for 20 min, washed twice in cRPMI and left to rest for 1 h before further staining with monoclonal antibodies as described in the ‘Multiparametric flow cytometry’ section.
FITC–LPS uptake assay
To assess the functionality of the TLR4–CD14 complex, 0.5 × 106–1 × 106 freshly isolated IHLs or stromal-cell-induced CD14-expressing CD8+ T cells (described in the ‘Isolation of primary human hepatic stellate cells and co-cultures with PBMCs and CD8+ T cells’ section above) were cultured in warm cRPMI in the presence or absence of 5 µg ml−1 fluorescently labelled LPS (Thermo Fisher Scientific) at 37 °C for 30, 60 or 120 min before flow cytometry staining to determine LPS uptake by the cell using flow cytometry assessment of the MFI of the FITC channel.
Measurement of supernatant cytokine profiles by Luminex assay
To functionally profile CD14-expressing CD8+ T cells, we used a multiplex array to profile stromal-cell-induced CD14±CD8+ T cells (described in the ‘Isolation of primary human hepatic stellate cells and co-cultures with PBMCs and CD8+ T cells’ section above). CD14±CD8+ T cells were sorted on the BD FACSAria II system to exclude MNPs by FSC-A and SSC-A, dead cells and doublets, then sorted for CD3+CD56−CD4−CD8+Vα7.2− and split in to CD14± based on an isotype control. Purified CD14±CD8+ T cells were seeded in 96-well round-bottom plates at a density of around 1 × 105 cells per well in 200 µl cRPMI and cultured at 37 °C for 24 h in the presence or absence of exogenous stimulation with about 0.3 × 106 ultraviolet-killed E. coli or 1 µg ml−1 immobilized anti-CD3 and 5 ng ml−1 soluble anti-CD28 antibodies (both eBioscience; Thermo Fisher Scientific). After incubation, cell culture supernatants were collected and stored at −80 °C for multiplex analysis. Cytokine expression profiles were determined using the Luminex array platform with the Human XL Cytokine Luminex 45-plex Performance Assay (BioTechne). The samples were acquired on the MAGPIX Luminex (Thermo Fisher Scientific) system running xPONENT v.4.2, and were analysed using a custom pipeline (Python v.3.6; https://github.com/ljpallett/pallettetal_2022_luminex).
In vitro analysis of phenotype and functionality
To assess the effect of in vitro bacterial LPS exposure on CD14 expression, 0.5 × 106–1 × 106 freshly isolated IHLs from human resected tissue were cultured in the presence or absence of around 0.3 × 106 ultraviolet-killed E. coli for 24, 48, 72 or 96 h in cRPMI. The phenotype of CD8+ T cells was assessed by flow cytometry as described above.
To assess the functionality of intrahepatic CD14-expressing T cells ex vivo, 1 × 106 IHLs were stimulated with 1 µg ml−1 immobilized anti-CD3 and 5 µg ml−1 soluble anti-CD28 antibodies for 4 h at 37 °C in the presence of 1 µg ml−1 brefeldin A (Sigma-Aldrich; Merck). Functionality was assessed by intracellular cytokine production as described in the ‘Multiparametric flow cytometry’ section.
To confirm the ability of stromal-cell-induced CD14±CD8+ T cells (described in the ‘Isolation of primary human hepatic stellate cells and co-cultures with PBMCs and CD8+ T cells’ section above) to secrete cytokines/chemokines, we used intracellular cytokine staining. Freshly isolated peripheral CD8+ T cells were first co-cultured with autologous MNPs 1:1 for 4 days. After co-culture with or without MNPs, CD8+ T cells were removed and reseeded into 96-well round-bottom plates in cRPMI supplemented with 100 IU ml−1 IL-2 and cultured at 37 °C for 24 h in the presence or absence of exogenous stimulation with around 0.3 × 106 ultraviolet-killed E. coli and brefeldin A. Strict gating, including an anti-αβTCR antibody, was used to exclude MNPs from the analysis.
CFSE labelling and dual CD14-labelling of leukocytes
Where indicated, freshly isolated intrahepatic leukocytes were prelabelled with CD14–APC (ReaAffinity; Miltenyi Biotech) and 2 µM CFDA (CFSE; Invitrogen; Thermo Fisher Scientific) for 15 min at 37 °C before culture for 3 d in the presence of around 0.3 × 106 ultraviolet-killed E. coli to determine cellular proliferation. After culture, leukocytes were further stained with CD14–PE (BioLegend) along with other surface monoclonal antibodies to determine the phenotype as described in the ‘Multiparametric flow cytometry’ section.
Human in vivo skin blister model
The human in vivo skin blister model was approved by UCL Research Ethics Committee (10527/001). Seven young healthy volunteers were recruited. Intradermal ultraviolet-killed E. coli (ultraviolet-killed E. coli; strain: NCTC10418; source: Public Health England) was used to model in vivo Gram-negative bacteria exposure using suction blisters to induce an acute, resolving inflammatory response for interrogation as previously described41. In brief, 1.5 × 107 ultraviolet-killed E. coli in 100 µl of sterile saline was injected intradermally into a marked site on the ventral side of the right forearm of participants. From each participant, the left forearm was used to raise a ‘naive’ unexposed blister to analyse the exudate (baseline blister). After 24 h, each participant returned, and a suction blister was used to remove the inflammatory exudate from the LPS-exposed injection site from the right forearm. Both blister exudates were centrifuged to concentrate the cells and analysed as described in the ‘Multiparametric flow cytometry’ section.
Generation of TCR-redirected T cells
Where indicated, genetically engineered CD8+ T cells with known TCR specificity were used to generate CD14-expressing T cells in vitro. For this, CD8+ T cells were modified to express a TCR specific for an immunodominant HLA-A*02-restricted epitope of the HBV envelope protein; HBVe183-91 (e183-specific CD8+ T cells; amino acid sequence: FLLTRILTI, provided by A. Bertoletti). For this, Phoenix amphotrophic packaging cells (ATCC, CRL-3213) were transiently co-transfected using FuGENE (Promega) with plasmids encoding the e183-specific TCR alongside an amphotrophic envelope protein. For these transfected phoenix cells, retroviral supernatants were collected for the further transduction of healthy donor CD8+ T cells obtained from freshly isolated PBMCs by MACS-bead isolation (Human CD8+ isolation kit). The Phoenix amphotrophic package cell line used in this study was regularly tested throughout for mycoplasma contamination by PCR (EZ-PCR Mycoplasm Kit, Biological Industries). Before transduction, isolated CD8+ T cells were stimulated with Human T-Activator CD3/CD28 Dynabeads (Thermo Fisher Scientific) in cRPMI supplemented with 5 ng ml−1 recombinant human IL-15 and IL-7 (PeproTech) and 100 IU ml−1 IL-2 for up to 48 h. For transduction, 1 × 106 activated CD8+ T cells per well were plated onto retronectin-coated (Takara) plates and mixed by centrifugation with previously collected and concentrated retroviral supernatants. Transduced CD8+ T cells were expanded in cRPMI supplemented with 5 ng ml−1 recombinant human IL-15/IL-7 and IL-2 as described above for up to 10 days and continually monitored for the expression of the engineered e183-specific TCR by staining for the murine constant region of the TCRβ chain by flow cytometry before co-culture with autologous MNPs/MDMs (as described in the ‘Isolation of MNPs and the in vitro generation of MDMs’ section).
After gene-modification and expansion, TCR-redirected CD8+ T cells were co-cultured with autologous MNPs/MDMs at a ratio of between 1:1 and 10:1 (T cells:MNPs; depending on MNP availability) for 4 days to allow for T cell acquisition of the CD14–TLR4 complex. After MNP/T cell co-culture, e183-specific TCR-redirected CD14±CD8+ T cells were sorted on the BD FACSAria II system to exclude MNPs by FSC-A and SSC-A, dead cells and doublets, then sorted for CD3+CD56−CD19−CD4−CD8+Vα7.2−murine TCRβ+ and split in to CD14± based on an FMO control.
preS1 HepG2 maintenance
An HLA-A*02 human HCC cell line, HepG2, transduced with a construct containing the preS1 portion of the genotype D HBV envelope protein (HepG2-preS1 cells) using the Lenti-X HTX packaging system, was provided by A. Bertoletti’s laboratory. HepG2-preS1 cells were cultured in DMEM (Invitrogen) supplemented with 10% HI-FBS, 100 IU ml−1 penicillin–streptomycin, 0.5 mM sodium pyruvate and MEM non-essential amino acids. HepG2-preS1 cells are capable of endogenous processing and presentation of HBV envelope protein, recognized by engagement of the e183-specific TCR, as previously described62. The HepG2-preS1 line used in this study was regularly tested throughout for mycoplasma contamination by PCR as described above.
Assessment of the functionality of TCR-redirected T cells
To evaluate the function of the TCR-redirected CD8+ T cells, HepG2-preS1 cells were plated and allowed to adhere at various cell densities (as indicated in the appropriate figure) for 6 h before use. Once adhered, FACS-sorted e183-specific TCR-redirected T cells with or without CD14 were added directly to the HepG2-preS1 cells in cRPMI supplemented with 100 IU ml−1 IL-2 for 16 h, either in the presence or absence of BFA/monensin. The CD8+ T cells cultured in the presence of BFA/monensin were collected after the 16 h incubation and assessed for cytokine production by intracellular cytokine staining as described in the ‘Multiparametric flow cytometry’ section. The cell-free supernatant was collected from the CD8+ T cells cultured without BFA/monensin and assessed for cytotoxicity using the ToxiLight Non-destructive Cytotoxicity BioAssay (Lonza) according to the manufacturer’s instruction. In brief, the assay evaluates the specific target cell lysis by measuring relative light units (RLU), using the following calculation: (RLU[effector + target] − RLU[effector] − RLU[target])/(RLU[100% target lysis] − RLU [effector] − RLU [target]) × 100.
Statistical analyses
Statistical analyses were performed in Excel (v.16.16.09), Prism (GraphPad v.7 or v.8) or Python (v.3.6) using the appropriate tests (Mann–Whitney t-tests, Wilcoxon signed-rank t-tests, Kruskal–Wallis tests (ANOVA) with Dunn’s post hoc test for pairwise multiple comparisons between each group) as indicated in the legends. These tests were performed as two-tailed tests, with significant differences marked on all of the figures. Where appropriate, Bonferroni correction for multiple testing was used. Error bars represent s.e.m. Significance levels are marked on all of the figures. Relevant visualizations were created using Python (v.3.6) and the seaborn statistical visualization package.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The source data used within the main figures is available in the Supplementary Information and at Figshare (https://doi.org/10.6084/m9.figshare.21623379). scRNA-seq data have been deposited in the European Genome–Phenome Archive under accession number EGAS00001006885. Further datasets generated during and/or analysed as part of this study are available from the corresponding authors on reasonable request. Source data are provided with this paper.
Code availability
Custom codes used in this study are available at GitHub. For the heat-map visualization used to display the soluble mediator profiles of CD14-expressing CD8+ T cells, the custom code created is available at GitHub (https://github.com/ljpallett/pallettetal_2022_luminex). For the image mass cytometry analysis, the custom code created is available at GitHub (https://github.com/ljpallett/pallettetal_2022_imc).
References
Crispe, I. N. Immune tolerance in liver disease. Hepatology 60, 2109–2117 (2014).
Protzer, U., Maini, M. K. & Knolle, P. A. Living in the liver: hepatic infections. Nat. Rev. Immunol. 12, 201–213 (2012).
Macpherson, A. J., Heikenwalder, M. & Ganal-Vonarburg, S. C. The liver at the nexus of host-microbial interactions. Cell Host Microbe 20, 561–571 (2016).
Wiest, R., Lawson, M. & Geuking, M. Pathological bacterial translocation in liver cirrhosis. J. Hepatol. 60, 197–209 (2014).
Tripathi, A. et al. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 15, 397–411 (2018).
Pallett, L. J. et al. IL-2high tissue-resident T cells in the human liver: sentinels for hepatotropic infection. J. Exp. Med. 214, 1567–1580 (2017).
Burel, J. G. et al. Circulating T cell-monocyte complexes are markers of immune perturbations. eLife 8, e46045 (2019).
Pallett, L. J. et al. Longevity and replenishment of human liver-resident memory T cells and mononuclear phagocytes. J. Exp. Med. 217, e20200050 (2020).
Fernandez-Ruiz, D. et al. Liver-resident memory CD8+ T cells form a front-line defense against malaria liver-stage infection. Immunity 45, 889–902 (2016).
Curbishley, S. M., Eksteen, B., Gladue, R. P., Lalor, P. & Adams, D. H. CXCR3 activation promotes lymphocyte transendothelial migration across human hepatic endothelium under fluid flow. Am. J. Pathol. 167, 887–899 (2005).
Liepelt, A. & Tacke, F. Stromal cell-derived factor-1 (SDF-1) as a target in liver diseases. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G203–G209 (2016).
Neumann, K. et al. Chemokine transfer by liver sinusoidal endothelial cells contributes to the recruitment of CD4+ T cells into the murine liver. PLoS ONE 10, e0123867 (2015).
Mazza, G. et al. Rapid production of human liver scaffolds for functional tissue engineering by high shear stress oscillation-decellularization. Sci. Rep. 7, 5534 (2017).
McQuitty, C. E., Williams, R., Chokshi, S. & Urbani, L. Immunomodulatory role of the extracellular matrix within the liver disease microenvironment. Front. Immunol. 11, 574276 (2020).
McNamara, H. A. et al. Up-regulation of LFA-1 allows liver-resident memory T cells to patrol and remain in the hepatic sinusoids. Sci. Immunol. 2, eaaj1996 (2017).
Benechet, A. P. et al. Dynamics and genomic landscape of CD8+ T cells undergoing hepatic priming. Nature 574, 200–205 (2019).
De Simone, G. et al. Identification of a Kupffer cell subset capable of reverting the T cell dysfunction induced by hepatocellular priming. Immunity 54, 2089–2100 (2021).
Wei, Y. et al. Liver homeostasis is maintained by midlobular zone 2 hepatocytes. Science 371, eabb1625 (2021).
Baumann, T. et al. Regulatory myeloid cells paralyze T cells through cell-cell transfer of the metabolite methylglyoxal. Nat. Immunol. 21, 555–566 (2020).
Crispe, I. N. The liver as a lymphoid organ. Annu. Rev. Immunol. 27, 147–163 (2009).
Huang, L. R. et al. Intrahepatic myeloid-cell aggregates enable local proliferation of CD8+ T cells and successful immunotherapy against chronic viral liver infection. Nat. Immunol. 14, 574–583 (2013).
Krenkel, O. & Tacke, F. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 17, 306–321 (2017).
Pallett, L. J. & Maini, M. K. Liver-resident memory T cells: life in lockdown. Semin. Immunopathol. 44, 813–825 (2022).
Legut, M. et al. A genome-scale screen for synthetic drivers of T cell proliferation. Nature 603, 728–735 (2022).
Kumar, B. V. et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20, 2921–2934 (2017).
Smith, L. K. et al. Interleukin-10 directly inhibits CD8+ T cell function by enhancing N-glycan branching to decrease antigen sensitivity. Immunity 48, 299–312 (2018).
Fioravanti, J. et al. Effector CD8+ T cell-derived interleukin-10 enhances acute liver immunopathology. J. Hepatol. 67, 543–548 (2017).
Schurich, A. et al. Dynamic regulation of CD8 T cell tolerance induction by liver sinusoidal endothelial cells. J. Immunol. 184, 4107–4114 (2010).
Tan, A. T. et al. Use of expression profiles of HBV-DNA integrated into genomes of hepatocellular carcinoma cells to select T Cells for immunotherapy. Gastroenterology 156, 1862–1876 (2019).
Frey, E. A. et al. Soluble CD14 participates in the response of cells to lipopolysaccharide. J. Exp. Med. 176, 1665–1671 (1992).
Komai-Koma, M., Gilchrist, D. S. & Xu, D. Direct recognition of LPS by human but not murine CD8+ T cells via TLR4 complex. Eur. J. Immunol. 39, 1564–1572 (2009).
Yoshimura, A. et al. Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J. Immunol. 163, 1–5 (1999).
Zanoni, I. & Granucci, F. Role of CD14 in host protection against infections and in metabolism regulation. Front. Cell Infect. Microbiol. 3, 32 (2013).
Sakai, N. et al. Interleukin-33 is hepatoprotective during liver ischemia/reperfusion in mice. Hepatology 56, 1468–1478 (2012).
Taub, R. Hepatoprotection via the IL-6/Stat3 pathway. J. Clin. Invest. 112, 978–980 (2003).
Taub, D. D., Anver, M., Oppenheim, J. J., Longo, D. L. & Murphy, W. J. T lymphocyte recruitment by interleukin-8 (IL-8). IL-8-induced degranulation of neutrophils releases potent chemoattractants for human T lymphocytes both in vitro and in vivo. J. Clin. Invest. 97, 1931–1941 (1996).
Gehring, A. J. et al. Licensing virus-specific T cells to secrete the neutrophil attracting chemokine CXCL-8 during hepatitis B virus infection. PLoS ONE 6, e23330 (2011).
Foussat, A. et al. Production of stromal cell-derived factor 1 by mesothelial cells and effects of this chemokine on peritoneal B lymphocytes. Eur. J. Immunol. 31, 350–359 (2001).
Albillos, A. et al. Increased lipopolysaccharide binding protein in cirrhotic patients with marked immune and hemodynamic derangement. Hepatology 37, 208–217 (2003).
Sierro, F. et al. A liver capsular network of monocyte-derived macrophages restricts hepatic dissemination of intraperitoneal bacteria by neutrophil recruitment. Immunity 47, 374–388 (2017).
Motwani, M. P. et al. Pro-resolving mediators promote resolution in a human skin model of UV-killed Escherichia coli-driven acute inflammation. JCI Insight 3, e94463 (2018).
Nowarski, R., Jackson, R. & Flavell, R. A. The stromal intervention: regulation of immunity and inflammation at the epithelial-mesenchymal barrier. Cell 168, 362–375 (2017).
Croft, A. P. et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature 570, 246–251 (2019).
Krausgruber, T. et al. Structural cells are key regulators of organ-specific immune responses. Nature 583, 296–302 (2020).
Gola, A. et al. Commensal-driven immune zonation of the liver promotes host defence. Nature 589, 131–136 (2021).
Bonnardel, J. et al. Stellate cells, hepatocytes, and endothelial cells imprint the Kupffer cell identity on monocytes colonizing the liver macrophage niche. Immunity 51, 638–654 (2019).
Seki, E. et al. TLR4 enhances TGF-β signaling and hepatic fibrosis. Nat. Med. 13, 1324–1332 (2007).
Zanin-Zhorov, A. et al. Cutting edge: T cells respond to lipopolysaccharide innately via TLR4 signaling. J. Immunol. 179, 41–44 (2007).
Seki, E. & Brenner, D. A. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology 48, 322–335 (2008).
Kucykowicz, S. et al. Isolation of human intrahepatic leukocytes for phenotypic and functional characterization by flow cytometry. STAR Protoc. 3, 101356 (2022).
Cossarizza, A. et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (third edition). Eur. J. Immunol. 51, 2708–3145 (2021).
Singh, H. D. et al. TRAIL regulatory receptors constrain human hepatic stellate cell apoptosis. Sci Rep. 7, 5514 (2017).
Daubeuf, S., Puaux, A. L., Joly, E. & Hudrisier, D. A simple trogocytosis-based method to detect, quantify, characterize and purify antigen-specific live lymphocytes by flow cytometry, via their capture of membrane fragments from antigen-presenting cells. Nat. Protoc. 1, 2536–2542 (2006).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis Nat. Methods 9, 676–682 (2012).
Schwabenland, M. et al. Deep spatial profiling of human COVID-19 brains reveals neuroinflammation with distinct microanatomical microglia-T-cell interactions. Immunity 54, 1594–1610 (2021).
McAdam, S. et al. Cross-clade recognition of p55 by cytotoxic T lymphocytes in HIV-1 infection. Aids 12, 571–579 (1998).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Popescu, D. M. et al. Decoding human fetal liver haematopoiesis. Nature 574, 365–371 (2019).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Pavesi, A. et al. A 3D microfluidic model for preclinical evaluation of TCR-engineered T cells against solid tumors. JCI Insight 2, e89762 (2017).
Acknowledgements
We thank all of the patients and control volunteers who participated in this study, and all of the clinical staff who helped with participant recruitment, including the members of the Tissue Access for Patient Benefit Initiative at The Royal Free Hospital; H. Stauss for his suggestions; D. Dixon for help with the cytospin microscopy; the support staff at the UCL Infection and Immunity Flow Cytometry Core Facility; and P. S. Chana for his help with the imaging cytometry analysis. This work was funded by Wellcome Investigator Awards (101849/Z/13/A and 214191/Z/18/Z), a Medical Research Council grant (G0801213), a CRUK Immunology grant (26603) and a Hunter Accelerator award to M.K.M.; a UKRI Future Leader Fellowship to L.J.P.; a Medical Research Foundation grant to L.S.; a Wellcome Clinical Research Training Fellowship to U.S.G. (107389/Z/15/Z); a European Research Council H2020 Starter grant (ERC-StG-2015-637304) and the Wellcome New Investigator award (104771/A/14/Z) to M. Dorner; and the Berta Ottenstein Programme and IMM-PACT-Programme for Clinician Scientists, University of Freiburg funded by the Deutsche Forschungsgemeinschaft 413517907 to M.S.
Author information
Authors and Affiliations
Contributions
L.J.P., A.S. and M.K.M. conceived the project. L.J.P., D.W.G., M. Dorner, B.B., A.S., B.M.C., M.H. and M.K.M. designed experiments. J.D., J.K.S., K.A.S., K.S., M. Diniz, A.R.B., I.U., O.E.A., E.S. and W.A.-A. prepared samples. L.J.P., L.S., M. Diniz, A.A.M., M.S., A.D.G., M.W., J.S., R.d.M., C.T., J.D., S.K., J.K.S., N.M.S., O.E.A., A.R.B., E.S., G.R., M.W., J.S., R.d.M., C.T., I.U. and A.M.O.-P. generated data. L.J.P., L.S., G.R., N.T., B.M.C., M.S., A.D.G., M.P., B.B., M.H. and M.K.M. analysed data. A.A.M., U.S.G., C.G., F.F., S.L., S.P.-D.-P., G.F., P.T.F.K., B.R.D., W.A.-A., G.M., M.N. and A.A. provided essential models or patient samples/clinical analysis. L.J.P. and M.K.M. prepared the manuscript and all of the other authors provided input to the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.K.M. and L.J.P. have received project funding from Gilead for research unrelated to this manuscript. M.K.M. has sat on advisory boards/provided consultancy for Gilead, Roche, GSK and VirBiosciences. L.J.P. has sat on advisory boards/provided consultancy for Gilead and SQZ Biotech. M.K.M. and L.J.P. have a patent application P116607GB filed through UCL-Business on the use of CD14+CD8+ T cells.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Confirmation of CD14+CD8+ T cells in human liver.
a Flow cytometric gating strategy defining CD14+CD8+ T cells in human liver with sequential exclusion of: debris and MNP by forward (FSC-A) and side scatter (SSC-A), doublets, dead cells, CD45−, CD3−, CD56+, CD19+, and CD3+CD4+ T cells. b Representative flow cytometric staining of peripheral and intrahepatic CD14+CD8+ T cells (using a different clone, and company than example shown in Fig. 1a). c Representative histograms of αβTCR and CD3ζ expression on intrahepatic CD14−CD8+ T cells (black outline), CD14+CD8+ T cells (blue filled) and classical MNP (large SSC-A, Lin−HLA-DR+CD3−CD14+; grey filled). d Summary data: cell size using FSC-A (n = 80) and CD45 expression (n = 28) on CD14−CD8+ T cells and CD14+CD8+ T cells. e CD14 percentage on CD3−CD56+ NK cells, CD3+CD8−CD4+ T cells and CD3+CD4-CD8+ T cells from the same donors (n = 28). f Summary data: percentage of CD69 (n = 89) and CD103 (n = 96) on intrahepatic CD14−CD8+ T cells or CD14+CD8+ T cells. g Representative plots showing the frequency of donor-derived (donor-HLA+) or recipient-derived (donor-HLA-) CD14−CD8+ T cells and CD14+CD8+ T cells. h Frequency of donor-derived CD14+CD8+ T cells in liver allografts explanted after 8 months-11 yrs from HLA-haplotype disparate recipients (n = 5) using the gating strategy in Fig. 1h. Each dot (or pair of dots) represent(s) a study participant, processed, stained and analysed independently; bars represent mean. e and f solid lines: median, dotted lines: IQR. p-values determined using either a Kruskal-Wallis test (ANOVA) with Dunn’s multiple comparisons test e; or two-tailed Wilcoxon t Test d,f.
Extended Data Fig. 2 Residency and transcriptional features of CD14+CD8+ T cells.
a Representative plots: %CD14 on peripheral CD8+ T cells after PBMC and pHSC co-culture (representative of four PBMC donors with 3 pHSC donors). b Summary data: in vitro induction of CD14+CD8+ T cells after PBMC and pHSC co-culture (4d; PBMC donors used with: pHSC#1 n = 15; pHSC#2 n = 30; and pHSC#3 n = 24). c Summary data: MFI of TLR4 (n = 4) and TLR2 (n = 7) of pHSC-induced CD14−CD8+ T cells (black outline) or CD14+CD8+ T cells (blue filled). d %CD14 on peripheral CD8+ T cells after PBMC cultured ± 3D de-cellularised human liver ‘extracellular matrix scaffolds’ (3 d; n = 12 PBMC donors; 3 experiments). e Frequency of peripheral CD14+CD8+ T cells after co-culture of purified CD8+ T cells with a varied ratio of autologous MNP (4d; n = 11). f Flow cytometric gating strategy used to isolate ex vivo or stromal-cell induced CD14+CD8+ T cells with the sequential exclusion of: debris and MNP by forward (FSC-A) and side scatter (SSC-A), doublets, dead cells, CD45−, CD3−, CD56+, CD3+CD4+ T cells and γδT-cells/Vα7.2+ T cells (strategy used for experiments shown in Fig. 4b and Extended Data Fig. 2g–i, 3g and 4b). g Assessment of CD14 mRNA by RT-PCR (relative to 18S control) of FACS-sorted Lin-HLA-DR+CD3-CD14+ MNP, ex vivo intrahepatic CD14−CD8+ T cells and CD14+CD8+ T cells (n = 5). h Percentage expression and relative levels of T-cell genes by scRNAseq transcriptome analysis of FACS-sorted intrahepatic CD14−CD8+ T cells, CD14+CD8+ T cells or Lin−HLA-DR+CD3−CD14+ MNP (n = 2 liver samples). i Histogram plot: CD14 mRNA transcript expression and UMAP depicting the transcriptomic profile of FACS-sorted intrahepatic CD14−CD8+ T cells (red), CD14+CD8+ T cells (green) or Lin−HLA-DR+CD3−CD14+ MNP (blue; n = 2 samples). j Frequency of peripheral CD14+CD8+ T cells after co-culture of purified CD8+ T cells ± MNP supplemented with nocodozole (4d; n = 10). k MFI of CD14 on CD14+CD8+ T cells after co-culture ± MNP supplemented with latrunculin B or nocodozole (n = 10). l Representative and summary data: Percentage CD14 expression on CD8+ T cells post co-culture ± MDM or MNP isolated from the same donor (n = 9). m Representative confocal images of purified CD8+ T cells co-cultured with MDM (4d; images representative of 2 donors) and summary data: comparison of MFI of CD14 on the T-cell population post culture with media alone or autologous MDM (defined by identification of the cell boundary segmented using the extent of CD8+ expression). n (left panel) CD14 signal intensity of MDM-derived exosomes from 3 donors and (right panel) percentage CD14 after co-culture of purified CD8+ T cell with concentrated MDM-derived exosomes or the autologous MDM from which exosomes were derived (4d; n = 3). o Representative plots: MFI of MD-2, TLR4, HLA-DR and TLR2 on peripheral T-cells co-expressing (or lacking) CD14 and the biotin-streptavidin complex after exposure to biotin-labelled MNP. p Summary data: frequency of intrahepatic CD14+CD8+ T cells as determined by imaging mass cytometry (IMC) (mean ±S.E.M. of percentage expression in two distinct regions of interest/liver). q Representative IMC image of macroscopically healthy liver tissue sections (2 regions of interest captured from n = 3 sections) as shown in Fig. 2n with the demarcation of three liver zones defined by intensity of CYP1A2 staining (scale bar: 200 µm; magnification: 50 µm); enrichment scores of the zonal localization of intrahepatic CD14−CD8+ T cells and CD14+CD8+ T cells. Each dot (or pair of dots) represent(s) a study participant, processed, stained and analysed independently; bars represent mean and error bars (where appropriate) represent ± S.E.M. b,e,j-m solid lines: median, dotted lines: IQR. p-values determined using either a Kruskal-Wallis test (ANOVA) with Dunn’s multiple comparisons test b (compared to media alone), j,k (compared to MNP + T-cells); or two-tailed Wilcoxon t Test c–d,l,m.
Extended Data Fig. 3 Nutrient transporter and immunomodulatory profile of intrahepatic CD14+CD8+ T cells and soluble mediator profile of stromal cell-derived CD14+CD8+ T cells.
Representative flow cytometric plots and summary data: MFI expression of a CD98 (n = 19) and CD71 (n = 14) and b Foxp3 (n = 32) and CTLA-4 (n = 19) of ex vivo intrahepatic CD14−CD8+ T cell (black outline) and CD14+CD8+ T cells (blue filled). c Heatmap showing relative expression (fold change) of immunomodulatory markers on CD14+CD8+ T cells compared to their CD14−CD8+ T cell counterparts. d Assessment of intrahepatic CD14+CD8+ T cell intracellular cytokine production ± 4 h anti-CD3/CD28 stimulation in the presence of brefeldin-A: IL-10 (n = 15) and IL-2 (n = 19) e Assessment of CD14+CD8+ T cell production of IL-10 (n = 15) and IL-2 (n = 19), and IFNγ (n = 19) andMIP1β (n = 12) afte 4 h anti-CD3/CD28 stimulation further defined by co-expression of CD69 and CD103. f Summary data: peripheral stromal cell-induced CD14−CD8+ T cell and CD14+CD8+ T cell populations post-stimulation with anti-CD3/CD28 by flow cytometric intracellular cytokine staining for IL-2, IFNγ, TNF (n = 15) and CD107a (n = 12) (4 h; upper panel) and by supernatant luminex detection of secreted IL-2, IFNγ, TNF, IL-10 and granzyme B from FACS-sorted stromal cell-induced populations (n = 5; 16 h; lower panel). g Example flow cytometry plots: gating strategy for the FACS-sorting of TCR-transduced CD8+ T cells with or without CD14 staining using the murineTCRβ (mTCR) region contained within the transgene. Each dot (or pair of dots) represent(s) a study participant, processed, stained and analysed independently; bars represent mean, error bars (where appropriate) represent ± S.E.M. d solid lines: median, dotted lines: IQR. p-values determined using either a Kruskal-Wallis test (ANOVA) with Dunn’s multiple comparisons test d,f; or a two-tailed Wilcoxon t Test a–b,e.
Extended Data Fig. 4 LPS-induced effects on CD14+CD8+ T cells in vitro.
a MFI of bound LPSAlexaFluor488 on stromal cell and MNP-induced CD14−CD8+ T cells or CD14+CD8+ T cells (n = 5) after culture for 120 min with LPSAlexaFluor488, or media alone. b UMAP: soluble mediator profiles produced by FACS-sorted stromal cell-induced CD14+CD8+ T cells ± stimulation with media alone (purple), 0.3 × 106 UV-killed E. coli (green) or anti-CD3/28 (orange). c Percentage CD14 on intrahepatic CD8+ T cells ± 72 h exposure to 0.3 × 106 UV-killed E. coli as a proportion of total CD8+ T cells (n = 14) or a proportion of total intrahepatic leukocytes (CD45+; n = 13). d CFSE dilution of intrahepatic CD14-labelled CD14+CD8+ T cells after 72 h culture in vitro in the presence of UV-killed E. coli and e the co-expression of intrahepatic CD14-labelled CD14+CD8+ T cells (APC; prelabelled on d 0) and further stained with CD14PE 72 h after in vitro culture in the presence of 0.3 × 106 UV-killed E. coli. f MFI expression of CD49b (n = 6) and CXCR4 (n = 8) on peripheral CD14 ± CD8+ T cells after co-culture in the presence of LPS. g Representative plots and summary data: MFI of TLR2 and TLR4 (n = 7) on ascitic CD14−CD8+ T cells (black outline) or CD14+CD8+ T cells (blue filled) ex vivo. h Frequency of CD14+CD8+ T cells isolated from human skin punch biopsies (n = 6; taken from the forearm of healthy controls) or from skin blister aspirates (n = 7; control blister). i CD14+CD8+ T cells as a proportion of total leukocytes (CD45+) from blood (n = 8) or blister exudates (n = 8) with or without prior UV-killed E. coli intradermal injection. j TLR4 expression on CD8+ T cells aspirated from skin blisters ± UV-killed E. coli injection (representative of n = 3 blister exudates). k Expression of CD38 on CD14−CD8+ T cells or CD14+CD8+ T cells from blister exudates ± UV-killed E. coli intradermal injection (n = 6). l Overview schematic of the functional role of CD14+CD8+ T cells. Each dot (or pair of dots) represent(s) a study participant, processed, stained and analysed independently; bars represent mean, error bars (where appropriate) represent ± S.E.M. a solid lines: median, dotted lines: IQR. p-values were determined using a Mann-Whitney U Test h; two-tailed Wilcoxon t Test c,g; or a two-tailed Kruskal-Wallis test (ANOVA) with Dunn’s multiple comparisons test f,i,k.
Supplementary information
Supplementary Tables
Supplementary Tables 1 and 2: details of antibody fluorochromes, clones, manufacturers and the dilutions used for flow cytometry (Supplementary Table 1) and spectral imaging (Supplementary Table 2).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pallett, L.J., Swadling, L., Diniz, M. et al. Tissue CD14+CD8+ T cells reprogrammed by myeloid cells and modulated by LPS. Nature 614, 334–342 (2023). https://doi.org/10.1038/s41586-022-05645-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05645-6
- Springer Nature Limited
This article is cited by
-
Immunology of human fibrosis
Nature Immunology (2023)