Abstract
Reconfigurable, mechanically responsive crystalline materials are central components in many sensing, soft robotic, and energy conversion and storage devices1,2,3,4. Crystalline materials can readily deform under various stimuli and the extent of recoverable deformation is highly dependent upon bond type1,2,5,6,7,8,9,10. Indeed, for structures held together via simple electrostatic interactions, minimal deformations are tolerated. By contrast, structures held together by molecular bonds can, in principle, sustain much larger deformations and more easily recover their original configurations. Here we study the deformation properties of well-faceted colloidal crystals engineered with DNA. These crystals are large in size (greater than 100 µm) and have a body-centred cubic (bcc) structure with a high viscoelastic volume fraction (of more than 97%). Therefore, they can be compressed into irregular shapes with wrinkles and creases, and, notably, these deformed crystals, upon rehydration, assume their initial well-formed crystalline morphology and internal nanoscale order within seconds. For most crystals, such compression and deformation would lead to permanent, irreversible damage. The substantial structural changes to the colloidal crystals are accompanied by notable and reversible optical property changes. For example, whereas the original and structurally recovered crystals exhibit near-perfect (over 98%) broadband absorption in the ultraviolet–visible region, the deformed crystals exhibit significantly increased reflection (up to 50% of incident light at certain wavelengths), mainly because of increases in their refractive index and inhomogeneity.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Main
The bonds between the building blocks of a periodic crystal are crucial in determining its intrinsic properties, including its thermal, chemical and physical stability, vibrational phonons, and electronic and magnetic conductivity11,12,13,14,15. Therefore, developing new types of linkages between the constituent building blocks in crystals and studying their novel bonding characteristics are topics of intense research in materials science and chemistry16,17,18,19. In particular, the strength, length and flexibility of the bonds within a crystal directly affect its responses to chemical and mechanical stimuli. For example, unusual elastic behaviours of molecular crystals, including hyper-expandable contraction, bending, twisting, coiling, jumping and self-healing, have been observed in single crystals by deliberately engineering the bonds to be both flexible and restorable1,2,5,6,7,8,9,10. Importantly, these findings have enriched the notion of bonding in crystalline materials from being rigid and static to flexible and dynamic.
However, the allowable restorable deformations in synthetic organic and inorganic crystals still do not compare well with highly flexible synthetic polymers and biological organisms20. In general, such crystals lose their macroscopic integrity and crystallinity when they experience heavy deformations, and they require time-consuming melting or dissolution and subsequent recrystallization or annealing processes to regain their initial crystallinity. Furthermore, because large-scale, reversible crystal deformation is not possible in these systems, the structure-dependent physical property changes that can be achieved with these structures in response to stimuli are limited.
Colloidal crystal engineering with DNA allows one to prepare an enormous number of crystal types, spanning over 70 different symmetries, with exquisite control over the type of bonding interactions (length and strength) between ‘programmable atom equivalents’ (PAEs: particles modified with short sequences of single-stranded DNA) within a given crystal16,21,22,23,24,25,26. In this study, the flexible and stimuli-responsive DNA bonds within such crystals enable polymer-like anisotropic but reversible self-adaptive responses to chemical and mechanical stimuli. To synthesize crystals capable of large deformation and recovery behaviour, a low-density body-centred cubic (bcc) crystal structure was designed and prepared using PAEs, synthesized from 5 or 10 nm Au nanoparticles (AuNPs) functionalized with radial arrays of DNA ligands (18 bases, more than 6 nm long) at high density (Fig. 1a,b and Supplementary Methods 1). Single crystals, large enough to exhibit mechanical behaviours that can be readily investigated, were synthesized using a range of slow-cooling rates (between 0.1 °C per 5 min and 0.1 °C per 120 min) during the crystallization process (Extended Data Figs. 1 and 2).
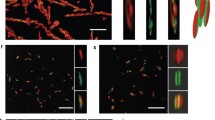
a, Schematic of an A-type PAE structure. Note, B-type PAEs have the same structure, but the complementary sticky end sequence is TTCCTT. b, A- and B-type PAEs are designed to interact with each other through DNA hybridization and assemble into non-close-packed bcc-type structures. c,d, Optical microscope images of crystals assembled from (c) 5 and (d) 10 nm PAEs. From left to right, the three columns indicate intact, dehydrated and rehydrated crystals. The inset images were collected with two times lower magnification to show the clear projection boundaries of the crystals. Scale bars, 20 and 50 µm for the top and bottom row, respectively. e,f, Bright (e) and dark-field (f) optical microscopy images of typical dehydrated crystals assembled from 5 nm PAEs. Scale bars, 20 µm.
These large crystals enable the in situ observation of their mechanical responses under an optical microscope (Fig. 1c–f and Supplementary Methods 2). Upon dehydration, both the 5 nm and the 10 nm PAE crystals experienced substantial morphological deformations that would not be observed in conventional crystals (Fig. 1c–f and Extended Data Fig. 3). Anisotropic dehydration of the crystals occurs starting from the sharp vertices and edges (Fig. 1c), which leads to the kinetically controlled, irregular shapes of the dehydrated crystals. The reduced water content in the crystals alters several physical and chemical properties, including temperature by evaporation of the solvent, dielectric constant, ionic strength, degree of solvation of DNA and pressure exerted by the surface tension of the water, which results in the DNA conformational changes that lead to crystal deformation. However, after rehydration, the initial rhombic dodecahedron morphology of the crystals was recovered within seconds (Fig. 1c,d). The dehydration–rehydration cycle was applied multiple times to the crystals composed of 5 nm PAEs, and some crystals maintained their morphological integrity, even after six cycles (Supplementary Video 1). It is likely that even though the double helix geometry of the hybridized DNA strands in the dehydrated crystals becomes substantially distorted or dissociated, the complementary strands are still located close enough to each other so that rapid rehybridization can occur upon rehydration. It is remarkable that the DNA not only accommodates such extreme deformations but essentially programs the reformation of the same crystal symmetry, habit and size. In other words, even when the hydrogen bonds are broken, the sequence and information content of the DNA ensures the path back to the original state.
To evaluate the degree of elasticity, semi-quantitative shape analysis based on the projected 2D morphology of the crystals was performed (Methods and Supplementary Methods 2). Two shape parameters commonly used to analyse the irregular morphologies of amorphous materials27 were adopted for this analysis: solidity (areareal/areaconvex hull) and convexity (perimeterconvex hull/perimeterreal), which are sensitive to morphological and textural roughness changes, respectively. The resulting solidity–convexity diagram reveals that the rehydrated crystals have values similar to those of the original crystals, indicating an almost full recovery of morphology and texture (Fig. 2a). Both parameters for the dehydrated crystals varied over a wide range (from 0.96 to 0.83), implying that the degree of deformation may be affected by the water evaporation rate. The average curvatures of the crystals increased approximately 1.8 times when they were deformed and, upon rehydration, returned to almost the same initial values (Fig. 2b). The slightly higher curvature (2.60 × 10−3 rad per unit length) of the rehydrated crystals compared to the initial intact crystals (2.53 × 10−3 rad per unit length) indicates the existence of minute deformations that were not reversible; however, considering the extent of the crystal deformation, the morphological recovery observed is remarkable (Fig. 1c–f and Extended Data Fig. 3).
a, Solidity–convexity diagram of the crystals. b, Average curvatures of the crystals. Error bars represent the standard deviations of the samples. c, Nanoindentation profiles. d,e, Deformation of model crystals with three different core sizes and the same angular deformation of the chain (θ0_d = 175°) (d) and accompanying volumetric solidity changes (e). f,g, Deformation of model crystals with three different core sizes and angular deformation of the chain (θ0_d = 175°, 120° and 90° for rcore = 20, 10 and 5 nm) (f) and accompanying volumetric solidity changes (g).
Based on the shape analysis and qualitative inspection of the optical microscopy images, crystals composed of 5 nm PAEs deform more significantly compared to those composed of 10 nm PAEs (Fig. 2a,b). To understand the differences in shape deformation, the reduced moduli of the original, intact crystals in 0.5 M phosphate buffered saline solution were measured using atomic force microscopy (AFM, Fig. 2c and Methods). The crystals assembled from 5 nm PAEs exhibited a less-steep force value change as compared to the crystals composed of 10 nm PAEs, indicating that they are softer. Moreover, the measured reduced moduli of the crystals assembled from 5 and 10 nm PAEs were 187 kPa and 241 kPa, respectively (the higher the reduced modulus, the more resistant the material is to deformation). These values are in the lower region of the previously reported theoretical and experimental range, from 97 kPa to 2.1 MPa, for a similar system28,29.
To understand the difference in stiffness based on PAE size, the number of DNA bonds in the lattices was evaluated (Supplementary Methods 1). The average number of DNA strands attached to the 5 and 10 nm PAEs is 59 and 179 strands, respectively; thus, approximately three times as many DNA strands are involved in interactions in crystals composed of 10 nm PAEs, qualitatively explaining the higher stiffness observed. The total viscoelastic volume fraction, which is different for different particle sizes, is also likely to be important in understanding crystal stiffness. To test this supposition, molecular dynamics (MD) simulation models were constructed and studied using the HOOMD-blue particle simulation toolkit30 (Methods and Supplementary Methods 3). By simulating crystal deformation with different PAE core sizes but the same bond length and stiffness, we confirmed that the crystals with the smallest core (thus higher viscoelastic volume fraction) experience more significant deformation (Fig. 2d,e), as measured by the volumetric solidity parameters, \({{\rm{volume}}}_{{\rm{real}}}/{{\rm{volume}}}_{{\rm{convex}}{\rm{hull}}}\). In another set of simulations, the bond stiffness was increased proportionally for larger cores, and it was found that the degree of deformation of crystals with the smallest core (5 nm) is the most pronounced (Fig. 2f,g). These results confirm that both the overall number of bonding interactions and the viscoelastic volume fraction affect the response of the crystals to mechanical stimuli and deformation.
To understand the internal structural changes that accompany the mechanical responses, the crystals were investigated using single-crystal X-ray diffraction (SXRD), as the large size of the crystals enabled a strong X-ray scattering signal. (Extended Data Figs. 4 and 5). A high-energy, high-flux X-ray beam can damage hybridized DNA, so four separate crystals at different points in the dehydration–rehydration cycle were analysed: (1) intact, (2) partially dehydrated (during mounting, partial evaporation of water from the intact crystals occurred), (3) fully dehydrated and (4) rehydrated. The images possessing the highest X-ray scattering intensities are shown (Fig. 3 and Extended Data Fig. 6). Upon dehydration, the higher-order reflections, such as (440), (420) and (400), which were observed for the intact crystals, disappeared; only the lowest order (110) reflections remained with the fully dehydrated crystals, indicating that they are in a nearly amorphous or paracrystalline state (Fig. 3a). However, the higher-order reflections reappeared in the patterns of the rehydrated crystals, indicating that the internal structures and morphologies of the crystals were recovered following rehydration (Fig. 3a and Extended Data Fig. 6). This was accompanied by a change in unit cell parameters (from 221 to 164 Å in 5 nm PAE crystals and from 286 to 189 Å in 10 nm PAE crystals) after dehydration (Extended Data Fig. 7). Such large unit cell changes would not be observed in electrostatically stabilized bcc crystals assembled with the same particles because of the relatively short and non-flexible Au–Au van der Waals interactions, of around 0.332 nm, between the particles.
a, SXRD patterns collected during a dehydration–rehydration cycle of crystals assembled from 5 nm PAEs. High-order, weakly observable reflections obtained from intact and rehydrated crystals are highlighted with white circles in the insets. b, (110) reflection profiles collected while rotating a dehydrated crystal by 3°. c, Diffraction images collected from a dehydrated crystal assembled from 5 nm PAEs exhibit fine fringes in the (110) reflection. d,e, (111) cross-section of model bcc crystal of N = 7,562 PAEs (d) before and (e) after deformation. Left images are simulation snapshots and right images are their diffraction patterns obtained by applying a fast Fourier transform to the simulated structures.
A closer examination of the diffraction patterns of the fully dehydrated crystals revealed the presence of unusual fine features in the reflections. Specifically, sporadic lines and delicate fringes that are closely related to the internal structural deformation and rapid recovery of the crystals were found (Fig. 3b,c). Typically, the reflections of mosaic crystals exhibit a relatively isotropic arc or ellipsoidal shape, which is an integral sum of all diffractions scattered from randomly misoriented mosaic domains in a single crystal. Here, the reflections were composed of sharp lines traversing the reciprocal space, implying that there is a strong positional correlation between the PAE domains in the highly deformed crystals.
To visualize the internal structures of the crystals, the (111) cross-sections of intact and deformed model crystals with flexible DNA bonds were simulated using MD and compared (Supplementary Methods 3). In contrast to the ordered PAEs in the original, intact crystals, the d-spacings and lattice orientations of the PAEs within the dehydrated simulated crystals gradually differed at different points within the crystal (Fig. 3d,e and Supplementary Methods 3). In particular, the lattices near the vertices of each crystal showed noticeably different d-spacings and orientations relative to one another. The diffraction patterns calculated from the simulated crystals contain sporadic, sharp lines similar to those observed in the experimental diffraction patterns, reinforcing that the line features are due to gradual changes in the lattice vector over a long range. By contrast, when another model crystal was simulated in which the interactions between PAEs are comparatively stiff, the crystal fractured under deformation into mosaic domains and exhibited the typical arc or ellipsoidal-shaped reflections (Extended Data Fig. 8). The aforementioned fringes were frequently observed in the (110) reflections of the dehydrated crystals composed of 5 nm PAEs (Fig. 3c and Supplementary Video 2). X-ray diffraction moiré fringes are generated by X-rays travelling through artificially arranged superimposed crystals with misfit lattice vectors, which is explained with dynamical X-ray diffraction theory31,32. Although the global geometries of these single crystals deviate from those that exhibit X-ray diffraction moiré fringes, it is possible that domains within a crystal may bear misfit adjacent lattices and thus generate fringes. Other potential origins of the fringes, such as defects and crystal shapes, were simulated using MATLAB codes (Supplementary Data Files 1 and 2), and the resulting patterns are discussed in Supplementary Methods 4. These two fine diffraction features are unusual in conventional rigid crystals and emphasize the notable flexibility of DNA-engineered colloidal crystals prepared with programmable atom equivalents.
Next, the optical properties of the crystals were investigated in response to mechanical deformation. The optical properties changed dramatically concomitant with the structural changes and ranged from near-perfect broadband absorption to enhanced reflectance and scattering (Fig. 4 and Extended Data Fig. 9). Indeed, the intact and recovered crystals composed of PAEs of both sizes exhibited near-perfect absorption (more than 98%) over a broad range of wavelengths (from 430 to 830 nm, Fig. 4b and Extended Data Fig. 9b). The dark, low-reflectivity surfaces of the crystals were observed in both dark-field (Fig. 1c,d) and bright-field modes (Fig. 4a), indicating high crystal absorptivity.
a, Bright-field optical microscopy images of crystals measured in situ. b, Absorption spectra of an intact and rehydrated crystal assembled from 5 nm PAEs. c, Enhanced reflection of the deformed crystals. Reflections of randomly chosen dehydrated crystals are indicated in pale blue. d–f, Simulated electric field maps (d), effective index (e) and absorption spectra (f) of single crystals considering only contraction, without distortion, of the unit cells. Scale bars, 20 µm.
Finite-difference time-domain (FDTD) simulations were utilized to calculate the effective refractive index of the intact crystals33 (Supplementary Methods 5). In these calculations, the intact crystals exhibit relatively low effective indices with pronounced oscillatory behaviours around the frequency of the localized surface plasmon resonance of Au (Fig. 4d,e). Such a low refractive index is indicative of a low reflection throughout the ultraviolet–visible region. Qualitatively, a substantial amount of the incident light penetrates the surface of the crystals owing to the relatively low volume fractions of Au within them (around 0.91% for the crystals with 5 nm PAEs and 2.3% for the crystals with 10 nm PAEs). To estimate the absorption of a 50-µm-thick crystal, the calculated refractive index was used in the transfer matrix method. The data show that the crystals do exhibit high absorption throughout the ultraviolet–visible region (Fig. 4f). The thick arrays of plasmonic AuNPs within the crystals absorb a significant portion of the penetrated light.
By contrast, the deformed crystals show significantly increased reflection (Fig. 4c and Extended Data Fig. 9c). The FDTD simulations considering only the unit cell parameter contractions with retained crystallinity indicate that both the real and imaginary components of the refractive index increase because of the enhanced plasmonic coupling among the closely packed AuNPs (Fig. 4d,e). The increase in the effective refractive index results in enhanced reflection and therefore lower absorption. Although the intensities and peak positions of the reflected spectra obtained from the experiments varied, probably attributed to the irregular unit cells and surface orientations of the crystals, the deformed crystals reflected more light than the intact and recovered crystals without exception (Fig. 4c and Extended Data Fig. 9c).
Although the contraction of the unit cells explained the increased reflection of the deformed crystals, noticeable discrepancies between the reflection intensities obtained from experiment and FDTD simulation were observed (the experimental values are approximately two to three times higher than the simulated values, Fig. 4c,f). The discrepancy may partly be explained by the fact that the FDTD simulation assumed the crystal to be periodic even in the deformed state; it is computationally prohibitive to perform an FDTD simulation with disordered crystals with such large dimensions. In addition, it is a well-known phenomenon that inhomogeneous materials, such as broken glasses and damaged opals, display increased reflection and scattering34: the inhomogeneous internal and external structures of the deformed crystals increase the number of reflection and scattering events within them and therefore increase the overall reflection intensities.
In conclusion, this work shows that preparing crystals held together with macromolecular bonds is a viable strategy for creating shape memory materials that can be deliberately engineered to exhibit a wide range of reversible structure and property changes simply not accessible with conventional crystalline architectures held together by other types of bonds. The synthetic method and crystal properties described herein, when combined with emerging methods to tailor crystal stability and DNA bond flexibility35,36, may lead to new stimuli-responsive properties of colloidal crystals that are useful for chemical sensing, optics and soft robotics.
Methods
Synthesis of bcc-type crystals with 5 nm PAEs
The DNA sticky end sequences (5ʹ-AAGGAA-3ʹ for A-type PAEs and 5ʹ-TTCCTT-3ʹ for B-type PAEs) were designed to be complementary to induce selective interactions between A- and B-type PAEs and thus form a non-close-packed bcc structure, as opposed to a close-packed face-centred cubic structure (Extended Data Fig. 1 and Supplementary Methods 1). A stock solution was prepared by combining concentrated solutions of the functionalized nanoparticles and linker strands (linker strands were added to 100 equivalents per particle) in 0.5 M NaCl, 0.01 M phosphate buffer solution (pH = 7.4) and 0.01 wt% sodium dodecyl sulfate, such that the final concentration of each particle type, A and B, was 200 nM. A thermocycler (ProFlex PCR System, Thermo Fisher Scientific) was used to slowly cool the reaction chamber containing the PAE solution from 65 °C (above the melting temperature, so therefore starting from dissociated free particles) to room temperature at the following rates: 0.01 °C per 5 min, 0.01 °C per 10 min, 0.01 °C per 30 min, 0.01 °C per 60 min, 0.01 °C per 90 min and 0.01 °C per 120 min.
Synthesis of bcc-type crystals with 10 nm PAEs
The same conditions as above were used for the synthesis of crystals consisting of 10 nm AuNPs, except that the 10 nm AuNPs were at a concentration of 50 nM; 400 equivalents of linker strand per particle was used. Small-angle X-ray scattering (SAXS) patterns of the resulting crystals were collected at the synchrotron beamline 5-ID with a wavelength of 1.0 Å (Extended Data Fig. 1). Both diffraction patterns match well with the simulated peak positions of the anticipated bcc structures (Im-3m space group).
Mechanical responses and shape analysis of the crystals
For a typical crystal shape characterization using shape factor analysis, optical microscope images were gathered. In brief, approximately 20 crystals were prepared on a slide glass. The crystals were then monitored in situ in dark-field mode throughout a drying and rehydration process. Except for the auto exposure time, all other configurations were maintained throughout a set of in situ measurements. Images were taken of the same crystals in the intact, dehydrated and rehydrated states. Unfortunately, during the dehydration and rehydration process, the crystals floated around on the slide glass, so the positions of the crystals changed slightly during this process. Within each batch of crystals, images were taken by adjusting the z-height of the microscope stage, and crystals were chosen for analysis from the images, in which they appeared most in focus. To generate the relevant shape parameters, the images of the crystals were manually thresholded in ImageJ36 and, from this, outlines of the two-dimensional projections of the crystals were generated using the computational software. Crystals that were too highly fused with others to draw a distinct boundary were rejected37.
AFM Young’s modulus
To characterize the elasticity of the crystals assembled with either 5 nm and 10 nm AuNPs, force–displacement curves were obtained using AFM (Bruker Dimension ICON). For each sample, a 500 × 62.5 nm2 area was scanned in the force–volume mode using SNL-10B (Bruker) cantilevers. The measurement was carried out on the (110) faces of the crystals in the buffer solution, and a spring constant of 0.125 N m−1 and deflection sensitivity of 64.03 nm V−1 were measured by calibrating the tip via a thermal tune process. The scan and ramp sizes (displacement) were set at 0.1 Hz and 700 nm, respectively. The resulting force curves were analysed and fitted to the Sneddon model to calculate the reduced Young’s modulus of each sample using Nanoscope Analysis v.1.9 software (Bruker). The average Young’s moduli of the crystals were 187 ± 12 kPa and 241 ± 22 kPa for 5 nm and 10 nm AuNPs, respectively. The Young’s moduli of the crystals are comparable to those of polyacrylamide hydrogels38 (around 240 kPa) and soft polydimethylsiloxane39 (around 580 kPa), which are often used for cellular studies; the crystals are much softer than the glass substrate.
Single-crystal X-ray diffraction
SXRD experiments were performed at two beamlines at Argonne National Laboratory. For the wide-angle measurements, the data were collected at the macromolecular crystallography beamline 21-ID-D with a wavelength of 1.0 Å (Extended Data Fig. 4). For the SAXS measurements, the data were collected at the SAXS beamline 12-ID-B with a wavelength of 1.2 Å (Extended Data Fig. 5). The unit cell parameters of the crystals measured with an SXRD goniometer were calculated based on the three-dimensionally reconstructed image frames (Extended Data Fig. 7). Both experiments were conducted at 100 K under a liquid nitrogen cold stream.
MD simulations
Simulations were conducted using MD implemented in the HOOMD-blue particle simulation package30 (https://github.com/glotzerlab/hoomd-blue). The performance of HOOMD-blue is highly optimized on graphics processing units, and most of the simulations in this work were performed on NVIDIA Tesla P100 or V100 graphics processing units40.
A simplified model of the PAEs was developed, in which PAEs were connected by flexible chains to simulate the large PAE crystals with high viscoelastic volume fractions in a reasonable timescale (Supplementary Methods 3). In this model, PAEs are represented as spherical cores (r = 5, 10 and 20 nm) surrounded by a set of eight anchoring points: (\(\frac{2}{\sqrt{3}}r\),\(\,\frac{2}{\sqrt{3}}r,\frac{2}{\sqrt{3}}r\)), (\(\frac{2}{\sqrt{3}}r\),\(\,-\frac{2}{\sqrt{3}}r,\frac{2}{\sqrt{3}}r\)), (\(-\frac{2}{\sqrt{3}}r\),\(\,\frac{2}{\sqrt{3}}r,\frac{2}{\sqrt{3}}r\)),(\(-\frac{2}{\sqrt{3}}r\),\(\,-\frac{2}{\sqrt{3}}r,\frac{2}{\sqrt{3}}r\)), (\(\frac{2}{\sqrt{3}}r\),\(\,\frac{2}{\sqrt{3}}r,-\frac{2}{\sqrt{3}}r\)), (\(\frac{2}{\sqrt{3}}r\),\(\,-\frac{2}{\sqrt{3}}r,-\frac{2}{\sqrt{3}}r\)), (\(-\frac{2}{\sqrt{3}}r\),\(\,\frac{2}{\sqrt{3}}r,-\frac{2}{\sqrt{3}}r\)), (\(-\frac{2}{\sqrt{3}}r\),\(\,-\frac{2}{\sqrt{3}}r,-\frac{2}{\sqrt{3}}r\)) where the centre of each core is at (0, 0, 0) in xyz coordinates. The direction from the centre of the core to each anchor corresponds to the direction from the centre of the core to eight nearest neighbouring cores in a bcc crystal, which allows the crystal structure to be stabilized with the minimum number of chains. The anchoring points were connected by flexible chains to the adjacent cores. Each chain between two PAEs consists of 16 nodes and 2 anchors, and they are bonded to each other by a harmonic potential, \({V}_{{\rm{bond}}}=\left(1/2\right)k{(r-{r}_{0})}^{2}\), where \(k=330\varepsilon /{\sigma }^{2}\), and by an angular potential, \({V}_{{\rm{angle}}}=(1/2){k}_{\theta }{(\theta -{\theta }_{0})}^{2}\), where kθ = 10ε and θ is the angle between three consecutive nodes. The quantities ε and σ are energy and length units in the simulation. The r0 and θ0 of the undeformed chain are 0.706 nm and 180°, respectively, but they can vary during the crystal deformation process, as discussed below. A purely repulsive Weeks–Chandler–Andersen potential was applied to the spherical cores to avoid overlap with chains and other cores41.
As an initial configuration, an ideal bcc crystal (\({N}_{{\rm{core}}}={\rm{7,825}}\), where Ncore is the number of spherical cores) with a well-faceted rhombic dodecahedron shape and a large (111) cross-section (\({N}_{{\rm{core}}}={\rm{7,562}}\)) were constructed (Supplementary Fig. 9). The constructed crystal was placed in a large simulation box (\({\varphi }_{{\rm{core}}} < 0.01\)), where \({\varphi }_{{\rm{core}}}={N}_{{\rm{core}}}\times {v}_{{\rm{core}}}/{V}_{{\rm{box}}}\) and \({v}_{{\rm{core}}}\) is the volume of a core. All simulations were conducted under periodic boundary conditions. The NVT (constant number of particles, volume and temperature) integrator and a constant temperature (\({T}^{* }=0.2\)) were applied to all systems. An MD simulation timestep \(\triangle t\) = 0.002 was used for all systems. Chains were included between the cores in the initial crystal structure, and then the crystal was deformed as described in Supplementary Methods 3.
Data availability
The data that support the findings of the study are available from the corresponding author, C.A.M., upon reasonable request.
Code availability
The MATLAB codes used to generate the model diffraction patterns in the Supplementary Information can be found in the Supplementary Data Files.
References
Taniguchi, T. et al. Walking and rolling of crystals induced thermally by phase transition. Nat. Commun. 9, 538 (2018).
Naumov, P., Chizhik, S., Panda, M. K., Nath, N. K. & Boldyreva, E. Mechanically responsive molecular crystals. Chem. Rev. 115, 12440–12490 (2015).
Mason, J. A. et al. Methane storage in flexible metal–organic frameworks with intrinsic thermal management. Nature 527, 357–361 (2015).
Koshima, H. Mechanically Responsive Materials for Soft Robotics (Wiley, 2019).
Ahmed, E., Karothu, D. P., Warren, M. & Naumov, P. Shape-memory effects in molecular crystals. Nat. Commun. 10, 3723 (2019).
Kobatake, S., Takami, S., Muto, H., Ishikawa, T. & Irie, M. Rapid and reversible shape changes of molecular crystals on photoirradiation. Nature 446, 778–781 (2007).
Naumov, P., Sahoo, S. C., Zakharov, B. A. & Boldyreva, E. V. Dynamic single crystals: Kinematic analysis of photoinduced crystal jumping (the photosalient effect). Angew. Chem. Int. Edn 52, 9990–9995 (2013).
Rai, R., Krishnan, B. P. & Sureshan, K. M. Chirality-controlled spontaneous twisting of crystals due to thermal topochemical reaction. Proc. Natl Acad. Sci. USA 115, 2896–2901 (2018).
Skoko, Ž., Zamir, S., Naumov, P. & Bernstein, J. The thermosalient phenomenon. “Jumping crystals” and crystal chemistry of the anticholinergic agent oxitropium bromide. J. Am. Chem. Soc. 132, 14191–14202 (2010).
Zhang, L., Bailey, J. B., Subramanian, R. H., Groisman, A. & Tezcan, F. A. Hyperexpandable, self-healing macromolecular crystals with integrated polymer networks. Nature 557, 86–91 (2018).
Pauling, L. The Nature of the Chemical Bond and the Structure of Molecules and Crystals: An Introduction to Modern Structural Chemistry (Cornell Univ. Press, 1960).
Hoffmann, R. Solids and Surfaces: A Chemist’s View of Bonding in Extended Structures (Wiley, 1989).
Field, J. E. & Pickles, C. S. J. Strength, fracture and friction properties of diamond. Diamond Relat. Mater. 5, 625–634 (1996).
Desiraju, G. R. & Steiner, T. The Weak Hydrogen Bond: In Structural Chemistry and Biology (Oxford Univ. Press, 2001).
Maldovan, M. Phonon wave interference and thermal bandgap materials. Nat. Mater. 14, 667–674 (2015).
Laramy, C. R., O’Brien, M. N. & Mirkin, C. A. Crystal engineering with DNA. Nat. Rev. Mater. 4, 201–224 (2019).
Diercks, C. S. & Yaghi, O. M. The atom, the molecule, and the covalent organic framework. Science 355, eaal1585 (2017).
Leunissen, M. E. et al. Ionic colloidal crystals of oppositely charged particles. Nature 437, 235–240 (2005).
Novoselov, K. S. et al. Electric field effect in atomically thin carbon films. Science 306, 666–669 (2004).
Egan, P., Sinko, R., LeDuc, P. R. & Keten, S. The role of mechanics in biological and bio-inspired systems. Nat. Commun. 6, 7418 (2015).
Mirkin, C. A., Letsinger, R. L., Mucic, R. C. & Storhoff, J. J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382, 607–609 (1996).
Nykypanchuk, D., Maye, M. M., van der Lelie, D. & Gang, O. DNA-guided crystallization of colloidal nanoparticles. Nature 451, 549–552 (2008).
Park, S. Y. et al. DNA-programmable nanoparticle crystallization. Nature 451, 553–556 (2008).
Jones, M. R., Seeman, N. C. & Mirkin, C. A. Programmable materials and the nature of the DNA bond. Science 347, 1260901 (2015).
Lin, H. et al. Clathrate colloidal crystals. Science 355, 931–935 (2017).
He, M. et al. Colloidal diamond. Nature 585, 524–529 (2020).
Liu, E. J., Cashman, K. V. & Rust, A. C. Optimising shape analysis to quantify volcanic ash morphology. GeoResJ 8, 14–30 (2015).
Lequieu, J., Córdoba, A., Hinckley, D. & de Pablo, J. J. Mechanical response of DNA–nanoparticle crystals to controlled deformation. ACS Central Science 2, 614–620 (2016).
Lewis, D. J., Carter, D. J. D. & Macfarlane, R. J. Using DNA to control the mechanical response of nanoparticle superlattices. J. Am. Chem. Soc. 142, 19181–19188 (2020).
Anderson, J. A., Glaser, J. & Glotzer, S. C. HOOMD-blue: a Python package for high-performance molecular dynamics and hard particle Monte Carlo simulations. Comput. Mater. Sci. 173, 109363 (2020).
Chu, X. & Tanner, B. K. Bragg case X-ray Moiré patterns observed in GaAlAs/GaAs laser structures. Mater. Lett. 5, 153–155 (1987).
Lang, A. R. X-ray moiré topography of lattice defects in quartz. Nature 220, 652–657 (1968).
Smith, D. R., Vier, D. C., Koschny, T. & Soukoulis, C. M. Electromagnetic parameter retrieval from inhomogeneous metamaterials. Phys. Rev. E 71, 036617 (2005).
Chýlek, P., Grams, G. W. & Pinnick, R. G. Light scattering by irregular randomly oriented particles. Science 193, 480–482 (1976).
De Fazio, A. F. et al. Light-induced reversible DNA ligation of gold nanoparticle superlattices. ACS Nano 13, 5771–5777 (2019).
Lee, S., Zheng, C. Y., Bujold, K. E. & Mirkin, C. A. A cross-linking approach to stabilizing stimuli-responsive colloidal crystals engineered with DNA. J. Am. Chem. Soc. 141, 11827–11831 (2019).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Sunyer, R., Jin, A. J., Nossal, R. & Sackett, D. L. Fabrication of hydrogels with steep stiffness gradients for studying cell mechanical response. PLoS ONE 7, e46107 (2012).
Park, J. Y. et al. Increased poly(dimethylsiloxane) stiffness improves viability and morphology of mouse fibroblast cells. BioChip J. 4, 230–236 (2010).
Towns, J. et al. XSEDE: accelerating scientific discovery. Comput. Sci. Eng. 16, 62–74 (2014).
Chandler, D., Weeks, J. D. & Andersen, H. C. Van der Waals picture of liquids, solids, and phase transformations. Science 220, 787–794 (1983).
Wang, S. et al. Colloidal crystal “alloys”. J. Am. Chem. Soc. 141, 20443–20450 (2019).
Acknowledgements
This material is based upon work supported by Air Force Office of Scientific Research FA9550-17-1-0348, FA9550-16-1-0150 and FA9550-18-1-0493. It was also supported as part of the Center for Bio-Inspired Energy Science, an Energy Frontier Research Center (CBES) funded by the US Department of Energy, Office of Science, Basic Energy Sciences award DE-SC0000989. This work made use of the EPIC facility of Northwestern University’s NUANCE Center, which has received support from the SHyNE Resource (NSF grant no. ECCS-2025633), the International Institute for Nanotechnology (IIN) and Northwestern’s MRSEC program (NSF grant no. DMR-1720139); the IIN; the Keck Foundation; and the State of Illinois, through the IIN. This research used resources of the Advanced Photon Source, a US Department of Energy Office of Science User Facility operated for the US Department of Energy Office of Science by Argonne National Laboratory under contract no. DE‐AC02‐06CH11357. X-ray diffraction experiments were carried out at the Dupont–Northwestern–Dow Collaborative Access Team beamline at Sector 5, 12-ID-B, and sector 21 at the Advanced Photon Source at Argonne National Laboratory. H.A.C. acknowledges support by the National Science Foundation Graduate Research Fellowship Program grant (DGE-1842165). K.A. acknowledges partial support from the Office of Naval Research Young Investigator Program (ONR-YIP) Award (no. N00014-17-1-2425). S.L. (UM) and S.C.G. acknowledge support from CBES (grant no. DE-SC0000989). MD simulations were supported in part through computational resources and services supported by Advanced Research Computing at the University of Michigan, Ann Arbor and also used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant no. ACI-1548562 and XSEDE award no. DMR 140129.
Author information
Authors and Affiliations
Contributions
C.A.M. and Seungkyu Lee conceived the work and designed experiments. Seungkyu Lee and H.A.C. synthesized the single crystals. Seungkyu Lee conducted the experiments to determine the mechanical behaviours of the crystals. Sangmin Lee and S.C.G. designed the MD simulation studies. Sangmin Lee developed the model, performed the MD simulations and simulated diffraction pattern calculations. Seungkyu Lee and B.L. collected the X-ray diffraction data and analysed the results. E.O. conducted the nanoindentation, Oligreen and inductively coupled plasma (ICP) experiments and analysed the results. Seungkyu Lee, W.H. and K.A. measured the optical properties of the crystals and analysed the results. All authors wrote and edited the manuscript together.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Melting temperature curves for the synthesis of the crystals and their SAXS patterns.
a, Melting temperature curves for solutions of A and B-type PAEs. A cooling rate of 0.1 °C/120 min was applied over the crystallization temperature range (shaded region) that encompasses the aggregation temperatures of the PAEs. b,c, SAXS patterns of BCC crystals (b) with 5-nm PAEs and (c) with 10-nm PAEs. In case of the crystals prepared with 5-nm PAEs, a smaller fraction of the crystals that have a slightly larger unit cell parameter, 292.9 Å, were detected in the diffraction profile. The two phases are likely to occur during crystallization owing to the polydispersity of the 5-nm particle cores. Size exclusion occurs more readily during crystallization of the 5-nm PAEs compared to the 10-nm PAEs, as a result of the larger relative size difference between small and large particles in the 5-nm batch. This is a well-documented behaviour for this class of materials42.
Extended Data Fig. 2 Crystal size dependency on the cooling rate during the crystallization process.
a, b, Optical microscopy images of crystals assembled from 5-nm PAEs at the cooling rates indicated (scale bar: 50 µm), and crystal size distribution histogram. Inset, average crystal sizes as a function of the cooling rate. c, d, Optical microscopy images of crystals assembled from 10-nm PAEs at the cooling rates indicated (scale bar: 50 µm), and crystal size distribution histogram. e, Optical microscopy images of >100-µm single crystals assembled from 5-nm and 10-nm PAEs, respectively.
Extended Data Fig. 3 Electron microscope images of the dehydrated crystals.
a,b, Crystals assembled from (a) 5- and (b) 10-nm PAEs. Scale bars: 5 µm.
Extended Data Fig. 4 SXRD data of the intact crystals assembled with 5- and 10-nm PAEs.
a, A crystal assembled with 5-nm PAEs (a: 223 Å). b, A crystal assembled with 10-nm PAEs (a: 274 Å). In the high-angle area of both data, FCC structure powder rings from randomly oriented AuNPs in the crystals were observed. The majority of the reflections of the BCC superlattices are blocked by the beam stop owing to the typical short sample-detector distance of the instrument configuration of a wide-angle beamline.
Extended Data Fig. 5 A goniometer installed on the SAXS beamline.
a, A single crystal mounted on a fibre loop. b, Tilt motor to orient the crystal. c, Y-axis control motor. d, X-axis control motor. e, X-ray beam collimator.
Extended Data Fig. 6 Single crystal X-ray diffraction data of a crystal assembled with 10-nm PAEs.
The SXRD patterns were collected during a dehydration-rehydration cycle. High-order, weakly observable reflections obtained from intact and rehydrated crystals are highlighted with white circles in the figure insets.
Extended Data Fig. 7 Three-dimensionally reconstructed diffraction patterns of the crystals during a deformation and recovery cycle.
a,b, The unit cell parameters were determined by overlapping the simulated BCC reciprocal matrixes (grey dotted circles) with the reconstructed diffraction patterns of the crystals assembled with (a) 5-nm PAEs and (b) 10-nm PAEs, respectively. In the case of heavily deformed crystals, precise unit cell parameter determination was impossible because of the limited number of reflections. The unit cell parameters of the heavily deformed crystals were estimated by measuring the average distances between the centers of (110) reflections and the origin of the reciprocal space and converting the average distances to the direct space values.
Extended Data Fig. 8 Deformation of a crystal with relatively stiff PAE-PAE interactions.
a, MD simulation snapshot after the crystal deformation. The crystal was fractured into many domains with different sizes and orientations. b, Effective pair potential between A-type PAEs and B-type PAEs in this model (Supplementary Methods 3). c, Diffraction pattern obtained from the (111) direction of the fractured crystal.
Extended Data Fig. 9 Optical property changes of the crystals assembled with 10-nm PAEs.
a, Schematic view of the simulation region used in the FDTD simulation, highlighted by the bounding box. b, Bright-field optical microscope images of the crystals measured in situ and absorption spectra of the intact and rehydrated crystals assembled with 10-nm PAEs. c, Enhanced reflection of the deformed crystals. Reflections of randomly chosen dehydrated crystals are indicated in pale blue. d, Simulated effective index plot for the crystals assembled with 10-nm PAEs, considering only contraction, without distortion, of the unit cells.
Supplementary information
Supplementary Information
Supplementary Methods 1–6, Figs. 1–16, Tables 1–11 and references.
Supplementary Data File 1
Source MATLAB code used to generate the diffraction fringes.
Supplementary Data File 2
Source MATLAB code used to plot the diffraction data.
Supplementary Video 1
Colloidal crystals composed of PAEs during dehydration-rehydration cycles. Crystals composed of 5 nm PAEs were dehydrated and rehydrated on glass slides and their response was recorded by an optical microscope. Some crystals were able to recover their initial morphology even after six dehydration–rehydration cycles.
Supplementary Video 2
Reflections of dehydrated crystals during single-crystal X-ray diffraction. Dehydrated crystals composed of 5 nm PAEs frequently exhibit sharp lines and streaks in their (110) reflections during single-crystal X-ray diffraction experiments.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lee, S., Calcaterra, H.A., Lee, S. et al. Shape memory in self-adapting colloidal crystals. Nature 610, 674–679 (2022). https://doi.org/10.1038/s41586-022-05232-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-05232-9
- Springer Nature Limited
This article is cited by
-
Macroscopic photonic single crystals via seeded growth of DNA-coated colloids
Nature Communications (2023)
-
Spherical Packing Superlattices in Self-Assembly of Homogenous Soft Matter: Progresses and Potentials
Chinese Journal of Polymer Science (2023)
-
Accurate computational design of three-dimensional protein crystals
Nature Materials (2023)